JOURNAL OFVIROLOGY, Feb. 1991, p. 1046-1052 0022-538X/91/021046-07$02.00/0
CopyrightC)1991, AmericanSociety for Microbiology
Expression and
Characterization
of the
Thymidine Kinase
Gene of African Swine Fever Virus
A. M. MARTINHERNANDEZ AND E. TABARES*Departamentode Microbiologia, Facultad de Medicina, UniversidadAutonoma de Madrid,
Arzobispo
Morcillo4,
28029Madrid,
Spain
Received5July1990/Accepted 26 October 1990
The thymidinekinase(TK)geneof African swine fever virus (ASFV)waslocatedwithinthe viralgenomeby using two degenerate oligonucleotide probes derived from sequences of the vaccinia virus and cellular TK genes. The TKgenewasmappedwithin a0.72-kbpBglll-XhoI fragment (0.242to0.246mapunits) derived
froma23.9-kbpSal-Bfragment oftheASFVgenome.Identification ofthisregionastheASFVTKgene was confirmed by expression ofTK inEscherichia coliand by the synthesis ofactive TK in a cell-free system programmedwithRNA synthesized invitro.Thesequencedgenefor TK includesanopenreadingframe of588
nucleotidesencodingaproteinof 196 amino acids. The deduced amino acidsequenceshows32.4% identity with
the TKof vaccinia virus.
Thethymidine kinase (TK) marker hasprovenvaluable in
the establishmentof vaccinia virus and herpes simplex virus cloningvectorsandforthedevelopment ofsurrogate
genet-ics in these viruses (18, 21). Therefore, it is of particular interest toidentify andcharacterize the African swine fever virus (ASFV) TKgene as apotentialtarget site for genetic manipulations. ASFV is an icosahedral cytoplasmic DNA virus withpropertiescommontoiridoviruses and poxviruses (30, 34). It causes a highly contagious and generally fatal disease ofpigs (8, 9, 16, 23, 34, 35). Productive infection is accompanied byaweakshutoff of hostprotein synthesisand
the appearance of44 virus-specific polypeptides ranging in
molecularweight from 9,500to243,000 (5, 27, 29, 30). These polypeptideshavebeenclassifiedasimmediateearly, early, and late (5, 33). Increases in the levels oftwo enzymatic activitieswhich maybe involvedin DNAreplication, those
ofaTK(19)andaDNApolymerase (20),havebeen detected followingASFV infection. ASFV infection ofbaby hamster
kidney (BHK) cells may induce the formation of a TK different from thatfound in normal cells(19). This activity canbe induced in BHK cellsdeficient in TKas an
immedi-ate-early protein, and we have generated ASFV TK-
mu-tants by bromodeoxyuridine mutagenesis (datanot shown).
These results strongly suggested that the TKinduced after infection is encoded byASFV. Herewe reportthe identifi-cation, characterization, and expression of the ASFV TK gene as aninitialsteptoward the geneticmanipulation of this
virus.
Mapping of the ASFV TK gene. The use of degenerate oligonucleotides to locate the TK genes of Shope fibroma
virus and avipoxvirus (26, 32) suggested that such
oligonu-cleotides couldalso be usedfor ASFV, given the
phyloge-netic proximity between ASFV and the poxviruses. Oligo-nucleotide pools 1 [GG(A/G/T/C)CCCATGTT(T/C)TC(A/G/ T/C)GG] and 2 [GA(T/C)GA(G/A)GG(G/A)CA(G/A)TT(T/C) TT], representing conserved regions inthe 3' portion of the
vaccinia virus, human, and mouse TK genes (32), were synthesized by F. Barahona in Centro deBiologia Molecu-lar, Madrid, Spain. ASFV DNA isolated from MS cells
*Correspondingauthor.
infectedwith ASFV strainE70MS44 (31)wasdigestedwith ClaI, SalI,andSmaI andelectrophoresedinanagarosegel.
The DNA fragmentswere denatured in thegel, transferred
to a nitrocellulose filter, and hybridized tothe
32P-oligonu-cleotideprobes (32). Probe 1 annealed with theSall-A, -B, -H', and -E fragments (see Fig. 4). This indicated possible viral sequences related to proteins that contain an
ATP-binding domain, since probe 1 represents a consensus se-quenceforsucha nucleotide-binding site. Probe 2 annealed onlywith theSalI-BandSalI-Efragments (datanotshown).
