0022-538X193/106239-07$02.00/0
Copyright © 1993, AmericanSociety forMicrobiology
Inhibition of
Bombyx
moni
Nuclear
Polyhedrosis Virus
(NPV)
Replication by the Putative
DNA
Helicase Gene of
Autographa
califomica
NPV
SHIZUO GEORGE KAMITAANDSUSUMU MAEDA*
Department of Entomology, University of California, Davis, California 95616-8584
Received 29 March1993/Accepted 12July 1993
Coinfection ofBombyx mori nuclear
polybhedrosis
virus(BmNPV) withAutographa californicaNPV(AcNPV)in theBmNPV-permissiveBmNcellline resulted in thecomplete inhibition ofBmNPVreplication. Coinfected
BmN cells exhibited an atypical cytopathic effect (CPE) and synthesis of viral and host proteins was
dramatically
attenuated by 5 h postinfection (p.i.) and nearlycompletely
blocked by 24 h p.i. Viraltranscription, however, appearedto occur
normally
during bothearly
(5-h-p.i.) and late (24-h-p.i.) stages ofinfection. Superinfection of BmN cells with AcNPVat 5 and 12hpost-BmNPV infection resulted inlimited
inhibition of BmNPVreplication. BmN cells singly infected with AcNPV also showed similarCPE, premature
inhibitionof viral and host protein synthesis, and
apparently
normal viraltranscription. BmNPV replicationoccurred
normally
following coinfection ofBmNPVand eh2-AcNPV,anAcNPVmutantidenticaltoAcNPVexcept fora572-bp regioninitsputativeDNAhelicase geneoriginatingfrom BmNPV(S. Maeda, S. G. Kamita,
andA. Kondo, J. Virol.67:6234-6238, 1993). Furthermore,atypical CPEandpremature attenuationof host
andviralprotein synthesiswere notobserved. These results indicated that the inhibition of BmNPVreplication
was causedeither
directly
orindirectly
at the translational level by the putative AcNPV DNAhelicasegene.Nuclear polyhedrosis viruses (NPVs), a genus of the
familyBaculoviridae, arecharacterized by circular
double-stranded DNA genomes. Recombinant NPVs are efficient
vectors for the high-level expression of foreign genes for
basic research and medicalapplications (15,16, 25). Recent
advances in genetic engineering have resulted in
recombi-nantNPVsthat holdpromiseaspotent yetsafealternatives
to traditional chemical insecticides (5, 24, 31, 33). NPVs
haveonly beenisolated from arthropods (mainly insects) and
areconsidered to have narrowhost ranges, i.e., they
repli-cate in only a few species of the same or similar families.
Previous host range studies have aimed to identify a
com-monregion in the genomes of baculoviruses with low DNA
homology but whichcan infect the samehost. The general
mechanism(s) of baculovirus hostspecificity, however,
re-mains undetermined.
Bombyx
mon NPV(BmNPV) and Autographa califomnica NPV(AcNPV) arehighly homologousatthegenomiclevel(23),yettheyexhibitnonoverlappinghostspecificities;e.g.,
BmNPV replicates in BmN cells, but not in SF-21 cells,
whereas AcNPVreplicates in SF-21, but not in BmN. We
have demonstrated that BmNPVreplicationcanbeinduced
in SF-21 cells coinfected with AcNPV (12). This
phenome-non was shown to be the result of (i) a helper function of
AcNPVtowards BmNPV replication and (ii) the appearance
of AcNPV-BmNPVhybridswith expanded host ranges. By
backcross infection of one of these hybrid NPVs with
AcNPV,wegenerated eh-AcNPV, abaculovirus capable of
replicating in both BmN and SF-21 cells and possessing
DNA restriction endonuclease patterns nearly identical to
those of AcNPV.By cotransfection andnucleotide
sequenc-inganalysis,a572-bpregion was hypothesized to be
respon-sible for the hostrange-expanded phenotype of eh-AcNPV.
This572-bpregionof eh-AcNPVwasidenticaltothe
corre-*
Corresponding
author.sponding 572-bp region of BmNPV and was localized within the putative DNA helicase gene. Cotransfection of the 572-bp BmNPV fragment with AcNPV genomic DNA
gen-erated the hostrange-expanded virus eh2-AcNPV,
confirm-ing that DNA helicase playsarole inhost range expansion
(20).
