Copyright01977 American Societyfor Microbiology Printed in U.S.A.
Synthesis
of mRNA Guanylyltransferase and mRNA
Methyltransferases in Cells Infected with Vaccinia Virus
ROBERT F. BOONE, MARCIA J. ENSINGER,1 AND BERNARD MOSS*Laboratory ofBiology of Viruses,National Institute ofAllergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20014
Received for publication 12July 1976
Guanylyltransferase and methyltransferases that modifythe 5'-terminalsof viral mRNA's to form the structuresm7G(5')pppAm- and m7G(5')pppG'n- appear tobesynthesized after vaccinia virus infection of HeLa cells. Elevations in these enzyme activities weredetected within 1 h after virus inoculation and increased
15- to 30-fold by 4 to 10 h. Increases in the guanylyl- and methyltransferase
activities were prevented by cycloheximide, an inhibitor of protein synthesis, but not by cytosine arabinoside, an inhibitor of DNA synthesis. The latter results suggest that the mRNA guanylyl- and methyltransferases are "early" or prereplicative viral gene products. The guanylyltransferase and two methyl-transferases, a guanine-7-methyltransferase and a nucleoside-2'-methyltrans-ferase, were isolated by column chromatography from infected cell extracts and found to have properties similar or identical to those of the corresponding enzymespreviously isolated from vacciniavirus cores.Incontrast, enzymeswith
thesepropertiescouldnotbeisolated from uninfected cells.
mRNAsynthesized in vitro by enzymes pres- ular weights of 95,000 and 31,400
ent in vaccinia virus cores contain the se- (14, 15). The nucleoside-2'-methyltransferase,
quences m7G(5')pppGm- and m7G(5')pppAm- which catalyzes reaction (iii), has been par-(abbreviations: m7G, 7-methylguanosine; Gm, tially purified and its characterization is in
2'-O-methylguanosine; Am, 2'-O-methyladeno- progress(unpublished data).
sine) at their 5'-terminals (22, 26, 27). Three Foravariety ofreasons wesuspectedthat the activitiesresponsible for the 5'-terminal modi- guanylyl- and methyltransferases packaged fications, an mRNA guanylyltransferase, an within vaccinia viruscores arecoded forby the mRNA (guanine-7-)-methyltransferase, and large viral DNA genome. Experiments de-an mRNA (nucleoside-2'-)-methyltransferase, scribed here support this view; they indicate have been extracted from vaccinia virus cores that these 5'-terminal modification enzymes (4).The enzymes appeartoactinthe following canbeisolatedfrom infected HeLa cells butnot
order: from uninfected cells. In addition, elevations
in the enzyme activities occur
prior
to viral pppG +ppN- G(5')pppN- + PPi (i) DNAreplication, suggesting that theenzymes should be classifiedas"early"orprereplicativeG(5')pppN- + AdoMet viral products. Elsewhere we will describe the
-.
m7G(5')pppN-
+ AdoHcy (ii) purification and characterizationof a HeLa cellm7G(5')pppN- + AdoMet RNA (guanine-7-)-methyltransferase that spe-m7G(5')pppNm- + AdoHcy (iii) cifically methylates G(5')pppN- terminals, but thatisdistinct from thecorresponding vaccinia to modify nascent RNA represented here as enzyme (5).
ppN- (4, 16).(S-adenosylmethionineand
S-ade-nosylhomocysteine are abbreviatedasAdoMet MATERIALS AND METHODS
andAdoHcy, respectively.) Theguanylyltrans- Infection of cells. HeLa S-3 cells weregrownin ferase andguanine-7-methyltransferase, which suspension in Eagle medium, concentrated to 5 x
catalyze reactions (i) and (ii), have been iso- 108perml, and mixed atzerotime with 15 PFU of lated as a 127,000-molecular-weight complex purified vaccinia virus per cell. After 15
min
at37°Ccontainingtwomajo polypeptids wh
m.
theinfected cells were diluted 10-fold and theincu-bation
wascontinued.
Mock-infected
cells were XPresentaddress:DepartmentofMicrobiology, College treated in the same manner.ofPhysicians and Surgeons, Columbia University, New Preparation of cell lysates. At specified times York,N Y 10032. after infection, approximately 1.5 x 107 cells were