We studied theSall-B fragmentbecauseSalI-Eencodes late
proteins (22) and the TK of ASFV is an immediate-early protein.ASFVDNAisolated from strainE70MS14wasused intheconstructionof therecombinantplasmid pRPEM513,
and theEcoRI-Kfragmentwas subcloned inpUC18 (Fig. 1) bystandardtechniques (15). Theseplasmidswere subjected
torestrictionenzymeanalysis,and the DNAfragmentswere hybridized to 32P-labeled oligonucleotides in order to map the TKgene more precisely. Aregionofhomology with the TKgenewaslocalized ina390-bpPstI-HindIII fragmentof
pRPEM204 (Fig. 2, lanes 2). Sail-B represents unique
se-quences on theviral genome because this segment
hybrid-izesonlywithfragments SmaI-A,ClaI-B, -F, -P,and-R,and
Sall-B (reference31 and datanotshown), indicatingthatthe TKgeneisa single-copygene.
Identificationof the ASFV TKgene.The TKgene andits
flanking regions were sequenced by the dideoxynucleotide
chain termination method(24),with recombinant pUC18or pGEM3Z as the template. The nucleotide and deduced
protein sequences are presented in Fig. 3. The TK gene mapsatcoordinates 0.242to0.246(Fig. 4)inE70MS14DNA
(31).ThepredictedASFV TKprotein comprises 196 amino
acids, with a calculated molecular weight of 22,394. The
translation initiation codon of the open reading frame has been assigned on the basis of homology of the amino-terminal domain to that of other TK proteins. Also, the
proposedinitiator AUG is flankedbynucleotides character-istic ofpreferred eucaryoticinitiationsites,whereas thenext ATG codon, located 93 nucleotides downstream, is not flankedby consensustranslation initiation sequences(25).
Characterization and expressionof the TK gene in
Esche-1046
Vol.65, No. 2
on November 10, 2019 by guest
http://jvi.asm.org/
E
B
p
x
H/EaBEn /Bom HI
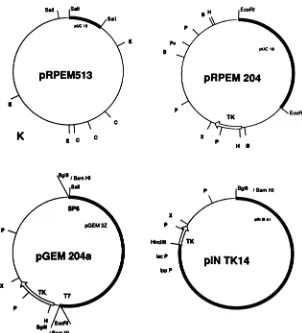
FIG. 1. Physicalmapofplasmids containing the TK gene. Viral DNA was digested withSalIandcloned in plasmid pUC18 (2,686 bp), and recombinant plasmids were obtained upon transformation of E. coli NM522. One of these plasmids, pRPEM513, contained the SalI-B (23.9-kbp) and
SaII-J
(0.9-kbp) fragments (31). After digestion withEcoRI, the EcoRI-K fragment (1) was subcloned in pUC18 to obtain plasmidpRPEM204. The largerBgIIIfragment and theHindIII-BglIIfragment ofthis plasmid were cloned in pGEM3Z(2,743bp)(Promega Biotec, Madison, Wis.)andpINIIIAl(7.4kbp) (kindly supplied by E.Garcia,Madrid, Spain) (4, 12), respectively, toyieldpGEM204aand pINTK14, respectively. RestrictionenzymesitesBglII(B),ClaI(C),EcoRI(E),HindlIl (H),PstI(P),PvuI(Pv), andXhol(X) are indicated.richiacoli cells. TheHindIII-BglII fragment was cloned into the HindIll and BamHI sites ofpINIIIAl (4, 12) to yield
plasmidpINTK14 (Fig. 1).Inthis clone, the N-terminal five
amino acids of ASFV TK protein are replaced by the five
A
B
1
23456789
1
2345 6789
amino acids encoded bylinker DNA from pINIIIAL. Plas-midpINTK14wasused totransformE. coliKY893 (TK-). Untransformed E. coli KY893 TK- is unable to grow on
selection mediumcontaining5-fluorouracil (11).Onlyclones containing viral TK from pINTK14, which complemented the cellular TKdefect,survived on the drugselectionplates. It was important to confirm the TK complementation of KY893 TK- cellsharboringtheplasmidpINTK14by
exam-ining incorporation of [3H]thymidineinto the bacterial cells. TheKY893mutantofE. coli transformed with theplasmids was cultured overnight in peptone-glucose medium (10)
containing (per ml) 50 ,ug ofampicillin, 25 ,ug of
5-fluorou-FIG. 2. Mapping of the ASFVTK genewithinplasmid pRPEM 204.(A)Lanes 1to8, pRPEM204digestedwithHindlIl plus EcoRI, HindlIl plus PstI, XhoI plus PstI, BgIIH plus PstI, BglII plus HindIII, BglII plus HindlIl plus EcoRI, EcoRI,andBamHI, respec-tively;lane9,plasmid pUC18digested with RsaI.Thegelwas0.75% agarose(31),and the DNAwasstained withethidiumbromide.(B)
Southern blot of thegelshown inpanelAprobedwithend-labeled oligonucleotide pool (map positions of the various restriction
en-zymes are shown in Fig. 1). The asterisk indicates the 390-bp
PstI-HindIIIfragmentofpRPEM204.
