0022-538X/92/084639-08$02.00/0
Copyright©D 1992, AmericanSociety forMicrobiology
Transcription
of
the
E6
and
E7
Genes
of
Human
Papillomavirus
Type 6 in
Anogenital
Condylomata
Is
Restricted
to
Undifferentiated Cell
Layers
of
the Epithelium
T.
IFTNER,1
M.OFT,'
S.BOHM,'
S. P.WILCZYNSKI,2
AND H.PFISTER`*
Institutfur KlinischeundMolekulare Virologie, Friedrich Ale-xander Universitat
Erlangen-Nurnberg,
Schlossgarten 4, 8520Erlangen,
Germany,'
andDepartmentof Pathology, City ofHope,National Medical Center, Duarte, Califomia 910102
Received 21January1992/Accepted13April 1992
The E6 and E7 genes of human genital papillomaviruses (HPVs) appear to transform cells by different mechanisms. They seem to act
synergistically
but are not equally important when tested under diverse experimental conditions. We were therefore tempted to investigate the E6- and E7-specific transcription pattern inHPV6-infected condylomas separately, byinsituhybridization. Recent studies have identified three promoters within the E6-E7 region of HPV6 andHPVll by applyingSi,exonuclease VII, and cDNAanalyses.
On the basis ofthesedata,weclonedsubgenomicfragments of HPV6 intoplasmidpBS toobtainriboprobes thatdifferentiated between transcripts starting upstream oftheE6 andE7 openreadingframes, respectively. These different species of mRNAs were analyzed in serial thin sections of eight HPV6-positive anogenital condylomas. The E6 probe (nucleotides 7862to241) led to weaksignals within the basallayer.Inthree cases, rather strongsignals were confined to a few basal cells. The E7 probe (nucleotides 242 to 534) gave riseto a morepronounced labeling of all cells within the two to three lowest epidermal layers.Insituhybridization with a riboprobe for human c-fos revealed an expression pattern similar tothat observed with the E7 probe.In contrast tothepreferentialexpression of the transforming E6 and E7 genesinthe lowerepithelium, themajor transcriptional activityof the virus was detectedin themiddle and upper thirdby probes colinear with the 3'
moiety
of theearly
region.The oncogenic potential of human papillomaviruses
(HPVs) is reflected in vitroby their ability to immortalize
keratinocytes and to change their differentiation behavior
(17). Genetic experiments identified the E6 and E7 genes of
HPV16 asboth necessary andsufficient for this activity (9, 11). However, under certain experimental conditions, E6 or E7 wasable to immortalize humanprimary epithelial cells by itself (1, 8). It was recently suggested that genital HPVs induceproliferation, at least in part, through interaction of the viraloncoproteinsE6 and E7 withthetumorsuppressor
proteins p53 andRb, respectively (7, 12,23).
The E6 and E7 genes were shown to be retained and transcribed in most cervical carcinomas (24), which may indicate that they contributetotheproliferative potential of the cancer cells. The activity of these genes in benign HPV-inducedtumorsisnotyetclearly elucidated. Northern
(RNA)blot studies of HPV16 RNA revealed nodifferences
in the amount of E6 and E7 transcripts in premalignant versus malignant lesions (19). In situ hybridization with HPV6-, HPV11-, and HPV33-infected condylomas showed a differentiation-linked gene expression,however, and itwas
surprising to note that E6 and E7 transcripts were almost
undetectable in the proliferating lower layers (2, 21). The separateexpressionof E6 and E7was notassessed because ofuncertaintiesconcerningthestructureof thegene-specific transcripts.
Recently, two promoters were identified within the E6 open reading frame(ORF) of HPV6 and HPV11 (20). Initi-ationatpromoter P2(Fig. 1) leadsto atranscript with E7as thefirst translatable ORF.Transcripts derived fromP1 (Fig.
* Correspondingauthor.