475
on November 10, 2019 by guest
http://jvi.asm.org/
476 BOONE, ENSINGER, AND MOSS J. VIROL.
sedimented, washed three times with isotonic sa- 1.5 mM MgCl2-1 mM dithiothreitol. All operations line, and suspended in cold hypotonic buffer con- from this point on were performed at0to4°C. The taining 10 mM Tris-hydrochloride, pH 9.0, 10 mM cellswereallowedtoswell for10minanddisrupted NaCl, and 1.5 mM MgCl2. To prepare whole-cell by Dounce homogenization, and the nuclei were re-lysates, the suspension was adjusted to contain0.1 moved bycentrifugation. The cytoplasmic superna-MTris-hydrochloride, pH 8.3, 0.3 M NaCl, 50 mM tant was brought to 0.1 MTris-hydrochloride (pH dithiothreitol, and 0.1% sodium deoxycholate and 8.4)-0.25MNaCl-0.05Mdithiothreitol-0.1% sodium sonically treated four times for 15-sintervals with deoxycholate-2 mM EDTA. After 15 min, the ex-the microtip of a Branson sonifier. Cytoplasmicex- tract was sonically treated as described and then tracts were obtained by Douncehomogenization of centrifuged at 100,000 xg for 1h. The supernatant the cells swollen in hypotonic buffer followed by wasrecovered,glycerol and Triton X-100 were added centrifugation to remove nuclei. Protein concentra- to 10 and 0.1%, respectively, and the mixture was tions weredetermined by the method of Lowry et al. then passed through a DEAE-cellulose column (0.9 (12) with a bovine serum albumin standard and by 7 cm)equilibrated with 0.1 MTris-hydrochloride were between 1 and2 ,g/ml. (pH 8.4)-0.25 NaCl-10% glycerol-0.1% Triton X-Guanylyltransferase and methyltransferase as- 100-1 mMdithiothreitol-2 mM EDTA. Thematerial says. Total cell lysates orcytoplasmic extracts were that did not bind to the column was diluted to 0.05 M assayed for guanylyltransferase and methyltrans- NaCl in 0.15 M Tris-hydrochloride (pH 8.4)-10% ferase activity in 0.1 ml containing 50 mM Tris- glycerol-0.1% Triton X-100-2 mM EDTA-1 mM hydrochloride, pH 7.6, 1 mM dithiothreitol, 2.5 mM dithiothreitol (buffer A). The material was then
MgCl2, 10 ,ug of diphosphate-terminated poly(A) applied to apoly(U)-Sepharose column (0.7 by 8 cm)
[pp(A)n]synthesized aspreviouslydescribed (14), 10 equilibrated with buffer A containing 0.05 M NaCl. to 20
gg
ofprotein extract,andindicated amounts of After washing the column with 40 ml of the same GTP and AdoMet. Specific assays for guanine-7- buffer, the activity was eluted with a gradient of methyltransferase utilized 50 mM Tris-hydrochlo- NaCl from 0.05 to 1 M in buffer A. Fractions (4 ml) ride, pH 7.6, 1 mMdithiothreitol, 2mMEDTA, 10 were collected at a rate of 2 ml per h and assayed,tgofG(5')ppp(A)n prepared asdescribed (14), and for methyltransferase activity using either
un-0.5 ,uMS-adenosyl[methyl-3H]methionine. Afterin- methylated vaccinia mRNA or pp(A)n as acceptor. cubation at 37°C for 30 min, the mixtures were ap- Source of materials. Nucleotides, methylated nu-plied to Whatman DE-81 filters and washed five cleosides, and unmodified poly(A) were from P-L times with 5% anhydrous, dibasic sodium phos- Biochemicals. P1 nuclease was from Yamasa Shoyu phate, once with water, twice with ethanol-ether Co., T2 RNase was from Sigma Chemical Co., and (1:1), and once with ether, dried, andcounted in a snake venom phosphodiesterase and bacterial al-toluene-based scintillation fluid (1). kaline phosphatase were from Worthington Bio-Enzyme digestions. Digestion with2U ofRNase chemical Corp. Poly(U)-Sepharose and DEAE-cellu-T2 per ml wasin10 mMammonium acetate, pH 4.5, lose were purchased from Pharmacia Fine Chemi-for 16 h at37°C. P1 nuclease digestion was carried cals and Whatman, Inc., respectively. S-adeno-outin 10mMsodium acetate, pH 6.0, at an enzyme syl[methyl-3H]methionine, [3H]GTP, and [a-concentration of 0.25 mg/ml for 2h at 37°C. Com- 32P]GTP were products of New England Nuclear bined digestions with snake venom phosphodiester- Corp.
ase(0.25mg/ml) and Escherichia colialkaline
phos-phatase (0.1 mg/ml) were in 50 mM Tris-hydrochlo- RESULTS ride, pH 8.5, and 5 mM MgC12.
Column chromatography ofpolyribonucleotides Guanylyltransferaseandmethyltransferase
anddigestion products.Poly(U)-Sepharose chroma- activities after vaccinia virus infection.
Pre-tography was carried out in columns (0.7 by 5 cm) as vious studies (14) indicated that the mRNA
follows. Polyribonucleotides were applied in 10 mM guanylyltransferase isolated from vaccinia
vi-Tris-hydrochloride (pH 7.6)-0.12 M NaCl-2 mM rus cores catalyzes the transfer of GMP resi-EDTA-0. 1%sodiumdodecyl sulfate. The column was dues from GTPtosynthetic poly(A) containing
washed withthe above buffer and theneluted with
duerminal tosphte
pp(A)Thenrod
75%formamide-10mMTris-hydrochloride (pH 7.6). a 5-terminal diphosphate
[pp(A)d
n Theprod-DEAE-cellulose chromatography of oligonucleo- uct of this reaction was identified as
tideswas carried out withcolumns (0.9 by 15 cm) as ppp(A)n. Accordingly, [3H]GTPwasusedasthe
follows. Samples were applied in 7 M urea-10 mM labeled donor and
pp(A),
wasusedastheaccep-Tris-hydrochloride (pH 7.6) and eluted with linear 0 tor in assays designed to detect the
guanylyl-to0.3 MNaClgradients. Peak fractions were pooled transferase in lysates of uninfected and infected
anddesalted by adsorption to smallDEAE-cellulose cells. A rise inactivity was detected at 1 hafter
columns thatwerewashed with 5 mM ammonium
acetateand eluted with 1 M ammonium acetate.