a&
1769bp
676bp
241bp
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.152.454.70.403.2] [image:2.612.64.288.552.724.2]1048 NOTES
MA
p x
GCRARRRGTR
CCGCRRRTRR
RRRRRCRRCG
RRGGGCTCCT CCRRRTCTOG TTCCTCCRGR
GGCCRCRCCGOCRRRRCCCR
TGCTTCTTCG
TCCRTGCRTT CCGGGRTGCT CTRTRRRGRT
Bgl
URTGGTRRRTR
TTGCTRfiRTC
TRGRGGCRTT CCGRTTTRCC
RGRRTGGRTC
GCGTCTTRCT
RBS
fidmRRRRGTGRRT
TGGRGRRRRRRRTRRRCffILCRM
RTG RRT RTR RTT RGGfiflCtL
M N I I R K L
RCR RTT
RGC
CTTT I S L
RTT
CRT
TGC RTTI H C I
RRR
TCT
RCC RRRK S T K
CAG
CTR CGR CCCo L R P
GTG GGR
TCT CTCV G S L
TTT
GRC GRT TTRF D D L
RTT CTT GCG GGR
I L A G
GTT
COT
RTT TTTV R I F
RTG RRR TGT RRC
M K C N
RRG
RCO
CTT RTCK T L I
RRC
TGT
CTR RRRN C L K
GTG
CTO
GGR CCCV L G P
TRC RTG CTC
ORR
Y M L E
RRC RCC
CGR GRC
N T R D
RRG
CRR TGT RRRK Q C K
RCC GRT RTC CRT
T D I H
RTR RRR TGC CGC
I K C R
CTC
RRT OCT
TCCL N A S
PsI
CCT
TRC
RAC
TGCP. y C S
CRR CRT RRT
OCR
Q H N A
CTT
GCG GORGOR
L A G G
RRT
RCRTTT RTT
N T F I
RTG TTT
0CC
GOC
M F A G
CGT
TTG
GRR RRRR L E K
RRR
ACT RTT
RRRK T I K
RTC RTR GRR RGC
I I E S
GCR GTT
GTC
GTR
A V V V
RCC TGG GCR GRG
T W A E
TTC GRG CRG RRR
F E Q K
RRR RCT K T RRR GTR K V RCR CRC T H
RCA
CR0
T O GRT GRR D E GRR GRR E ERTO
TTT
M FTOG OTT RRG TRT RTT GOC
W V K Y I G
TGC TTT RRT GTG COT RRG
C F N V R K
RGT GRR CTO TRC OTR RCR
S E L Y V T
RRG CR0 TTG CRR CCT RTT
K Q L Q P I
ARRTCTTRTR CRRTRRTOGR TCRTTRTCTT RRRRRRTTRC RRGRTRTTTR
XhOI
[image:3.612.132.494.89.185.2]TRCGRRGALfLQGGGGCCRTC CCTTTCTTTT TROCCCGTCG RRRRCCRRTO RRRRRGRGTT TRTTRCTCTO CTRRRCCROG CCTTGGCCTC RRCGCROCTT TRCCOCRGCR TRCRRCRACT GTTTTTRRCG RTOTATRROC TRGRTCCCRT TGOGTTTRTT RRCTRTRTTR RRRCGOGTRR RCRROROTRT TTATGCCTGT TORTTARTCC TRRACTCGTT RCTRRGTTTT TARRRATAAC
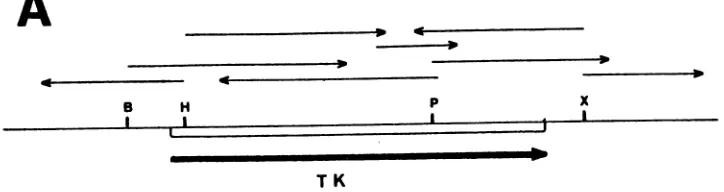
FIG. 3. NucleotidesequenceoftheASFVTKgeneand flanking regions. (A) The PstI-EcoRI 1.4-kbp TKgenefragmentfrompRPEM204was
inserted inpUC18toyield pRPEM207. Then the PstI-HindIII390-bp,HindIII-EcoRI 1.1-kb, andPstI1.1-kbpTKgenefragmentsweresubcloned
toyieldplasmids pRPEM211, pRPEM210, andpRPEM213,respectively. These plasmidsweresequenced, andtheopenreading framesof both
strandsweredetermined. Arrows show the direction andextentofsequencedeterminationfrom eachrestriction site. Restrictionenzymecleavage
sitesBglII(B), HindlIl (H), PstI (P), and XhoI (X)areindicated. (B) Nucleotidesequenceand predicted amino acidsequenceof the TKprotein (single-letter amino acid code). Ter, Termination codon. Locations of cleavage sites BgII, HindIII, PstI, and XhoI and the ribosome-binding site
areunderlined.