In this study, we found that BmNPV replication was
completely inhibited in BmN cells coinfected with BmNPV
and AcNPV. The coinfected BmN cells also exhibited a
unique cytopathic effect (CPE), and synthesis of both host
and viral proteins was prematurely attenuated. Viral
tran-scription, however, appearedto occurnormally during both
the early and late stages of infection. BmN cells coinfected
with eh-AcNPV and BmNPV oreh2-AcNPV and BmNPV
didnotexhibit any abnormal CPEorprematureattenuation
of viral or host protein synthesis, and viral replication
occurred normally. These analyses suggested that the
puta-tive DNA helicase gene of AcNPV(23)caneitherdirectlyor
indirectly cause the inhibition of BmNPV replication in
BmNcells.
MATERIALSANDMETHODS
Cell lines and viruses. BmN(BmN-4)and SF-9 cellswere
maintained in TC-100 medium supplemented with 10% fetal
bovineserumand inEx-Cell 400(JRH Biosciences,Lenexa,
Kans.) supplementedwith2.5% fetal bovine serum,
respec-tively, as described previously (17, 23). The BmNPV T3
isolate (21) and the AcNPV mutants, eh-AcNPV and
eh2-AcNPV (20), were propagated on BmN cells. The
AcNPVOT2 isolate(23)was propagatedonSF-9 cells.
Viral infection andplaque assay.BmN or SF-9cellswere
infected with BmNPV, AcNPV, eh-AcNPV, and/or
eh2-AcNPVat amultiplicityof infection (MOI)of 10(exceptas
noted) for each virusasdescribedpreviously (12,17). In all
experiments, time zero was defined as the point at which
freshmediumwas added following the 1-hviral
adsorption
6239
on November 9, 2019 by guest
http://jvi.asm.org/
200 ,u ofmethionine-freeculturemediumat 1,5, 12,24, 36,
48, and 72 h p.i. as described previously (11). Cell pellets
were dissolved in 100 pl of sample buffer, and 6
RI
wassubjectedtosodiumdodecyl sulfate-polyacrylamide gel
elec-trophoresis (SDS-PAGE) as described by Laemmli (13) on
10% acrylamide gels (0.4% bis-acrylamide). Gels were
treated with an enhancer (Resolution; EMCorp, Chestnut
Hill, Mass.), dried under avacuum, and exposed to X-ray film (Eastman Kodak Co., Rochester, N.Y.). Low-range SDS-PAGE molecular weight standards
(14.4
to 97.4 kDa)werefromBioRad Laboratories
(Richmond,
Calif.).Isolation of
poly(A)+
RNA. Poly(A)+ RNAwas isolatedfrom approximately 9 x 107 BmN cells which were either
mock infectedorinfected withvirus(es)asappropriate.Cells
were harvested at either 5 or 24 h p.i., washed once in
ice-cold phosphate-buffered saline, and oligo(dT) cellulose
wasusedto extractpoly(A)+ RNA asdescribedpreviously (19). The concentration of the poly(A)+ RNA was
deter-minedbymeasuring
A260-DNAprobes. Twoprobescontainingsequencesfromearly
and latebaculovirus geneswerelabeled with
[a-32P]dATP,
using a random primed DNA
labeling
kit (United StatesBiochemical, Cleveland, Ohio). The probe OT2EH was
derived from the AcNPV 0T2 EcoRI H
fragment
whichcorresponds to the EcoRI H fragment
(79.8
to 86.4 mapunits)in the conventional AcNPV map(foranexample, see
reference 28). This fragment contains the open reading
frames (ORFs) gp64, ORF1, ORF2, PE34, ORF4, ORF5,
and 94K,whichareconsidered
immediate-early
and/orlatebaculovirus genes (4, 27, 34) (see Fig. 4A). In this region,
BmNPV and AcNPV have over 95% nucleotide sequence
identity excluding the 94,000-Da gene (94K gene) (18). A gene corresponding to the AcNPV 94K gene is
lacking
inBmNPV(11).