1) could encode both E6 and E7, but reinitiation of transla-tionatthe E7AUG ishighly unlikely (10, 14); therefore,this
mRNAisprobably E6specific. On thebasis of these data,
we designed probes for in situhybridization which specifi-cally detect the E6 and E7 transcript of HPV6. Care was taken to avoid any overlap with the major E1-E4 mRNA class, which initiatesaround nucleotide(nt)700 in HPV6or betweennt 674 and 714 in HPV11 (Fig. 1) (4, 20).
MATERIALSANDMETHODS
Biological specimens and HPV typing. Specimens from randomly selectedpatientsofdifferentsexand age(Table 1)
were snap-frozen immediately after surgery and stored at
-70°C until processed. High-molecular-weight DNA was
extracted from the frozen tumor tissue. The polymerase chain reaction for HPV classification was performed as described previously (3). Eight condylomas from different sites of the anogenital region (Table 1) thatwereproven to contain HPV6 DNAwere chosen for in situ hybridization
analysis. Normal ectocervical mucosa was obtained from a
patientwithahistoryof breastcancerandwasproventobe
HPVnegative.
Construction ofspecific probes. For the synthesis of spe-cificRNAprobes,subgenomic restrictionfragments (Fig. 1) of HPV6b (6) were cloned into the SmaIsite of the vector
pBS+ (Stratagene) orpGEM1 (Promega). Theidentity and
orientation of each fragmentwere confirmed byrestriction enzyme analysis. The construct togenerate antisense ribo-probes for thec-fos gene is available fromAmersham.
Depending on the choice of the promoter, the in vitro-synthesized RNAs were either in the same polarity as
mRNA (sense) or in the opposite polarity
(antisense).
To4639
on November 9, 2019 by guest
http://jvi.asm.org/
4640 IFTNER ET AL.
00CD~~~~~~~~~cc>LOOtr~~~~-- O
123 4
HPV 6
probes
0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 kb
*
Pl1P2 P3 SA
_______
_______
C
-~~~~~~
-FG. 1. Genetic and transcriptional map of the earlyregion of HPV6andlocalization of themRNA-specific probes 1 to 4 used in thisstudy.The ORFs(shaded boxes) and the nucleotide positions of the probe boundaries (at the top) correspond to the published sequence (18). Vertical lines within the ORFs represent the first translational start codon. Themajor transcriptsof theearly region (a toc) aredepictedatthe bottom aslines with differentthicknesses correspondingtotheir relative abundances determined inprevious investigations (13, 20). The promoters areindicatedasP1toP3, and themajorspliceacceptoris indicated asSA.
generateantisenseprobes,thepBS+-based RNA expression vectors were each linearized with HindIll cutting in the multiple cloning site distal from the T3 bacteriophage pro-moter used for transcription. The HPV6 DNA fragment cloned in pGEM1to synthesizeprobe4(Fig. 1) spanned nt 2792 to 4029. After cleavage with AvaIl at nt 3491, the appropriate antisense RNA was transcribed from the T7 promoter. In vitro RNA synthesis in the presence of
[35S]UTP, resultingin probeswithaspecific activityof 3 x
108 cpm/pg, was followedby alkaline hydrolysis to reduce
theprobelength to approximately200nt (5). Thequalityof thetranscribed RNAswastestedbyagarosegel electropho-resis. The shortestprobe (probe 1,withaprobe complexity of 143nt)wasapplied at aconcentrationof 6 x 107cpm per
ml of hybridization mixture (0.15 ,ug/ml/kb of probe
com-plexity), and the others were normalized in relation to the probelength.