B H
I I1IiT
T K
B
RRG
CCT
GGRK P G
RCG TTT CTT
T F L
OTC
TTC RTRV F I
TCC
GGT
RTRS G I
TTR TCT
GRC
L S D
GCG CRT TTT
A H F
RRR
ATT
RTTK I I
CCG CCC RTC
p p I
CGC RCC
TGT
R T C
RRC GCR
GRC
N A D
TGT TGT
RRCC C N
RRR
TRT TRRK Y Ter
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
NOTES 1049
.2
B I
.4
l l
.6 .a
4 156 Kbp
K
C I
I C IH'i B F'I A . I IHI E
IGI~
E C C CE
ES
I I-Ia I .. 1
E BH P X
a I
p a E
I I aWa
TK
FIG. 4. Summary of the mapping and precise location ofthe TKgenein the ASFVgenome. Thesizeofthe DNAfromASFV growing
inMS cells is 156 kbp(31).Sallfragmentsareshown(boxed). Thelocationof the .Kgenewithin theSalI-Bfragment is indicated.Restriction
enzymesBglII (B), ClaI (C),EcoRI (E),HindIll (H), PstI (P),Sall(S),andXhol (X)areindicated.
racil, 2.5 ,ug of uridine, 5 ,ug of thymidine, 250 ,ug of
deoxyadenosine, and 1 ,uCi of[3H]thymidine (82.2 Ci/mmol; New England Nuclear Corp., Boston, Mass.). The cells were lysed with 2% sodium dodecyl sulfate-0.2 N NaOH
ABC D
P73
1P23.5
[image:4.612.85.526.73.239.2]IP16 ._
FIG. 5. Sodiumdodecyl sulfate-polyacrylamide gel electropho-resisofpolypeptides translated from different RNA preparations.
TheRNAresulting from in vitro transcription by T7 RNA
polymer-asefrom plasmid pGEM204a linearized with XhoI (pGEM204aX) (lane C) was translated in a rabbit reticulocyte lysate system
(Promega), with [35S]methionineasthelabeled aminoacid. Labeled proteins were analyzed by 12% polyacrylamide-sodium dodecyl
sulfategelelectrophoresis, and the labeled polypeptideswere
visu-alizedby autoradiography of the dried gel. Lane D, No RNA; lane A, polypeptides from uninfected MS cells; lane B, infected cells labeled with[35S]methioninefrom 20to22hpostinfection (28). IP, Polypeptides from infected cells; numbersaremolecularmassesin kilodaltons.
and precipitated with ice-cold 10% trichloroacetic acid.
Precipitates werecollected by filtration on glass fiber filters andcounted in toluene-based scintillationfluid. When these cellsweregrownin the presence of
isopropyl-p3-D-thiogalac-topyranoside (IPTG), the incorporation of [3H]thymidine
was increased about 25 times inrelation to that in parallel cultures in the absence ofIPTG (Table 1). Sincethe pINI-IIAl vectorcontains the lac promoter-operator region, the
expression of the viral TK gene can be induced by a lac inducer such as IPTG. The increase of[3H]thymidine
indi-catesthat theexpression ofthe ASFV TK gene isregulated by alac promoter-operator region.
Cell-free translation of synthetic RNA.Additional evidence
that the BglII-XhoI fragment contains the TK gene was
obtainedby the synthesis ofactive TKproteinin acell-free
system programmedwith RNA synthesized in vitro, using
plasmid pGEM204a (Fig. 1)and T7orSP6RNApolymerase in the presenceof 0.5 mM
m7G(5')ppp(5')G.