virs
inoctin
andlevels nearly
15times
Purification ofguanylyl-andmethyltransferases thoseof
uninfected
cell lysateswerereached byfromvacciniavirus-infected HeLa cells. At 6 h after 4 to 10 h (Fig. 1A). Activity was dependent on
infection, approximately 2 x 108 cells were har- pp(A)n since no significant incorporation was
vested by centrifugation, washed twice with phos- detected in its absence. Therise in
guanylyl-phate-buffered saline, and then resuspended in 8 ml transferase activity appeared to precede the
of10 mMTris-hydrochloride (pH7.2)-10 mM NaCl- synthesis of vaccinia DNA, which occurs from 2
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 21, 1977 mRNA GUANYLYL- AND METHYLTRANSFERASES 477
. ,.''. i§ i s were treated with 100 ,ug of cycloheximide per A , ~
oml,
vacciniainfection didnotcauseanyrisein@B
ffi
guanylyltransferase activity. Controlexperi-2- Infected ments carried out by mixing the appropriate
amount of inoculum virus with uninfected cell 8 /5/s
lysates
also indicated that enzyme containedz
1
infected AraC within the inoculum could not account for a significant portion of the activity of infected cells.zx
^ ,JfUninfected
Combinedassays forguanylyl-
andmethyl-transferases were carried out by using pp-=a§I I I§ ' a
(A),,
unlabeled GTP, andS-adenosyl[methyl-0
B
,3H]methionine
as the methyl donor. Increased-R 12 _ levels of activity were detected in lysates
ob-(L
;
6tained
from cells within2hafterinfection,andW. Infected
</values
30timesgreater
than those of uninfectedcells were reached by 4 to 10 h (Fig. 1B). When
4 /either theacceptor pp(A)n or GTP was omitted,
>//Infted+AxaC no significant difference in incorporation
be-> /,' _ tween infected and uninfected cells was
de-_2
1(/
- tected. As found for assays measuring the/,}> Uninfected
guanylyltransferase
alone, inhibition of DNAo' f. _ __
____
synthesis
by cytosine
arabinoside had littleor0 2 4 6 8 10 no effect on the levels of methyltransferase
ac-HOURSAFTERINFECTION tivity (Fig. 1B).
FIG. 1. Time course of guanylyltransferase and In the above experiment, incorporation of
methyltransferase activities after vaccinia infection. methyl groups was dependent on both
pp(A)"
Cell lysates from uninfected (U), infected (-),and and GTP since
G(5')ppp(A)n,
the product of theinfected cytosine arabinoside-treated (0) cells were guanylyltransferase, is the actual methyl
ac-assayed with 90 M[3H]GTP for guanylyltransfer- ceptor. Underthe latter assayconditions,a rise
ase(A)andwith 2.5mMunlabeled GTP and 0.5 M in methyl incorporation might simply reflect an
S-adenosyl[methyl-3H]methionine for methyltrans- increase in the rate-limiting
guanylyltrans-ferase activity (B) as described in the text. In each
erease mathe
heck.this
posblity,
case
pp(A),
was used as theacceptor,and eachpoint ferase reaction. To check this possibility,represents the average of duplicate assays. Cells were G(5')ppp(A)n was synthesized from pp(A)n by treated with 40pgof cytosine arabinoside per ml 15 using purified guanylyltransferase (14).
G(5')-min prior to infection, and the inhibitor was main-
ppp(A)n
was then used as the methyl acceptortained at this concentration during subsequent ma- in the absence of GTP and MgC12.
Further-nipulations. more, assays were carried out in the presence
of 2 mM EDTA toprevent the
guanylyltrans-to4hafter infection(9,20).Evidence that viral
ferase,
which isdependent
ondivalentcations, DNAreplication is notrequired for this effect from using any endogenous GTP. The results was obtained using cytosine arabinoside, an ofthis experiment (Fig. 2) indicate that the inhibitor of DNA synthesis. As shown in Fig. rise in methyltransferase activity is indepen-1A, asimilar riseinGMPincorporating activ- dent of andparallelsthe rise in guanylyltrans-itywas observedinthepresence orabsenceof ferase. An elevation in methyltransferase ac-cytosinearabinoside. That the inhibitor worked tivity preceded viral DNA synthesis and waseffectively was demonstrated by assaying the unaffected
by cytosine
arabinoside.same lysatesfor nucleoside triphosphate phos- Although all of the above experimentswere phohydrolase I, an enzyme also packaged carried out with total cell
lysates,
similarre-within vaccinia cores butpreviously shown to sultswereobtainedusing cytoplasmicextracts
besynthesized after viralDNAreplication(18). thatcontained the bulkof the activities.
A rise in phosphohydrolase activity was de- Since the previous methylation assays
pri-tectedonlyin lysates ofvaccinia-infected cells marily were designed to measure guanine-that hadnotbeen treated withcytosine arabi-
7-methyltransferase,
a specific nucleoside-2'-noside (datanotshown).methyltransferase
assaywas also devised. This Although viralDNAsynthesis isnotneeded involved theuseof brome mosaic virus (BMV)for the elevation inguanylyltransferase activ- RNA, which has 5'-terminal m7G(5')pppG (3) ity, protein synthesis is required. When cells as a substrate in astandard reaction mixture
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.501.53.242.56.318.2]478 BOONE, ENSINGER, AND MOSS J. VIROL.
containing S-adenosyl [methyl-3H]methionine crease was obtained using extracts from cells
but lacking GTP and Mg2+. Extracts of cells 8 h after infection (not shown). The
activity
at 1 h after infection had more activity than was dependent on added BMV RNA sincein-extracts ofuninfected cells, and a fourfoldin- corporation was reduced by more than 90%
in its absence. As with the other activities, cytosine arabinoside added prior to infection
o hadlittle effecton the timecourse.
x3 Analysis of reaction products. Thus far we
.D / haveassumed that the assays used in the
pre-o _-\ A / vious section specificallymeasure the
5'-termi-E nalmodification of added substrates. Direct
ev-o w infected idencetosupportthiswasobtained
by analysis
of the
products.