[image:2.612.58.297.73.205.2]RNA in situ hybridization. Serial cryostat sections of biopsy material, mounted on aminopropylsilane-coated slides, were fixed in 4% paraformaldehyde in phosphate-buffered saline anddehydratedthrough gradedethanols. The dry sectionswerethenacetylatedin 0.1M triethanolamine-0.25% acetic anhydride for 10 min, washed in 0.2x SSC(lx SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and preincubated for 2 h at 45°C covered by solution I (50%
TABLE 1. Clinical data
Case Localization Patient data
Pretreatment
no. oflesion Age (yr) Sex'
1 Genital 2 F None
2 Vagina 22 F None
3 Perianal 22 M Unknown
4 Perianal 31 M None
5 Vulva 16 F None
6 Perianal 28 F None
7 Genital 2 M None
8 Penis 41 M None
aF,female; M,male.
formamide, 0.6 M NaCl, 2.5x Denhardt's solution, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.1% sodium dodecyl sulfate, 0.15 mg of tRNA per ml). After removal of this prehybridizationsolution, the tissue sections were incubated for 16 h at 45'C with 35S-labeled antisense riboprobes in solution I containing 10% dextran sulfate. The incubated slides were washed in 50% formamide-lx SSC at 500C, treated with RNase A(5 ,ug/ml), washed for 1 h each in0.2x SSCat50°C and0.lx SSC at60°C, dehydrated,and finally coated with Kodak NTB-2 emulsion. All slides from one specimenweredeveloped after the same exposure time (10 to 14 days) to allow direct comparison of signals obtained with different probes. Hematoxylin-and-eosin-counter-stained sections were observed and photographed with a Reichart & Jung Polyvar microscope by usage of a dark- or bright-field condenser. The specific detection of RNA by in situ hybridization was established by using sense ribo-probes,which gave only background signals, or by pretreat-mentof the tissue withDNase-free RNase before hybridiza-tion. The intactness of the RNA in the tissue sections could be demonstrated by hybridization with aprobe specific for anepithelial cell adhesion molecule (E-cadherin [16]).
RESULTS
Toobtain riboprobes specific for different mRNA classes ofHPV6, weclonedsubgenomic viral DNAfragments into the RNAexpression vectorpBS+ (Stratagene) or pGEM1
(Promega). Probe 1 is specific for E6 mRNAs starting at
promoterP1(Fig. 1),whereasprobe 2 recognizes all mRNAs starting at promoter P2 or upstream of it. The net signal resulting from the E7 mRNA startingatP2canbeobtained
by subtracting signals generated by probe 1 from signals
generated by probe 2. Probe 3 hybridizes only to mRNA
specieswith thecodingcapacity forafull-length E2protein
(Fig. 1). Antisense probe 4 is complementary to all known
mRNAspecies of HPV6 exceptfor the L2 mRNA.
With the helpof theseprobes,weanalyzed thepatternof HPV6 RNA expression in adjacent serial sections of eight
condylomataacuminata fromdistinctsites of theanogenital
region, whichwereprovenby polymerase chain reaction and in situ hybridization to contain HPV6 DNA (data not
shown). Each of the four different riboprobes resulted in
highly characteristic signal patterns (Fig. 2), which were consistent for allcondylomas tested.
Fivecondylomas hybridized with theE6 probe(probe 1) revealed homogeneously distributed weak signals in the
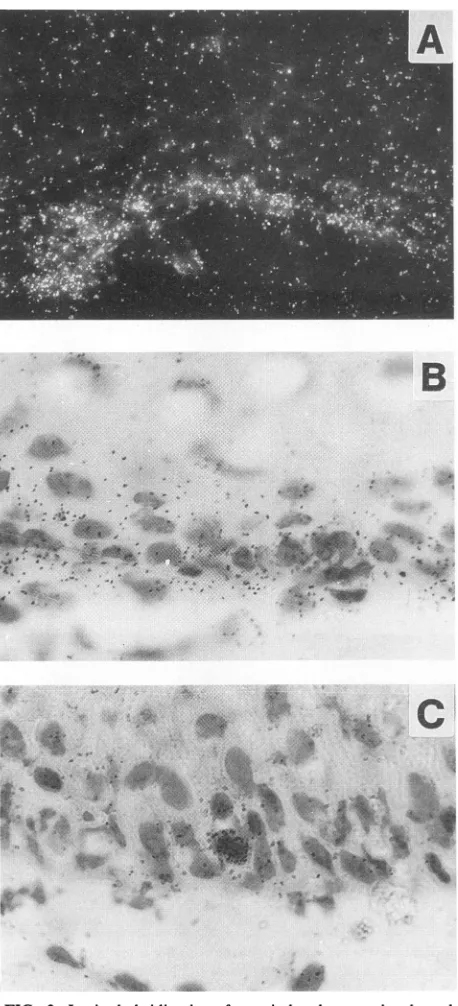
cytoplasmof basal cells(Fig.3A andB),whereas three other
condylomasexhibited onlyafewheavilylabeled cell nuclei
within the basallayer(Fig. 2B and 3C). In some cases, weak signalswerealso discernible insuprabasal cells.