The RNA was translated in a reticulocyte cell-free system, and samples from the translation mixture were then assayed for TKactivity.Positive controlswereearlyRNAsfrom BHK TK-cells(kindlysupplied byB.Roizman, UniversityofChicago)
at14hafter infection with ASFV(strainSpainM0/MS44[31])
in the presence of 0.1 mM cycloheximide. Total RNA was
[image:4.612.117.234.374.607.2]isolated from infected and mock-infected cellsbyguanidine
isothiocyanate-CsCl
extraction (15). Active TK was ob-tained upon translationofRNAfrominfected cells(Table 2).TABLE 1. Incorporation of[3H]thymidineinto DNAof
plasmid-transformedTK- E. coli(KY893 mutantstrain)a
Bacterialstrain . cpm/mlof
(culturesupplementb) asma bacterial culture
KY893 37
NH522 1,877
KY893(FUdr) pINIIIAl 37
KY893(FUdr + IPTG) pINIIIAl 58
KY893(FUdr) pINTK14 166
KY893(FUdr+ IPTG) pINTK14 4,110
aForselectionof TKexpression,theTK-deficient(TK-)strain KY893(10)
of E. coliwas used.TK activity was assayed bymeasuring theuptakeof [3H]thymidineasacid-precipitable radioactivity.
bFUdr,
Fluorodeoxyuridine.
PRPEM 513
pRPEM204
_"
.. VVOL.65, 1991
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.317.557.610.691.2]1050 NOTES
fHIIRKLKP6TISLULGPfFRGKTTFLIHCIYMLERLEKKUUFIKSTKHTR
-OK
MSC
INLPTULP6SPSKTRGQIQUILGPMFSGKSTELIRRURRFQIROYKCLUIKYRKOTRY----SS
MSYIHLPTULPSSPSKTRGQIQUILGPnFSGKSTELIRRURRFQIRQYKCLUIKYRKDTRY----SK
IHCLTUPGUHPGSPGRPRGQIQUIFGPfFSGKSTELIRRURRFQLRQYRCLLUKYRKDTRYC---TT
1SSGSIHUITGPnFSGKTSELURRIKRFnLSNFKCIIIKHCGONRYNDEDIN
nHGGHIOLIIGPnFSGKSTELIRRURRYQ1RQYKCUTIKYSNDNRYG
----TINGGHIQLIIGPMFSGKSTELIRRURRYQIRQYKCUTIKYSHDHRYG----T
IHGGHIQLIIGPIFSGKSTELIRRURRYQIRQYKCUTIKYSHDHRYG----T
n1
IGHIHLIIGP1FRGKSTELIRLURRYQIRKHKCLUUKYEKDIRYG----N
G I +
GPfF+GK+++L
+++ + + +++K+
+ R+TIKTHSGIQLRPKQCKIIESTQLSDU--GSLTO--IHRUU-UDERHFFODLIK-CRTUREEEKIIIL
SFCTHD.---RHTERLPRCLLRDURQERL----6URUIGIDEGQFFPDInEFCERnRHRGKTUIU
SFSTHD---RHT1DRLPRCfLRDUTQELL----GURUIGIOEGQFFPDIUDFCE1IRHEGKTUIU
GUSTHD---RHTMERRPRCRLQDUYQERL----GSRUIGIDEGQFFPDIUEFCEKIRHTGKTUIU
KUYTHD---LLFMERTRSSHLS-ULUPTLLHD-GUQUIGIDERQFFLDIUEFSESMRHLGKTUIU
GLUTHD---KHHFRRLEUTKLCDU----LERITDFSUIGIDE6QFFPDUUEFCERMRHEGKIUIU
GLUTHD---KHHFERLERTKLCDU----LESITDFSUIGIDEGQFFPDIUEFCERMRNEGKIUIU
GLUTHD---KNHFERLERTKLCDU----LERITDFSUIGIDEGQFFPOUUEFCERMRHEGKIUIU
GUCTHD---NS
ITRUCTPSLDK
I----DSURENREUII6OEGQFFPNIRTFCERRtHRGKUL
I UTH+
+ L ++ +U+++DE++FF++
+++oR+
+K+1+
AGLHRSFEQKMFPPIURIFPYCSIUKYIGRTCIKCHQHHRCFHURKHRDKTLILRGGSELYUTCCHH
RARLDGTFQRKPFGRILHLUFLRESUUKLTRUCIECF-RERRYTKRLGTEKEUEUIGGRDKYHSUCRL
RRLDGTFQRKRFGSILHLUPLRESUUKLTRUCMECF-RERRYTKRLGLEKEUEUIGGRDKYHSUCRL
RRLOGTFQRKRFGSILHLUPLRESUUKLHRUCfECY-RERSYTKRLGREREUEUIGGRDKYHSUCRR
RRLHGDFKRELFGHUYKLLSLRETUSSLTRICUKCY-CDRSFSKRUTENKEUMDIGGKDKYIRUCRK
RRLDGTFQRRPFHHILHLIPLSEMUUKLTRUCMKCF-KERSFSKRLGTETEIElI6GHODYQSUCRK
RRLOGTFQRKPFN
ILHLIPLSEIUUKLTRUCMKCF-KERSFSKRLGEETEIElIGGIDIYQSUCRK
RRLOGTFQRKPFHHILOLIPLSEfUUKLTRUCMKCF-KERSFSKRLGTETKIEIIGGHDIYQSUCRK
RRLDGTFORKPFSHISELIPLRENUTKLNRUCfYCY-KHGSFSKRLGDKMEIEUIGGSDKYKSUCRK
R+L+++F+++ F + + ++ + +
+++C+
C +*R++
+ + GG + Y++C+
ASFV CLKHTFIKQLQPIKY
Human
CYFKKRSGQPRPDONKEICPUPOKPGERURRRKLFRPQQILQCSPRH
Mouse
CYFKKSSRQTRGSONKN-CLULGQPGERLUURKLFRSQQULQYHSRH