Theprobe BE284BE was derived from the 1.3-kbp
frag-mentofthe BmNPV transfervectorpBE284
(17)
whichwasdigestedwith BamHI and EcoRI. BE284BE contains the 3'
nontranslatedregion of thepolyhedringene and1,340bpof
the 3' end of theORF1629codingregion(2.4to3.4 map units
in the BmNPV genome [22]) (18, 30) (see Fig. 4B). The
polyhedrin geneandORF1629 areconsidered very late and late baculovirus genes, respectively (30). Nucleotide
se-quences in this region are greater than 97% identical for
BmNPV andAcNPV(10, 18, 30).
Northern(RNA) blothybridization.Poly(A)+ RNAs (3 ,ug
per lane) were electrophoresed (1.5 h) in a formaldehyde denaturing gel (1% agarose) in morpholinopropanesulfonic
acid(MOPS) (0.02 M) buffer. Molecularweightwas
deter-mined by comparison with coelectrophoresed molecular
weightmarkers(3 ,ug of 0.24-to9.5-kbRNAladder; GIBCO
1-> 7
6 5
0 24 48 72
[image:2.612.372.515.69.374.2]HOURSPOSTINFECTION
FIG. 1. Growth curves of BmNPV(0)followingcoinfectionof BmNcells with BmNPVand AcNPV(O) (A), superinfection of BmNPV-infected BmN cells with AcNPV at 5(0)or12(-)hp.i. (A), andcoinfection of BmN cells with BmNPV andeh2-AcNPV (0,eh2-AcNPV;El, BmNPVplus eh2-AcNPV)(B).
BRL, Inc., Gaithersburg, Md.). Following electrophoresis,
RNAs were denatured, transferred, and fixed to a nylon
membrane (Zeta-Probe; Bio-Rad Laboratories)bya
modifi-cation of themethod describedpreviously(19).
The DNAprobesweredenatured inboilingwater(10min)
and hybridized to the mRNAs at 42°C as described
previ-ously (23). Filters used forhybridizationwerewashed twice
in 2x SSC (lx SSC is 0.15 M NaClplus 0.015 M sodium
citrate)-0.1% SDS and once in 0.1x SSC-0.1% SDS and
thensubjectedtoautoradiographyusing X-rayfilm(Eastman
KodakCo.). The filterswerereprobed after the firstprobe
wasstripped from the mRNAsbyboilingin 10 mMTris(pH
7.5)-0.1% SDS for 5 minfollowedbyincubation at80°Cfor 1 h.
RESULTS
Inhibition of BmNPV replication by coinfection with
Ac-NPV.OnBmNcells, BmNPV showedatypical viral growth
curvewhich reached aplateau maximum of about 2 x 108
PFU/ml at 72 h p.i. (Fig. 1). AcNPV doesnot replicate in
BmNcellsasreportedpreviouslyby Maedaetal.(23).When
BmN cells were coinfected with BmNPV and AcNPV,
BmNPV replication was completely inhibited (5,000-fold
from 2 x 108 PFU/ml to a residual level of about 4 x 104
PFU/ml) (Fig. 1A). BmNPV replication was also inhibited
1,000-fold and2,500-fold whencoinfected with AcNPVat2
and4MOI,respectively (datanotshown). Superinfectionof
BmNPV-infected(5MOI)BmNcells with AcNPV(20MOI)
on November 9, 2019 by guest
1:,
FIG. 2. The morphology ofBmN cells at 72 hp.i.followingBmNPV and AcNPVcoinfection(A), BmNPVinfection(B),AcNPV infection (C), andmock infection(D).Magnification, x400.
at 5 or12 h post BmNPV infection resulted in 16-fold and
2-foldinhibition, respectively (Fig. 1A).
BmN cell morphology and growth. The morphology and
growth characteristics (data not shown) of mock-infected
BmNcells was typical ofhealthy cultures during a log phase
ofgrowth (Fig. 2D). BmN cellgrowth was not detected in
anyof thevirally infected BmN cells. At 72 h p.i.,
BmNPV-infected BmN cells exhibited CPE typical of wild
type-infected cultures, including rough cell surface, detachment
from the culture surface due to rounding, nuclear expansion,
andformation ofpolyhedra(Fig. 2B).In contrast,coinfected
BmN cells did not exhibit rounding, remained strongly
attachedtothe culture surface during all stages of infection,
and did not produce polyhedra; however, they did exhibit
nuclear expansion (Fig. 2A). These characteristics were
initiallyobserved around 12 hp.i.and were maximized by 48
hp.i. The morphology of AcNPV-infected BmN cells (Fig.