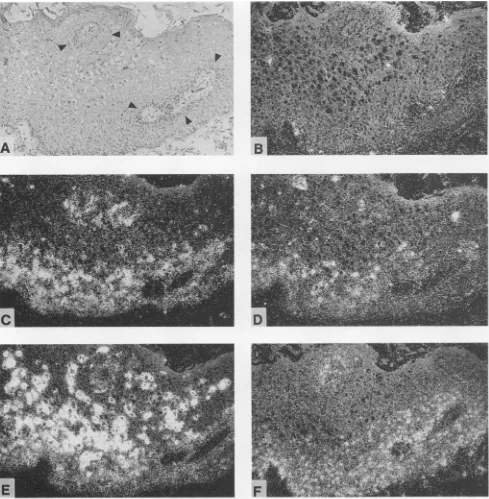
The E7 probe (probe 2) gave rise to a rather uniform cytoplasmic labeling of all cells within the two to three lowestlayers of thestratum germinativum (Fig. 4A, C, and E). No signals could be observed inhigher layers, with the exception that inonecondyloma,labelingextended through-outthe lower third(Fig. 2C).Thesignals in Fig.2C that seem to appearin the upper third oftheepidermis arelocated in undifferentiated basal cells around a crosswise-cut papilla, as canbeseenbycomparisonwith thebright-field histology
micrograph (Fig. 2A).When adjacent sectionsofall
condy-lomas were analyzedfor transcription of thec-fos gene by hybridization with an antisense probe, the distribution of
signalswassimilartothatobserved with theE7probe(Fig.
J. VIROL.
on November 9, 2019 by guest
http://jvi.asm.org/
VOL.66, 1992
..t..\
.Sv*
*.1;t8> , .4 8@- t*&
t2;
TRANSCRIPTION OF HPV6 E6 AND E7 GENES 4641
E
_
F
_
1
FIG. 2. Insituhybridization analysis ofananal condyloma whosehistopathology is shown by light microscopy after hematoxylin-and-eosinstaining (A). Adjacent tissue sectionswerehybridizedtoHPV6-specific probes1(B),2(C), 3(D), and4(E) (Fig. 1) and withahuman c-fosantisenseriboprobe(F).Thesilvergrainsgenerated in the film emulsion afterexposuretothe
35S-labeled
probesare seen aswhitegrains under dark-fieldillumination (BtoF). Black arrowheads indicatethelocations oftwocrosswise-cutpapillas.2F and 4). Investigation of normal HPV DNA-negative ectocervical mucosa revealed an identical c-fos expression
pattern, whereasthe HPV6 E7probe didnothybridize(data
not shown).
Hybridizationwith the E2probeyieldedsignals dispersed
through thelower and middle parts of the epithelium, with
somestrongsignalsin thenuclei ofkoilocytes(Fig. 2D and
5A).