Chicken
CYFQKRPQQL-GSEHKENUPIGUKQLDfPRSRKIFRS
FPV CFFSH
MPV CYIDS
VV CYIDS
VarV CYIDS
SFV CYEE
Homology C+
FIG. 6. Alignment of amino acidsequencesof TK from ASFV, humans (H) (3), mice (M) (13), chickens (Ch) (17), vaccinia virus (VV) (37), variolavirus (Var V) (6), monkeypox virus (MPV) (6), fowlpox virus (FPV) (2), and Shope fibroma virus(SFV) (32). Theconsensussequenceand
thepositionsatwhich identicalresiduesareobserved inseven ormoreof the nine alignedsequences areindicated (+) below the alignment.The
maximumidentity with vaccinia virus TKwaslocated in thenucleotide-binding siteatresidues 17to25 inthe ASFV TKgene,whichcorrespond
toresidues 11 to19in the vacciniavirus TKgene (7),and in thepossiblenucleoside-binding siteat residues 88to 122in ASFVTK,which
correspondtoresidues 77to116in thevaccinia virus TKgene.
The polypeptide synthesized had a molecular weight of
about22,400 (Fig. 5, lane C), in goodagreementwith thesize predicted from the open reading frame gene sequence. In conclusion, we have identified andexpressed, both in
bac-terial cells and by cell-free translation, the TK gene of
ASFV.
Comparison of the ASFV TK gene with poxvirus and cellular TK genes. In addition to providing a tool for the
TK
ASFV
Human
Mouse
Chicken
FPV
MPV
vV
Var V
Homology
ASFV
Human
Mouse
Chicken
FPV
MPV
VVVar V
SFV
Homology
ASFV
Human
Mouse
Chicken
FPV
MPV
VV
Var
V
SFH
lHomology
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
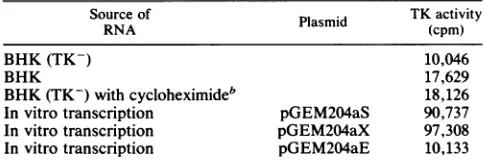
[image:5.612.137.504.77.590.2]TABLE 2. TK activityincellextract aftertranslationsa
Source of Plasmid TKactivity
RNA (cpm)
BHK(TK-) 10,046
BHK 17,629
BHK (TK-)withcycloheximideb 18,126
Invitro transcription pGEM204aS 90,737
In vitrotranscription pGEM204aX 97,308
Invitro transcription pGEM204aE 10,133
aThe level of TKactivitywasassayed byconversionof
[3H]thymidine
into [3H]thymidylate. Plasmid pGEM204a (Fig. 1) was linearized with Sail (pGEM204aS)orXhoI(pGEM204aX), transcribed withT7RNApolymerase orfurther linearizedwithEcoRI(pGEM204aE),and then transcribed with SP6 RNApolymerawe.About 3S.gof RNA transcribed in vitroandabout35 ,ug of totalRNAfrominfected oruninfectedBHKcellsweretranslated for4 h in a micrococcal nuclease-treated reticulocyte lysate, and the TKactivity was assayed in thepresenceof50p.Mthymidine tostabilizenascent TK activity (36).b Cells infected with ASFV.
development of ASFV genetics, the TK gene is of interest
for further defining phylogenetic relationships among large DNA viruses. The nucleotide sequence of the ASFV TK gene includes an open reading frame of 196 codons, with a calculated molecular weight for ASFV TK of 22,394, only
slightly higherthan that ofthe vaccinia virus TK, which is 20,102 (37). Homology analysis of the deduced ASFV TK
gene protein sequence indicates a relationship with both
poxvirus andcellularTKproteins (Fig. 6).The percentages ofidentityin TK sequencesaligned bythe FASTP program (14) were shown to be 28% for human, 29.5% for mouse,
26.1% for chicken, 26.2% for fowlpox virus, 31.3% for
monkeypoxvirus, 32.4%forvaccinia virus, 31.8% for vari-olavirus,and29% for Shope fibroma virusTKpolypeptides.