2C)was similar to thatofBmNPV and AcNPVcoinfected
cells. BmN cells superinfectedwith AcNPV (at 5 and 12 h
post BmNPV infection) did not exhibit cell morphology
specificfor AcNPV infection.
Protein synthesis. In order to examine the mechanisms behind the AcNPV-induced inhibition of BmNPV
replica-tion,proteinsynthesiswasinitiallyanalyzed by SDS-PAGE.
Mock-infected BmN cells showed auniform increase in all
protein synthesis correspondingtonormal cellproliferation
(data not shown). BmNPV-infected BmN cells showed a
pattern ofprotein synthesis typical of baculovirus-infected
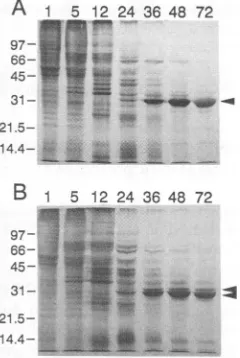
cells(Fig.3A),asreportedpreviously (32).Between5 and 24
h p.i., the synthesis of two or three putatively virally
encodedorvirally induced proteinswas detected. By24 h
p.i., the expression of host- and virus (e.g., the
38-kDa)-specific proteins was retarded or terminated. Between 24
and 72 h p.i., nearly all host- and virus-specific protein
synthesis was terminated or highly retarded except for a
highly expressed 30-kDa protein(indicated bythe arrowhead
inFig. 3A),whichwasconsideredtobeBmNPV-expressed
polyhedrinonthe basis of its size and time ofexpression.
In BmNPV-andAcNPV-coinfected BmNcells, synthesis
of both host- and virus-specific proteins was prematurely
anddramaticallyinhibited(Fig.3B).By 5 hp.i.,totalprotein synthesis droppedtoabout20%of thelevels foundfollowing
BmNPVsingleinfection, andby24 hp.i.,protein synthesis
was nearlycompletely inhibited. Between 36 and 72 h p.i.,
three putatively virally encoded bands (approximately 30,
38, and 42kDa)were slightlydetectable. The 30-kDa band
(indicated bythearrowheadinFig. 3B)wasconsideredtobe
polyhedrin expressed byBmNPV or AcNPV.
To examine whether the premature attenuation ofprotein
synthesis was induced by the interaction of BmNPV and
AcNPVor wassolelythe result of AcNPVinfection, protein
synthesis of AcNPV-infected BmN cells was analyzed. A
dramatic and premature drop in protein synthesis nearly
identicalto thatfollowing BmNPVandAcNPVcoinfection
was observed in BmN cells singly infected with AcNPV
(Fig. 3C). At 12 h p.i., at least one virus-specific band
on November 9, 2019 by guest
http://jvi.asm.org/
21.5-1 5 12 24 3648 72
VW -.
97-66-i
45-31-_ 21
.5-FIG. 3. SDS-PAGE of pulse-labeled proteins from BmN cells thatwereinfectedwith BmNPV(A),BmNPVand AcNPV(B),or AcNPV(C).Proteinswerepulse-labeledwith[35S]methionineat1,
5, 12, 24, 36, 48,or72 hp.i.Theaffowheadsindicate themigration
of the putative polyhedrins. The approximate sizes of marker proteinsareindicated totheleftinkilodaltons.
A Bm
Bm
BmcmB
eh-Aceh-Ac Ac m 5 24 524 524 524 5 24 5 24
I. I.
coinfected SF-9 cells was only about 1/10 that found in
AcNPV-infected SF-9 cells at 72 h p.i. Furthermore, the
normalattenuationof hostproteinsynthesisdid notseem to
occur as quickly in eh-AcNPV-infected SF-9 cells as in
AcNPV-infected cells (datanotshown). These results
indi-cated that the dramatic inhibition of host and viralprotein synthesis was induced by the AcNPV DNA helicase in
conjunctionwith ahost factor(s), e.g., interaction ofDNA
helicasewithhostproteinsor nucleicacids.