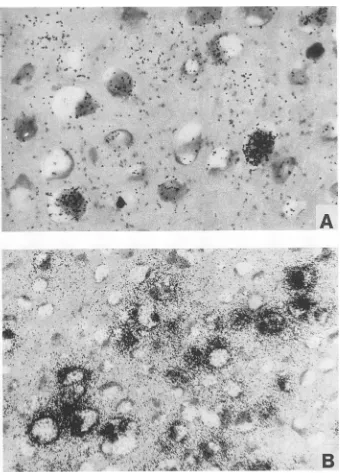
The major transcriptional activity of the virus wasdetected withprobe4 (Fig. 1)inthe middle and upper third
of theepithelium. The strongestsignalswere seenin koilo-cyte clusters with a perinuclear
cytoplasmic
distribution(Fig.2Eand
5B).
As therewere noE6-orE7-specificsignals
detectable in these differentiated areas, this massive label
probably representsthehighly abundantE1-E4 transcripts,
which are initiated downstream from the 3' end of the E7
probe (4), and latetranscripts havingashort exonfrom the
E4region
(15).
Hybridizationwithprobe4alsoledto asignalwithin the lower third of theepithelium, whichwas
on November 9, 2019 by guest
http://jvi.asm.org/
[image:3.573.46.535.73.572.2]4642 IFTNER ET AL.
I
w-.
FIG. 3. In situhybridizationofagenitaland twoperianal condy-lomata with the E6-specific riboprobe 1 demonstrated by light microscopy. Shown are details of the basallayersin dark-field(A) andbright-field (BandC) illumination.
rable tothat obtained with the E7 probe in both localization and signal strength (Fig. 2E). This finding was not unex-pected, because E7 mRNAs are known to contain a second exon largely colinear with probe 4 (20). The spatial separa-tion of the E7-specific mRNA (detected by probes 2 and 4) and the E1-E4 mRNA(detected by probe 4 only) becomes particularly obvious in Fig. 6, in which the strong signals generated by probe 4 appearedtobeunusually focal.
DISCUSSION
The infectious cycles of all DNA viruses appear to be
sequentially organized. Early proteins ofpapovavirusesand
adenoviruses drive the host cell into the S phase to allow
viral DNAreplication and then the expression of structural proteins. This activity of the early proteins may lead to an
oncogenic transformation ofabortively infected cells.
Inthe caseofpapillomaviruses, the multiplication cycle is
particularly complicated because of its strong dependence
on thedifferentiation state of the host cells. It starts witha
latent infection of the basal keratinocyte, proceeds with
ongoing cell maturation, and ends with the production of
virus particles in the uppermost layers of the epithelium.
Thisprocess is reflectedbyacell differentiation-dependent
geneexpression, whichcouldbe demonstratedby
morphol-ogy-basedmRNA detectioningenital condylomas (2, 21).
Witheach of fourearly-gene-specific probes,weobserved
a characteristic differentiation-dependent transcription
pat-tern that was consistent in eight condylomas from four distinct sites of the anogenital region. The majority of
transcripts of the early region appeared in the middle and
upperthird of theepithelium.Inaccordancewithpreviousin
situ hybridization studies with subgenomic probes (2, 21),
thebulk of thesignalin this area mostlikelyrepresents the E1-E4 mRNA, whichwasidentifiedpreviously byNorthern
blot hybridization as the most abundant transcript (13). A
minor percentage of thesignalin themidstratumseems tobe
due to mRNA with a coding potential for a full-length E2
protein.