Mixturescontaining
pp(A)n,
0- /
/Mg2+,
[a-32P]GTP,
andS-adenosyl[methyl-o y / 3H]methioninewereincubated with anextract
infected+AraC prepared from HeLa cells 6 h after infection.
O /t The
poly(A)
wasthenpurified
from the reactioncc 1 1l _ mixture
by
adsorption
topoly(U)-Sepharose
z anddigestedwith P, nuclease, an enzyme that
hydrolyzesall 3',5'-phosphodiester linkages (6).
Upon
DEAE-cellulose
chromatography
in 7M uninfected urea,all of the labeled material eluted withthe net charges expected for P, nuclease-resistant 0 2 4 6 8 10 5'-terminal structures (Fig. 3). The second of HOURSAFTER INFECTION the two peaks, which contained 58% of the in-corporated 32pbut was notmethyllabeled, had FIG. 2. Time course ofmethyltransferse activity anetcharge
ofapproximately
-3andwascon-measuredindependently ofguanylyltransferase. As-
sidered
tobe
G(5')pppA.
The otherpeak,
which
says were carried out as described in the legend to conaiedto ofthe
The
labelakthich
Fig.lB,except thatG(5')ppp(A), was the acceptor, contained all of the methyl label and the
re-GTPand MgCl2were omitted, and 02 mMEDTA mainder of the32plabel, had a lower net charge
was added as described in the text. attributed to methylation at the 7 position of
? -1 -2 -3 4 -5 -6
14O 14
012 12 _X
I
lo ZO
4 40
-J X.~~~~~~~2
0 10 20 30 40 50 60 70 80 90 100 110 120
FRACTIONNUMBER
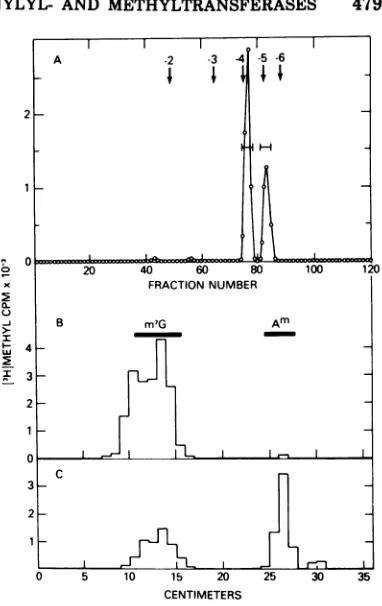
FIG. 3.Analysis of a P, nuclease digest of poly(A) labeled with [a-32P]GTP and
S-adenosyl[methyl-3H]methionineby an infected celllysate.Lysatesobtained from cells 6 h after vaccinia infection were usedto modify pp(A)n intwoseparate reaction mixtures. One contained 2.5 iM [a32PJGTPand 5uMunlabeled S-adenosylmethionine, and the other contained 2.5 mM unlabeled GTP and 0.5 AM
S-adenosyl[methyl-3H]methionine.After a30-min incubation at 37°C, the two reaction mixtures were combined and heated for 2
min at90°and poly(A) was purified by poly(U)-Sepharose and G-50 Sephadex chromatography andthen digestedwithP, nuclease. The digest was chromatographed on DEAE-cellulose in 7 M urea. The elution position andnetcharge of oligonucleotide markers are indicated.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.501.62.254.120.323.2] [image:4.501.113.402.388.579.2]479
guanineand was consideredto be m7G(5')pppA
and/orm7G(5')pppAm. A -2 -3 4 -5 -6
Discrimination between m7G(5')pppA and I
m7G(5')pppAm was achieved in the following 2 experiments. The methylated product from re-actionmixtures containingpp(A)n, Mg2+, unla-beled GTP, S-adenosyl[methyl-3H]methionine,
andextracts fromcells that had been infected 1 _
with vaccinia virus for 6 h was digested with RNase T2. In contrast to P1 nuclease, RNase T2 releases nucleoside-3'-phosphates and cannot
hydrolyze phosphodiester linkages when there ° 2 6 - b 1J0
is a 2'-methyl substitution. Accordingly, two x FRACTION NUMBER
labeled digestion products, m7G(5')pppAp and ,
m7G(5')pppAmpAp, were anticipated. Upon 5 B m7G Am DEAE-cellulose chromatography, two methyl- 44
labeled peaks with net charges of -4.5 and EI3
-5.5 were detected (Fig. 4A). The first peak T 3
wasidentified as m7G(5')pppAp since, after fur- 2 ther digestion with snakevenomphosphodies- 1 _
terase and alkaline
phosphatase, only
labeledl [image:5.501.251.442.51.354.2]m7Gwas found (Fig. 4B). The second peakin c
Fig. 4A was identified as m7G(5')pppAmpAp 3
since, after snake venom phosphodiesterase 2
-and alkaline phosphatase digestion, similar r
amounts ofm7Gand Am wererecovered (Fig. 1
4C).