Northern blot
analysis.
Northern blot analysis wasper-formedtoexaminewhether the inhibition ofprotein
synthe-sis inducedbyAcNPVinfectionwasduetothe lack of viral
transcripts.Poly(A)+RNAscollectedat5and24hp.i.from
mock- and virus-infected BmN cells wereanalyzedwith the
probes BE284BE and OT2EH, which were derived from
BmNPVandAcNPV,respectively (Fig. 4).Ingeneral,viral
transcription of both earlyand late (5- and
24-h-p.i.)
genesoccurred normally in AcNPV-infected and BmNPV- and
AcNPV-coinfected BmNcells.
OT2EH (gp64, ORF1, ORF2, PE34, ORF4, ORF5, and
94K) did not hybridize with any transcripts in the
mock--_9.5 7.5
-4.4 4.2_
-2.4 2.2 _
-1.4 1.3
I--0.24
Bm eh-Ac Bm Bm Ac m
ehSAc S4 2Ac
5 24 524 524 524 5 24 5 24
- 9.5
7.5
-4.4
-2.4
-1.4
-0.24
gp64
79.8 ORF--- ORPE34EzRl
EcoRI ORF2jOHF4ORFs probe (OT2EH)
94K Xbal
_ 1.9 I .-. ORF1629
86.8mapunits l - PoFyherin 3.4 mapunits
EcoRi -- BamHl
mp_ 1
probe (BE284BE)
FIG. 4. Northern blot analysis of mRNAcollected from BmN cells at 5 and 24 h following infectionwith BmNPV and eh-AcNPV (Bm/eh-Ac),eh-AcNPV(eh-Ac),BmNPVand AcNPV(Bm/Ac),BmNPV(Bm), AcNPV(Ac),orfrommock-infectedcells(m)probed with OT2EH(A)orBE284BE(B).Sizestandardsareindicatedtothe right inkilonucleotides. Major hybridized transcripts are indicated to the
leftbythearrowheads.Thecompositionsof theprobesareindicatedbelowthehybridizations.BE284BEcontainsthe 3'nontranslatedregion
of the BmNPVpolyhedringeneand1,340 bpof thecoding regionofORF1629. OT2EHcontainssequencesencoding gp64,ORF1, ORF2, PE34, ORF4,ORF5,and 94K.
on November 9, 2019 by guest
[image:4.612.124.238.80.339.2] [image:4.612.139.494.465.662.2]infected BmN cultures at either 5 or 24 h p.i. (Fig. 4A). At 5 h p.i., OT2EH hybridized to a 2,000-nucleotide (nt)
tran-script in BmNPV-infected, AcNPV-infected, and
BmNPV-andAcNPV-coinfected BmN cells. A 2,800-nt transcript was
also veryweakly hybridized at 5 h p.i. in AcNPV-infected
and in BmNPV- andAcNPV-coinfected BmN cells. At 24 h
p.i., OT2EH hybridized to 2,000-, 3,600-, and 4,300-nt
transcripts in BmNPV-infected, AcNPV-infected, and
BmNPV- and AcNPV-coinfected BmN cells. A 2,800-nt
transcript was also recognized by OT2EH in AcNPV-in-fected BmN cells but not in BmNPV- and
AcNPV-coin-fected cells. The 2,800- and 2,000-nt transcripts were most
likely derivedfrom the 94K (4) and gp64 (34) genes,
respec-tively,onthebasis oftheir size, expression at the early stage ofinfection, and relative abundance. The 2,800-nt transcript
mayalso result from the activation of other (non-early gene)
promoterscoveringPE34, ORF4, or ORF5 (27). The
3,600-and 4,300-nt transcripts were similar to those described
previously (27).