Theidentification ofthe ASFV TK gene will now permit direct construction, by genetic manipulation, of TK- mu-tants to serve asrecipientsforreinsertion offoreigngenes. Oneimportantchange thatoccursin theadaptation ofASFV strain E70to monkey cells is the lossofpathogenicity (31). Consequently,ASFVcouldbe usedas amodel in studiesof
pathogenesis of persistent infections by inserting genes
presumably involvedinpathogenesis into the genome of the
avirulent E70MS14 strain (31).
We are grateful to S. Fernandez and J. Alvarez forexcellent technical assistance.
This workwassupported byagrantfromComisionAsesora para elDesarrollo delaInvestigaci6nCientificayTdcnica(Spain).
REFERENCES
1. Almendral, J. M., R. Blasco, V.Ley, A. Blasco,A.Talavera, and E. Vihuela. 1984. Restriction site mapof African swine fever virusDNA. Virology 133:258-270.
2. Boyle, D.B., B. E. H.Coupar, A.J. Gibbs,L.J.Seigman,and G. W. Both.1987. Fowlpoxvirusthymidinekinase: nucleotide sequence andrelationshipstootherthymidinekinases.Virology 156:355-365.
3. Bradshaw, H. D., Jr., and P. L. Deininger. 1984. Human thymidine kinase gene: molecular cloning and nucleotide
se-quenceofacDNA expressiblein mammaliancells. Mol. Cell. Biol.4:2316-2320.
4. Duffand, G. D., P. E. March, andM.Inouye.1987.Expression
andsecretion offoreign proteins in Escherichia coli. Methods Enzymol. 153:492-507.
5. Escribano, J. M., and E. Tabaires. 1987. Proteins specified by
Africanswinefever virus.V.Identification of immediateearly,
earlyand lateproteins.Arch. Virol. 92:221-232.
6. Esposito, J. J.,andJ.C.Knight. 1984. Nucleotidesequenceof the thymidine kinase gene region of monkeypox and variola virus.Virology 135:561-567.
7. Gentry, G. A. 1985. Locatinga nucleotide-binding sitein the
thymidinekinaseof vacciniavirusand ofherpessimplexvirus
by scoringtriply alignedprotein sequences. Proc.Natl. Acad. Sci. USA82:6815-6819.
8. Hess, W. R. 1971. African swine fever virus. Virol. Monogr.
9:1-33.
9. Hess, W. R. 1981. African swine fever: areassessment. Adv. Vet. Sci. Comp. Med. 25:39-69.
10. Hiraga, S., K. Igarashi, and T. Yura. 1967. Deoxythymidine
kinase-deficient mutant of Escherichia coli. I. Isolation and
someproperties. Biochim. Biophys.Acta145:41-51.
11. Igarashi, K.,S. Hiraga, and T. Yura.1967. Adeoxythymidine
kinase-deficient mutant of Escherichia coli. II. Mapping and transduction studieswithphage
480.
Genetics37:643-654. 12. Inouye,S.,and M.Inouye.1985. Up-promoter mutations inthelpp gene of Escherichia coli. Nucleic Acids Res. 13:3101-3110.
13. Lin, P.-F.,H.B.Lieberman,D.-B.Yeh,T.Xu,S.-Y.Zhao,and F. H.Ruddle.1985. Molecularcloningandstructuralanalysisof murinethymidinekinase genomic andcDNAsequences. Mol. Cell. Biol.5:3149-3156.
14. Lipman, D. J., and W. R. Pearson. 1985. Rapid andsensitive
proteinsimilaritysearches. Science227:1435-1441.
15. Maniatis, T., E. F. Fritsch,andJ. Sambrook. 1982. Molecular
cloning: alaboratorymanual. ColdSpringHarborLaboratory,
ColdSpring Harbor,N.Y.
16. Mebus, C. A. 1988. African swine fever. Adv. Virus Res. 35:251-269.
17. Merrill, G. F., R. M. Harland, M. Groudine, and S. L. McKnight.1984. Geneticandphysicalanalysisofthechicken tk gene. Mol. Cell. Biol. 4:1769-1776.