Thesmallamountof fastermigrating
ma- 0 5 10 2 £ terial shown inFig.4C may represent thedou- CENTIMETERSbly methylated
adenosine derivative N602'-di-blymethylatedadenosine
dhathasbeeriv div
he
0'-d
FIG. 4.Analysis ofanRNaseT2
digest ofpoly(A)termethyl-adenosie
thatHas been foNd
in2e
a labeled by aninfected
celllysate with
S-adeno-terminal structure ofHeLJa cell mRNA (24) as syl[methyl-3H]methionine. Lysates obtained from
wellas in vacciniamRNA synthesizedinHeLa cells 6 h after infection were used to modify pp(A)n in
cells (unpublished data). a standard reaction mixture containing 2.5 mM The methyl-labeled product obtained using GTP,2.5 mMMg2+,and0.5 M
S-adenosyl[methyl-BMV RNA as substrate for cell extract pre- 3H]methionine.Afterpurification by
poly(U)-Sepha-pared at 6 h after infection was analyzed by roseand G-50 Sephadex chromatography, the
prod-paperchromatography after P1 nucleasediges- uct was digested with RNase T2 and chromato-tion. Greater than 95% of themethyl labelco- graphed on a
DEAE-cellulose
column in 7 M ureamigrated
with markerM7G(5')pppGm.
Upon
(A). Each ofthepeaks
wasdesalted,
successivelycedigestiontonucleosidesby successive
digested
with
P, nuclease and amixture
ofsnakecompletedlgestlontonucleosldesbysuccesslve venom phosphodiesterase and bacterial alkaline
treatments with P1 nuclease, snake venom phosphatase, and then analyzed by descending
pa-phosphodiesterase, and alkaline phosphatase, per chromatography in isopropanol-water-NH4OH greater than 99% of the methyl label comi- (7:2:1) with indicated internal nucleoside markers. grated with Gm, proving that BMV RNAwas a (B) -4charge peak;(C) -5charge peak.
substrate for specific
nucleoside-2'-methyl-transferase presentinthe infected cell extract. Proceduresderived from thoseoriginally em-Isolation of guanylyl- and methyltransfer- ployedtopurify theguanylyl- and methyltrans-asesfrom infected cells. The resultspresented ferases from virus cores (15) were usedto iso-thus far indicated thatanincreaseinguanylyl- late the enzymes from infected cells. At 6 h and both methyltransferase activities occurs afterinfection, mock-infected and vaccinia vi-after vaccinia virus infection. In this section, rus-infected cells weredisruptedand the
cyto-we will demonstrate that the enzymes can be plasmic fractionsweretreatedwithsodium
de-isolated from extracts of infected cells, that oxycholate and dithiothreitol. Afterhigh-speed they have column chromatographic elution centrifugation to remove insoluble structural properties previously found for the guanylyl- proteins, thesupernatantwaspassed througha andmethyltransferases obtained fromvaccinia DEAE-cellulose column to remove nucleic virus cores, and that enzymes with similar acids. The material thatdidnotbindwasthen chromatographic propertiesare notdetectedin applied to a poly(U)-Sepharose column and
extracts of uninfected cells. eluted with a gradient ofNaCl. Column
on November 10, 2019 by guest
http://jvi.asm.org/
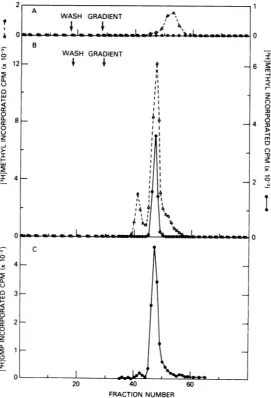
480 BOONE, ENSINGER, AND MOSS J. VIROL. tions wereassayed formethyltransferaseactiv- thereforeserve as anacceptor for the guanine-ity using two different methyl acceptors, syn- 7-methyltransferaseintheabsence of guanylyl-theticpp(A)11 and vaccinia mRNA made in vi- transferase activity. When material from unin-tro by vaccinia virus cores in the absence of fected cells was applied to the
poly(U)-Sepha-added
S-adenosylmethionine.
As previously rosecolumn (Fig. 5A), asingle peakofmethyl-mentioned, methylation of pp(A)n requires the transferase activity, eluting at 0.52 M
NaCl,
combined actions of guanylyltransferase and was detected using vacciniamRNA as theac-guanine-7-methyltransferase.
In contrast, vac- ceptor. The lack of detectable activity usingcinia mRNA contains guanylylated ends pp(A)n asacceptor inthepresence of GTP sug-[G(5')pppA- and G(5')pppG-] as well as free gested the absence ofan associated
guanylyl-diphosphate ends (ppA- and ppG-) and can transferase. Quite different results were
ob-2 A 1
WASH GRADIENT 0_
-:O 0
B
x
12
~WASH
GRADIENTt_6
q~~~~~~
x 1 - 6
I ~~~~~~~~~~~II
0-~~~~~~~~~~~~~~~~~~~~~~~~~-0
o ' ' 1
cr ~~~~~~~~~~~m
-J-cc 4 -M
2 >6
0~~~~~~~~~~~~~
CF
0
a-u
0
3-0
0 u
z
(2
0
20 40 60
FRACTION NUMBER
FIG. 5. Poly(U)-Sepharosecolumnchromatographyofguanylyltransferaseandmethyltransferases.