Followingthe initial hybridizations, OT2EH was stripped from the filter and the mRNAs were hybridized with
BE284BE (Fig. 4B). BE284BE (the 3' noncoding region of
polyhedrin
and coding region of ORF1629) recognized a2,200-nttranscript from BmNPV-infected, AcNPV-infected,
and BmNPV- andAcNPV-coinfected cultures at 5 and 24 h
p.i.,
albeit weakly at 5 h p.i. BE284BE also recognized a1,300-nt transcript in BmNPV-infected and in BmNPV- and
AcNPV-coinfected cultures. The 1,300- and 2200-nt
tran-scriptsmostlikely originated from ORF1629 and polyhedrin,
respectively,onthe basis of size and abundance during a late
stageofinfection. BE284BE alsohybridized weakly at 5 and
24 hp.i.to a2,200-nttranscript in mock-infected BmN cells. This hybridization, which was also found in virus-infected
BmN cells at 5 h p.i., was possibly due to nonspecific
bindingwithahost transcript. Probes specific toORF1629 of
AcNPV E2 show similar hybridization to host transcripts
isolated from uninfected larvae under stringent conditions
(29).
Effects ofeh2-AcNPV and eh-AcNPV on BmNPV
replica-tion and transcription. Infection of BmN cells with
eh2-AcNPVoreh-AcNPV resulted in typical viral growth curves
which reachedplateau maximums of about 4 x 108(Fig.1B)
and 6 x 107 PFU/ml (data not shown), respectively, at 72 h
p.i. Coinfectionof BmN cells with BmNPV andeh2-AcNPV
or eh-AcNPV also resulted in typical viral growth curves
which reachedplateaumaximums of about 7 x 108(Fig. 1B)
and3 x
108
PFU/ml (datanotshown), respectively.Theproteinsynthesispatternsof BmN cellsinfected with
eh2-AcNPV(Fig. SA), eh2-AcNPV and BmNPV (Fig. 5B),
eh-AcNPV (data not shown), or eh-AcNPV and BmNPV
(data
not shown) were essentially identical to those ofBmNPV-infected (Fig. 3A) BmN cells, as described above.
Theputative polyhedrin protein band of eh2-AcNPV
(indi-catedbythearrowheads in Fig. 5) and eh-AcNPV (data not
shown)
appeared slightlylarger (31 kDa) than the polyhedrinband of BmNPV (30 kDa). The 31-kDa band was also
observedbeginningat36 h p.i., whereas the 30-kDa putative
bandwasdetected beginning at 24 h p.i. (Fig. SB).
Asdescribedabove, transcription inAcNPV-infected and
in AcNPV- and BmNPV-coinfected BmN cells appeared to
occur normally and in a manner similar to that found in BmNPV-infected BmN cells. In order to analyze whether
transcriptionin eh-AcNPV-infected and in eh-AcNPV- and
BmNPV-coinfected BmN cellsoccurrednormally,Northern
blot analysiswas performed with the probes BE284BE and
OT2EH (Fig. 4). The transcription patterns of these
in-A 1 5 12 24 36 48 72
97-
66-
45-
.v-.-31-:.
21.5-14.4- -
-B 1 5 12 24 36 48 72
97-66-4
45- J
31- jg *
21.5-
14.4-FIG. 5. SDS-PAGE of pulse-labeled proteins from BmN cells that were infected with AcNPV (A) and BmNPV and
eh2-AcNPV (B). Proteins were pulse-labeled with [35S]methionine at 1,
5, 12, 24, 36, 48, or 72 h p.i. The arrowheads indicate the migration
of polyhedrin. The approximate sizes of marker proteins are indi-cated to the left in kilodaltons.
fections were very similar to those of BmNPV-infected,
AcNPV-infected, and BmNPV- and AcNPV-coinfected
BmN cells probed with OT2EH or BE284BE. The intensity of the OT2EH-probed patterns was, however,
approxi-matelyfive times higher than that of BmNPV-infected cells.
In eh-AcNPV-infected BmN cells, OT2EH hybridized to transcripts of 2,000 and 2,800 nt at 5 h p.i. and of 1,700,
2,000, 2,800, 3,600, 4,300, 7,000, and 8,000 nt at 24h p.i. In
eh-AcNPV- and BmNPV-coinfected BmN cells, OT2EH
hybridized to transcripts of 1,700, 2,000, and 2,800 nt at 5 h
p.i. and of 1,700, 2,000, 2,800, 3,600, and 4,300 nt at 24 h p.i. In eh-AcNPV-infected BmN cells, BE284BE hybridized to a 2,200-nt transcript (very weakly) at 5 h p.i. and to 1,300-, 2,200-, 3,300-, and 4,200-nt transcripts at 24 h p.i. In eh-AcNPV- and BmNPV-coinfected BmN cells, BE284BE hy-bridized to a 2,200-nt transcript (very weakly) at 5 h p.i. and to 1,300-, 2,200-, 3,300-, and 4,200-transcripts at 24 h p.i.