Previousinsituhybridizationstudies withaprobe colinear
with the growth-promoting E6 and E7 genes surprisingly revealed an expression pattern similar tothatof the E1-E4 mRNA, with almostno E6and E7transcripts detectable in
themitoticallyactive basal celllayer(2, 21). In contrast,we
found E6- and E7-specific mRNAs strictly confined to the lowest layers of the epithelium. These differences are
par-tially dueto the fact that the earlier in situ studies applied
ORF-specific probes, whereas
transcript-specific
probeswere used in this study. The ORF-specific probe for the detection of HPV6orHPV11 E6 and E7 mRNA(nt7694to
811 [21]) contains
approximately
100nt thathybridize withthe 5' end of the E1-E4 messages, which were shown to initiate between nt 674 and 714 (Fig. 1, P3 [4, 20]). It will thereforeprimarilydetect thehighlyabundant E1-E4 mRNA transcripts, whichare 50 to 100 times moreprevalent than theE6- andE7-specific transcripts in basal cells.Usingsuch
anORF-specific probefor theanalysisofourspecimens,we
observed prominent signals in the upper part of the
epithe-lium(datanotshown). Theyweresimilar in locationtothose
FIG. 4. Correlationof HPV6 E7 and c-fos expression in two perianal condylomas and one genital condyloma. In situ hybridization was carriedoutwithriboprobe2 inantisense (A, C, and E) or sense (G) orientation, a riboprobe for human c-fos(B,D, andF), and a riboprobe fortheepithelial cell-specificadhesion molecule E-cadherin (H). Details of bright-field photographs of the lower strata are shown as insets inpanelsA, C, andE.
J. VIROL.
Aft.1 im
on November 9, 2019 by guest
http://jvi.asm.org/
[image:4.612.66.295.106.609.2]on November 9, 2019 by guest
http://jvi.asm.org/
4644 IFTNER ET AL. J. VIROL.
at,~^s, Ss ,
r,
;iNO
MAKOX
^^
;
~~~~~*
'-^ ' * '*+ ;{'t '';;' ,A~~~~~~~~~4't' 4' a *'^z'FIG. 5. In situhybridizationofagenital condylomawithriboprobes3(A)and 4(B)asdemonstratedby bright-fieldillumination.Shown aredetails of the midstratum of theepithelium.
obtained with probe 4 although lower in intensity, as one would expectfrom the differentlengthsof the 5' and 3'exons of the E1-E4 mRNA. It is not quite clear why previous studies failed todetect major signals in the lowestlayersof
theepithelium,whichweredetected here both withprobe2
(Fig. 2C, 4, and 6) and the E6 and E7 ORF-specific probe mentioned above (datanotshown) aswell aswith probe4 (Fig. 2E and 6B), which recognizes the second exon of E6 andE7transcripts(Fig. 1[4,22]). Thisdiscrepancycould be due to different sensitivities obtained with frozen versus
paraffin-embeddedmaterial and
35S-
versus3H-labeled
ribo-probes. The exclusiveexpressionof thetransformingE6and E7 genes in the basal cell layers is fully in line with their
expectedrole earlyin the viral lifecycle.
The different signal intensities observed after hybridiza-tionwith eitherprobe 1 orprobe2pointtotheexistence of twoclasses of mRNAs from the E6-E7region. Themajority of thetranscriptsdonotcontain sequences betweennt7862 and241becausetheyare notdetectable withprobe1.These RNAscontain E7asthe first translatable ORF andarelikely
toinitiateat nt270,whichwasidentifiedas amajor capsite ofmRNAsfrom HPV6-infectedcondylomas (20). The
tran-scripts hybridizingtoprobe 1,whichwasdesignedtodetect
E6-specificmRNAs,weremuch less abundant. The relative
signalintensities obtained withprobes1and 2correspondto
the amounts of mRNAs initiated atpromoters P1 and P2,
respectively(20).
Arelatively sharp borderline above whichnoE7-specific
on November 9, 2019 by guest
http://jvi.asm.org/
[image:6.612.138.478.71.546.2]TRANSCRIPTION OF HPV6 E6 AND E7 GENES 4645
A
..'I.%
..
-If.
*A
A
.ti
Ap;A
.*f#~~ ~ ~ ~ ~ ~ ~ ~ ~
V@F
*' ,-;£si t 2gg ,
.: @ .. ... e
,. ,>. ...
*s>
,l,t,*.