Cyto-plasmic extracts ofuninfectedorvaccinia virus-infected HeLa cells were passed throughaDEAE-cellulose columnandthen appliedto apoly(U)-Sepharosecolumnasdescribedinthe text. Thecolumnfractions from theuninfected (A)orinfected (B) cell extracts were assayed formethyltransferaseactivityinthepresenceof2 mM eachof GTP andMg2+, using eitherunmethylated vaccinia mRNA (A)orpp(A)n(0)asacceptor.(C)The fractions from thepoly(U)-Sepharosecolumnoftheinfected cell extract were assayed forguanylyltransferase activity using5 MM[3H]GTP as donor andpp(A)" asacceptor.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.501.125.397.196.595.2]VOL. 21, 1977 mRNA GUANYLYL- AND METHYLTRANSFERASES 481
tained uponchromatography of material from particles contain trace amount of S-adenosyl-vaccinia virus-infected cells (Fig. 5B). There methionine. (Reovirus mRNA synthesized in weretwodistinctpeaks,notseenafter chroma- the absence of added S-adenosylmethionine tography of the uninfected cell extract, which also appearstocontain some methylated termi-elutedatapproximately0.27 and0.37 MNaCl nals [2].)The conclusion that the 0.27 Mpeak as well as a shoulder which corresponded in (fraction 41) is a nucleoside-2'-methyltransfer-level of activity and elution position with the ase which utilizes RNA molecules terminated methyltransferase ofuninfected cells. Of these by m7G(5')pppN- wasconfirmed using tobacco peaks, onlythesecond eluting at0.37Mcould, mosaic virus (TMV) RNA, which, like BMV in the presence of GTP, methylate pp(A)n as RNA, terminates inm7G(5')pppG- (11, 28) as a well as vaccinia mRNA (Fig. 5B). Evidence substrate. When the peak fractions from the that the latter result was duetoguanylyltrans- poly(U)-Sepharosecolumn of Fig. 5B were reas-ferase was obtained by assaying the column sayed using TMV RNA, only the 0.27 M en-fractions for their ability to incorporate zymepeak exhibited activity. In addition, the [3H]GTP into pp(A)n (Fig. 50). These results 0.27 MNaCl elution position of the enzyme is suggested that whereas the0.27 MNaCl peak similar to that of the nucleoside-2'-methyl-was amethyltransferase, the 0.37MNaCl peak transferase isolated from vaccinia virus cores was a guanylyltransferase methyltransferase (unpublished data).
complex. A guanylyltransferase*guanine-7- DI
methyltransferase complex isolated from vacci- SCUSSION
nia viruscoreswasfoundpreviouslytoeluteat Three activities,anmRNA guanylyltransfer-asimilar (0.36M) concentration of NaCl (15). ase,anmRNA(guanine-7-)-methyltransferase, To identify further the enzymesisolated from andanmRNA
(nucleoside-2'-)-methyltransfer-infected cells,weused eitherfraction41(0.27 M ase,whichareresponsible for the modification
peak) or fraction 47 (0.37 M peak) of the of the5'-terminals of vacciniamRNA,are pack-poly(U)-Sepharose column in Fig. 5B to label aged within the virus core (4, 14, 15). Since unmethylated vaccinia mRNA with S-adeno- vaccinia mRNA (27) and its host HeLa cell
syl[methyl-3H]methionine. The methylated mRNA (7, 23, 25) have similarly modified 5'-products were digested with nuclease P1 fol- terminal sequences, the possibility exists that lowed by alkalinephosphatase andanalyzedby the virus-associated enzymesare derived from
descendingpaperchromatography (Fig.6Aand the cell. Alternatively, thelarge poxvirus ge-B). In both cases, all of the methyl label was nome could easily code for these enzymes. foundinP1 nuclease-resistant structures, indi- The results presented here favor the second cating exclusively 5'-terminal methylation. alternative. Using synthetic or natural
poly-The product of the 0.37 M enzyme peak co- ribonucleotides containing ppA-, G(5')pppA-,or chromatographed with marker m7G(5')pppG m7G(5')pppG- terminals as specific acceptor and m7G(5')pppA, indicating that the enzyme substrates,arise inguanylyl-and both
methyl-is a guanine-7-methyltransferase (Fig. 6B) as transferase activitieswasdetectedinlysates of well as a guanylyltransferase. This was con- infected cells.Uponpoly(U)-Sepharose chroma-firmedby furtherdigestion of another portion tography of cytoplasmic extracts of infected of the labeled RNA and identification of m7Gas cells, the guanylyltransferase and
guanine-7-the soleproduct(Fig. 6D). TheP1nucleaseand methyltransferase eluted together as a com-alkaline phosphatase digestion product of the plex, and the nucleoside-2'-methyltransferase
0.27 Menzyme peakco-chromatographedwith eluted separately. Both the chromatographic
m7G(5')pppGm and m7G(5')pppAm (Fig. 6A), properties and substrate specificities of these suggesting the presence of both guanine-7- enzymes were similar or identical to those of methyltransferase and nucleoside-2'-methyl- thecorrespondingenzymesisolated from vacci-transferase. Nevertheless, uponfurther diges- nia virus cores (15). In contrast, similar en-tiononly9%ofthe labelwasfoundin7-methyl- zymes could not be isolated from uninfected guanosine and the remainderwasfound in 2'- cells.
O-methylguanosine and 2'--methyladeonsine A rise in guanylyltransferase, guanine-7-(Fig.
60).
Since incompletely methylated methyltransferase, andnucleoside-2'-methyl-G(5')pppG"' and G(5')pppAm had not been de- transferase activities was detected within 1 h tected in the previous analysis (Fig. 6A), the after infection, and thetime course was unal-unlabeled vaccinia mRNA used as substrate tered by cytosine arabinoside, an inhibitor of evidently contained some m7G(5')pppN- ter- DNA synthesis. These results suggested that mini, even though this RNAwas synthesized theseenzymesshouldbeclassifiedas an"early"
by virus particles in vitro in the absence of or prereplicative viral gene product. Similar
addedmethyldonor. We assume that the virus studies havesuggestedthat another
on November 10, 2019 by guest
http://jvi.asm.org/
482 BOONE, ENSINGER, AND MOSS J. VIROL.
A 2 46_C
A
6.0B 1 Dc~~ m'GGm Am_
1 3 5-
-1.00 0.8
m'G(5')pppGE m'G(5')pppAx
0.75 0.6
0.50 0.4
lb 0.25 -0.2
O 0 103
-J 2 4 6 0.