18. Piccini, A., and E. Paoletti. 1988. Vaccinia: virus, vector, vaccine. Adv. VirusRes. 34:43-64.
19. Polatnick, J., and W. Hess. 1970. Altered thymidine kinase
activity in culture cells inoculated with African swine fever virus. Am. J. Vet. Res. 31:1609-1613.
20. Polatnick, J., and W. Hess. 1972. Increaseddeoxyribonucleic
acid polymerase activity in African swine fevervirus-infected culture cells.Arch. GesamteVirusforsch.38:383-385. 21. Roizman, B., and F. J. Jenkins. 1985. Genetic engineering
of novel genomes of large DNA viruses. Science 229:1208-1214.
22. Salas,M.L., J.Rey-Campos,J.M.Almendral,A.Talavera,and E. Vinuela. 1986. Transcription maps of African swine fever virus. Virology152:228-240.
23. Sanchez Botia, C. 1982. Peste porcina africana. Nuevos de-sarrollos. Rev. Sci. Tech. O.I.E. 1:991-1029.
24. Sanger, F.,S.Nicklen,and A. R.Coulson. 1977. DNA
sequenc-ing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467.
25. Sargan,D.R.,S. P.Gregory,and P. H. W.Butterworth. 1982. Apossible novel interactionbetween the 3'-end of18 S ribo-somal RNA and the 5'-leader sequence of many
eukaryotic
messenger RNAs. FEBSLett. 147:133-136.
26. Schnitzlein, W. M., N. Ghildyal, and D. N. Tripatly. 1988. A
rapid method for identifying the thymidine kinase genes of
avipoxvirus.J.Virol. Methods 20:341-352.
27. Tabares,E.1987.Characterization ofAfricanswinefevervirus
proteins, p. 51-61. In Y. Becker (ed.), African swine fever. MartinusNijhoffPublishing, Boston.
28. Tabares, E.,M. A.Marcotegui,M. Fernandez,andC. Sanchez
Botija. 1980. Proteinsspecified byAfrican swine fever virus. I.
Analysis of the structural proteins and
antigenic
properties.
Arch. Virol. 66:49-59.29. Tabares,E., J.Martinez,F.R.Gonzalvo,andC. SanchezBotja.
1980.Proteinsspecified byAfrican swine fevervirus. II.
Anal-ysisofproteinsininfectedcells and
antigenic
properties.
Arch. Virol.66:119-132.30. Tabares,E.,J.Martinez,E.Martin,andJ.M.Escribano.1983.
on November 10, 2019 by guest
http://jvi.asm.org/
1052 NOTES
Proteins specified by African swine fever virus. IV. Glycopro-teins andphosphoproteins. Arch. Virol. 77:167-180.
31. Tabares,E.,I.Olivares,G. Santurde,M. J.Garcia, E.Martin,
and M. E. Carnero. 1987. African swine fever virus DNA: deletions and additions during adaptationtogrowthin monkey kidney cells. Arch.Virol. 97:333-346.
32. Upton, C., and G. McFadden. 1986. Identification and
nucleo-tide sequence of the thymidine kinase gene ofShope fibroma
virus. J. Virol. 60:920-927.
33. Urzainki, A., E. Tabares, and L. Carrasco. 1987. Proteins synthesized in African swine fever virus infected cells analyzed bytwodimensional gel electrophoresis. Virology 160:286-291. 34. Vifiuela,E. 1985. African swine fever virus. Curr.Top.
Micro-biol. Immunol. 116:151-170.
35. Wardley, R. C., C. M. Andrade, D. N. Black, F. L. Castro Portugal, L. Enjuanes, W. R. Hess, C. Mebus, A. Ordas, D. Rutili, J. M. Sanchez Vizcaino, J. D. Vigario, and P. J.
Wil-kinson. 1983. African swine fever virus. Arch. Virol.
76:73-90.
36. Weir, J. P., G. Bajszar, and B. Moss. 1982. Mapping ofthe
vacciniavirus thymidine kinasegeneby markerrescueand by
cell-freetranslation of selected mRNA. Proc. Natl. Acad. Sci. USA 79:1210-1214.
37. Weir, J. P., and B. Moss. 1983. Nucleotide sequence of the vaccinia virus thymidine kinasegeneand the natureof
sponta-neousframeshift mutations. J. Virol. 46:530-537.
J. VIROL.
![TABLE 1. Incorporation of [3H]thymidine into DNA of plasmid-transformed TK- E. coli (KY893 mutant strain)a](https://thumb-us.123doks.com/thumbv2/123dok_us/1317256.85253/4.612.117.234.374.607/table-incorporation-thymidine-dna-plasmid-transformed-mutant-strain.webp)