.j se. o- ws
' /' x ¢s;8J. ' sp
':Si.ii' tro
\. \% X
s a t. s i_y
* . g g
[image:7.573.129.437.56.708.2]tt + , # ;, ;. a,<3,i 1. s r*i
FIG. 6. In situhybridization ofan anal condyloma with riboprobe 4. The samesection is shownin light-field
(A)
and dark-field(B)
illumination.Theresultofhybridizationwithriboprobe 2 is visualizedbydark-fieldoptics(C). VOL. 66,1992
aa.,'
!",on November 9, 2019 by guest
http://jvi.asm.org/
4646 IFTNER ET AL.
transcripts could be observedwas aconsistent feature of all condylomas. That line also formed theboundaryof thearea showing c-fos expression. This coincidence doesnotreflect an influence of E7 on thetranscription of c-fos, because the c-fos expression pattern did notdiffer between condyloma-tous and HPV-negative, normal mucosa. However, one might speculate that only
c-fos-expressing
cellsprovidethe conditions allowing transcription from the E7 promoter. The lackof E6 and E7expression in the more differentiated cells of the condylomas points to a very effective control of the activity of promoters P1 and P2 (Fig. 1). Synthesis of the E6 mRNA seemstobeeven morerestrictedthanthat of theE7 mRNA.Previous studies noted that the total amounts of E6 and E7 mRNAs from HPV-infectedcondylomas differ by 1 order of magnitude (20). Our data provide the additional information that this difference is due not only to a lower abundance of E6 mRNA per cell but alsoto an expression strictly confined to undifferentiated cells of the basallayer. This highly restricted synthesis of the E6 mRNAindicates that the E6proteinactsveryearlyduring the multiplication cycle of the virus.ACKNOWLEDGMENTS
WethankA. Schmitt andM. Elbel for excellent technical assis-tance, G.Schmid forproviding biopsy material,W.Birchmeier for the E-cadherin plasmid, and T. Leist and P. Schirrmacher for helpful technicaladviceconcerning in situhybridization.
This researchwassupported bygrant 89.042.1from the Wilhelm-Sander-StiftungtoT.I.andby PublicHealth Service grant CA53005 toS.P.W.
REFERENCES
1. Band, V., J. A. De Caprio, L. Delmolini, V. Kulesa, and R. Sager. 1991. Lossofp53proteinin humanpapillomavirustype 16E6-immortalized human mammaryepithelialcells. J. Virol. 65:6671-6676.
2. Beyer-Finkler, E., M. H.Stoler, F. Girardi, and H. Pfister. 1990. Celldifferentiation-related geneexpressionof human papilloma-virus 33. Med.Microbiol. Immunol. 179:185-192.
3. Bloss,J. D., S. Y.Liao, S. P.Wilczynski,C.Macri, J. Walker, M. Peake, and M. Berman. 1991. Clinical and histological featuresofvulvar carcinomasanalyzedfor human papillomavi-rus status:evidencethat squamous cell carcinomas of the vulva has more thanoneetiology. Hum.Pathol. 22:711-718. 4. Chow, L.C., M. Nasseri, S. M. Wolinsky, andT. R. Broker.
1987. Humanpapillomavirustypes 6and 11 mRNA fromgenital condylomataacuminata. J. Virol. 61:2581-2588.
5. Cox,K.H.,D. V.DeLeon,L. M.Angerer, and R. C. Angerer. 1984. Detection of m-RNAs in sea urchinembryos by in situ hybridization using asymmetric RNA probes. Dev. Biol. 101: 485-502.
6. deVilliers, E. M., L. Gissmann, and H. zur Hausen. 1981. Molecularcloning of viral DNA from humangenitalwarts. J. Virol. 40:932-935.
7. Dyson, N., P. M. Howley, K.Munger,and E. Harlow.1989. The humanpapillomavirus-16E7oncoproteinis abletobindtothe retinoblastomageneproduct.Science243:934-937.
8. Halbert, C. L., G. W. Demers, andD.A.Galloway.1991.The E7 gene of human papillomavirus type 16 issufficient for
immor-talizationofhumanepithelialcells. J. Virol. 65:473-478. 9. Hawley-Nelson, P., K. H. Vousden, N. L. Hubbert, D. R. Lowy,
and J. T. Schiller. 1989. HPV 16 E6 and E7 cooperate to immortalize human foreskin keratinocytes. EMBO J. 8:3905-3910.