z B D mGG At
FIG6.0 3 5 8
(10:)._hearkrsere(1 m'G(5')pppA
m m'G(5')pppG
4.5 -6
3.0 --4
1.5 2
followedbyalkaln phshr Jnsnk eo hshdetrs ndaaye ydsedn ae
0 10 20 30 40 0 10 20 30 40
DISTANCE FROM ORIGIN(cm)
FIG. 6. Identification oftheproductsofthevacciniavirus-inducedmethyltransferasesbypaperchromatog-raphy.Fractions 41 (027MNaCi)and47(0.37MNaCL) from thepoly(U)-Sepharose column
chromatogra-phyofthe infectedcellextract (Fig.cB) were used to label unmethylated vaccinia mRNA with
S-adeno-sylDmethyl-3H]methionine. (A andB)Aportion ofeachproductwas digestedwith nucleaseP1followedby
alkalinephosphataseandanalyzed by descendingpaperchromatographyin isobutyricacid-0.5 MNH,OH
(10:6). The markers were: (1) m7G(5')pppG, (2) G(5)pppGm, (3) miG(SipppGm, (4) G(5)pppAm, (5) m7G(5')pppA,(6)m7GpppAm.(CandD)AsecondportionofeachproductwasdigestedwithRNaseAandT, followed by alkalinephosphatase andsnake venomphosphodiesterase andanalyzed by descending paper
chromatographyinisopropanol-water-NH4OH(7:2:1). (AandC)Digested productoffraction41.(BandD)
Digestedproductaeffraction47.
ciatedenzyme, thepoly(A)
polymerase,
is also ofsomeofthe third nucleosidesaremethylated,
an"early"enzyme
(2) but that core-associated andaninternalnucleoside ismethylated
onthe DNases(13, 19) and nucleosidetriphosphatases
base (R. F. Boone and B. Moss, Abstr. Annu.(18, 19) are "late" enzymes.
Although
the ma- Meet. Am. Soc. Microbiol. 1976,s227,
p. 242).jority
ofpolypeptides
that form the vacciniaAttempts
to characterizeenzymes
fromunin-virionare
synthesized only
after DNAsynthe-
fected HeLa cells thatcancarryoutthese addi-sis(8, 10, 17, 21),atleast four have been shown tionalmethylations
arein progress.Thepartial
to be
prereplicative
(8, 10, 17). Of these, two purification andcharacterization ofaguanine-designated
2Aand6B (10) haveelectrophoretic
7-methyltransferase
from uninfected HeLamobilities similar to the 95,000- and 31,400- cells will be
reported
elsewhere (5).molecular-weight polypeptides
ofthepurified
guanylyltransferase guanine-7-methyltrans- ACKNOWLEDGMENTS
ferasecomplex (15). WethankScottMartinforpp(A).andG(5')ppp(A)n,P.
Although
theenzymes
responsible
for the for- Kaesbergfor BMVRNA, A. Marcus for TMVRNA, andmation of m7G(5')pppAm- and M7G(5')pppG'- NormanCooperforgrowingvaccinia virusand HeLa cells. M. J. Ensingerwas therecipientof Public Health Ser-terminals appear to be specific viral enzymes vicepostdoctoral fellowshipF22A102274 from the National that are packagedwithin the virus cores, fur- Institute ofAllergyand Infectious Diseases.
ther modifications of vaccinia mRNA may be
carriedoutbycellularenzymes. Recentstudies LITERATURE CITED
indicate that vaccinia mRNA synthesized in 1. Blatti,S.P.,C.J.Ingles,T. J.Lindell,P. W.Morris,
infected HeLa cells has three additional
modlifi-
R.F.Weaver, F.Weinberg,andW. J. Rutter. 1970.cations: the 6-position ofsomepenultimate 2'- Structure and regulatory properties of eucaryotic
0-methladeosinersidue are mthylaedto RNApolymerase.ColdSpringHarborSymp.Quant.
O-methyladensineresiues are mehylated to Biol. 35:649-657.
form N6,02'-dimethyladeonsine, the 2'-position 2. Brakel, C., and J. Kates. 1974. Poly(A) polymerase
on November 10, 2019 by guest
http://jvi.asm.org/
fromvacciniavirus-infectedcells. I. Partialpurifica- Biol. Chem.250:9322-9329.
tionand characterization.J. Virol. 14:715-723. 16. Moss, B., A. Gershowitz, C. M. Wei, and R. F. Boone. 3. Dasgupta, R., F. Harada, and P. Kaesberg. 1976. 1976. Formation of the guanylylated and methylated Blocked5' termini inbrome mosaic virus RNA. J. 5'-terminus of vaccinia virus mRNA. Virology Virol. 18:260-267. 72:341-351 (also seeerratum 75:260).
4. Ensinger, M. J., S. A. Martin, E. Paoletti, and B. 17. Moss, B., E. N. Rosenblum, and C. F. Garon. 1973. Moss. 1975.Modification of the 5'-terminus of mRNA Glycoprotein synthesis in cells infected with vaccinia by soluble guanylyl- and methyltransferases from virus. III. Purification and biosynthesis of the virion vaccinia virus. Proc. Natl. Acad. Sci. U.S.A. 72:3385- glycoprotein. Virology 55:143-156.
3389. 18. Paoletti, E., N. Cooper, and B. Moss. 1974. Regulation 5. Ensinger, M. J., and B. Moss. 1976.Modification of the of synthesis of twoimmunologically distinct nucleic 5'-terminusof mRNAbyanRNA(guanine-7-)-meth- acid-dependent nucleoside triphosphate phosphohy-yltransferase from HeLa cells. J. Biol. Chem. drolases in vacciniavirus-infected HeLa cells. J.