10. Iftner, T., G. Sagner, H. Pfister, and F. 0. Wettstein. 1990. The E7 protein of human papillomavirus 8 is a nonphosphorylated protein of 17 kDa and can be generated by two different mechanisms. Virology 179:428-436.
11. Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. TheE6andE7 genesof the human papillomavi-rus type 16together are necessary and sufficient for transforma-tion of primary human keratinocytes. J. Virol. 63:4417-4421. 12. Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow,
and P. M.Howley.1989.Complex formationof human papillo-mavirusE7proteins with the retinoblastoma tumor suppressor geneproduct. EMBO J. 8:4099-4105.
13. Nasseri, M., R. Hirochika, T. R. Broker, and L. T. Chow. 1987. Ahuman papillomavirus type 11 transcript encoding anE1/E4 protein. Virology 159:433-439.
14. Peabody, D. S., S. Subramani, and P. Berg. 1986. Effect of upstreamreading frames on translation efficiency in simian virus 40 recombinants. Mol. Cell. Biol. 6:2704-2711.
15. Rotenberg, M. O., L. T. Chow, and T. R. Broker. 1989. Characterization ofrarehumanpapillomavirustype 11 mRNAs codingforregulatoryand structuralproteins, usingthe polymer-asechain reaction. Virology172:489-497.
16. Schipper, J. H., U. H. Frixen, A. Unger, K. Jahnke, and W. Birchmeier. 1991. E-cadherin expressionin squamous cell car-cinomas of head and neck: inverse correlation with tumor dedifferentiation and lymphnode metastasis. CancerRes. 51: 6328-6337.
17. Schlegel, R., W. C. Phelps, Y.-L. Zhang, and M. Barbosa. 1988. Quantitative keratinocyteassay detects twobiologicalactivities of humanpapilloma virusDNAand identifies viral types asso-ciatedwithcervical carcinomas. EMBO J. 5:3181-3187. 18. Schwarz, E., M. Durst, C. Demankowski, 0. Lattermann, R.
Zech, E. Wolfsperger, S. Suhai, and H. zur Hausen.1983. DNA sequence and genomeorganizationofgenitalhuman papilloma-virustype 6b. EMBO J.2:2341-2348.
19. Shirasawa, H., Y. Tomita, K. Kubota, T. Kasai, S. Sekiya, H. Takamizawa, and B. Simizu. 1988.Transcriptionaldifferences of the humanpapillomavirustype 16 genomebetween precancer-ouslesions and invasive carcinomas. J. Virol. 62:1022-1027. 20. Smotkin, D., H. Prokoph, and F.0.Wettstein. 1989.Oncogenic
andnononcogenichumanpapillomavirusesgeneratethe E6 and E7 mRNAbydifferent mechanisms. J. Virol. 63:1441-1447. 21. Stoler, M. H., S. M. Wolinsky, A.Whitbeck,T. R.Broker, and
L. T. Chow. 1989.Differentiation-linkedhumanpapillomavirus types 6 and 11transcriptioningenitalcondylomata revealed by in situhybridizationwithmessage-specificRNAprobes. Virol-ogy 172:331-340.
22. Ward, P., and P. Mounts. 1989. Heterogeneity in mRNA of human papillomavirus type-6 subtypes in respiratorytract le-sions.Virology168:1-12.
23. Werness, B. A., A. J.Levine,and P. M.Howley. 1990. Associ-ation ofhumanpapillomavirustypes 16 and 18 E6proteins with p53. Science248:76-79.
24. Wettstein,F.0.1990.State of viral DNA and geneexpressionin benignversusmalignanttumors, p. 155-179. In H.Pfister(ed.), Papillomavirusesand human cancer. CRCPress, Boca Raton, Fla.
J. VIROL.