Vi-251:5283-5291. rol. 14:578-586.
6. Fujimoto, M., A. Kuninaka, and H. Yoshino. 1974. 19. Pogo, B. G. T., and S. Dales. 1969. Regulation of the Substrate specificity of nuclease P. Agric. Biol. synthesis of nucleotide phosphohydrolase and neutral Chem. 38:1555-1561. deoxyribonuclease:two activities present within pur-7. Furuichi,Y., M. Morgan, A. J. Shatkin, W. Jelinek, M. ified vaccinia virus. Proc. Natl. Acad. Sci. U.S.A.
Salditt-Georgieff, and J. E. Darnell. 1975. Methyl- 63:1297-1303.
ated,blocked 5'-termini in HeLa cell mRNA. Proc. 20. Salzman, N. P. 1960.The rate of formation of vaccinia Natl. Acad. Sci. U.S.A. 72:1904-1908. deoxyribonucleic acid and vaccinia virus. Virology 8. Holowczak, J. A.,andW. K.Joklik. 1967. Studieson 10:150-152.
the structural proteins of vaccinia virus. II. Kinetics 21. Salzman, N. P., and E. D. Sebring. 1967. Sequential ofthe synthesis of individual groups of structural formation of vaccinia virus proteins and viral deoxy-proteins.Virology 33:726-739. ribonucleic acidreplication. J. Virol. 1:16-23. 9. Joklik,W.K.,andY. Becker.1964.Thereplicationand 22. Urishibara, T., Y. Furuichi, C. Nishimura, and K.
coatingof vaccinia DNA. J. Mol. Biol. 10:452-474. Miura. 1975. Amodified structure at the 5'-terminus 10. Katz, E.,and B. Moss. 1970. Vaccinia virus structural of mRNA of vaccinia virus. FEBS Lett. 49:385-389.
polypeptide derived from a high-molecular-weight 23. Wei, C. M., A.Gershowitz, and B. Moss. 1975. Methyl-precursor.formation andintegration into virusparti- ated nucleotides block 5'-terminus of HeLacell mes-cles. J.Virol. 6:717-726. senger RNA. Cell4:379-386.
11. Keith,J.,andH.Fraenkel-Conrat. 1975.Tobaccomo- 24. Wei, C. M., A.Gershowitz, and B.Moss.1975. N6,02'-saic virusRNAcarries5'-terminaltriphosphorylated dimethyladenosine, a novel methylated ribonucleo-guanosine blocked by 5'-linked methylguanosine. side next to the5'-termin4lof animalcelland virus FEBS Lett. 57:31-33. mRNAs. Nature(London) 257:251-253.
12. Lowry,0.H., N.J.Rosebrough, A. L.Farr, andR.J. 25. Wei, C. M., A. Gershowitz, and B. Moss. 1976. 5'-Randall. 1951.Protein measurement with the Folin terminal and internal methylated nucleotide se-phenol reagent. J. Biol. Chem. 193:265-275. quences in HeLa cell mRNA. Biochemistry 15:397-13. McAuslan, B. R., andJ. R. Kates. 1967. Poxvirus- 401.
induced aciddeoxyribonuclease: regulationofsynthe- 26. Wei, C. M., and B.Moss. 1974. Methylationofnewly sis; control ofactivity in vivo;purification andproper- synthesizedviral messenger RNAbyanenzyme in tiesoftheenzyme.Virology33:709-716. vaccinia virus. Proc. Natl.Acad. Sci. U.S.A. 71:3014-14. Martin, S. A., and B. Moss. 1975. Modification of 3018.
mRNA by mRNA guanylyltransferase and mRNA 27. Wei, C. M., and B.Moss. 1975.Methylated nucleotides (guanine-7-)-methyltransferase from vaccinia viri- block 5'-terminus of vaccinia virus messengerRNA. ons. J.Biol. Chem. 250:9330-9335. Proc. Natl. Acad. Sci. U.S.A. 72:318-322.
15. Martin, S. A., E.Paoletti, andB.Moss.1975.Purifica- 28. Zimmern, D. 1975.The5'-endgroupoftobaccomosaic tion of mRNAguanylyltransferase andmRNA(gua- virus RNA ism7G(5')ppp(5')Gp. Nucleic Acids Res. nine-7-)-methyltransferasefrom vaccinia virions. J. 2:1189-1201.
on November 10, 2019 by guest
http://jvi.asm.org/
![FIG. 1.feraseaseS-adenosyl[methyl-3H]methioninemethyltransferaseinfectedassayedcaseCellrepresentstreatedtainedmin Time course of guanylyltransferase and activities after vaccinia infection](https://thumb-us.123doks.com/thumbv2/123dok_us/1549021.107425/3.501.53.242.56.318/feraseases-adenosyl-methyl-methioninemethyltransferaseinfectedassayedcasecellrepresentstreatedtainedmin-guanylyltransferase-activities-vaccinia-infection.webp)
![FIG. 3.positionadenosylmethionine,digestedmodifymin3H]methionine3H]methionine. Analysis of a P, nuclease digest of poly(A) labeled with [a-32P]GTP and S-adenosyl[methyl- by an infected cell lysate](https://thumb-us.123doks.com/thumbv2/123dok_us/1549021.107425/4.501.62.254.120.323/positionadenosylmethionine-digestedmodifymin-methionine-methionine-analysis-nuclease-adenosyl-infected.webp)