The 2,000- and 2,800-nt transcripts which hybridized to OT2EH and the 1,300- and 2,200-nt transcripts which hy-bridized to BE284BE were putatively transcribed from gp64,
94K, ORF1629, and polyhedrin, respectively, as described
above. The other transcripts hybridized by OT2EH (1,700, 3,600, 4,300, 7,000, and 8,000 nt) and BE284BE (3,300 and 4,200 nt) were in general similar to those examined previ-ously (27, 30).
DISCUSSION
In previous studies, BmN cells coinfected with BmNPV and AcNPV were found to exhibit a unique morphological response, i.e., atypical CPE (12). In the present study, we found that exchange of a 572-bp (or smaller) region of a single AcNPV gene with the corresponding region of Bm-NPV can eliminate these effects. Nucleotide sequence and physical map comparisons indicated that this gene putatively encodes a DNA helicase (20) which is expressed at an immediate-early stage of infection (14). Superinfection of BmN cells with AcNPV at 5 or 12 h post-BmNPV infection did not induce the unique CPE found following BmNPV and AcNPV coinfection, nor did it significantly affect BmNPV
on November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.373.493.77.256.2]BmNcells, especially when probe OT2EH was used (Fig. 4A). This might be explained by (i) a higher efficiency of
AcNPVtranscriptional factors and/or(ii) a higher efficiency
of viralreplication by AcNPV-encoded polypeptides. These
effects might have been masked in AcNPV-infected or
AcNPV-BmNPV-coinfected BmNcells because of the CPE induced by AcNPV. These results indicated that the
prema-turedrop in proteinsynthesis was not due to the lack of viral
transcripts and that the inhibition of viral replication proba-bly occurred at the translational level.
A premature drop in protein synthesis and active viral
transcription has also been reported in
non-AcNPV-permis-sive gypsy moth (Lymantria dispar) cells infected with
AcNPV(7). Thiswasspeculated to be due to (i) the inability
of L. disparcells to translate theabundantlyexpressed viral
mRNAsand/or(ii)interferenceinthetranslational processes
by a viral structural polypeptide(s) in the infecting viral
particles. The normal transcription of late and very late
genes, e.g.,ORF1629 andpolyhedrin of AcNPV, indicated
thatbaculovirus early gene products required for the
trans-activation (e.g., IE-1 [6]) and transcription (e.g., a
virus-specificRNApolymerase[9])ofbaculoviruslate genes were
efficiently transcribed and translated. Furthermore, since translation of eh-AcNPV and eh2-AcNPV (Fig. 5A)
oc-currednormally in thenon-AcNPV-permissiveBmNcells, it
appears that an inability of the BmN cell translational
machinery to translate mRNAs from AcNPV isnotafactor
in the inhibition phenomena. This leads to the hypothesis
that the AcNPV DNA helicase gene itself may cause the
BmNtranslational machinerytobecome dysfunctional. Coinfectionof BmNPV and the AcNPVisolateLiin BmN
cells did not result in the complete inhibition of BmNPV
replication (BmNPV replicated to about 106 PFU/ml [data
not shown]), indicating that AcNPV isolates may vary in
their abilities to inhibit BmNPV replication. Morris and
Miller (26) have recently reported that a recombinant
AcNPV Li isolate carrying achloramphenicol
acetyltrans-ferase(CAT)genedrivenbyanAcNPV late gene promoter
canexpress CAT in BmN cells. Infection of BmN cells with
arecombinant AcNPV
Li
expressing the CAT gene under avery late gene promoter, however, was shown to result in
very low CATactivity. Although directcomparison between
theexpression of CAT (by a viral promoter) and expression
of virus- and host-derived proteins by SDS-PAGE is
diffi-cult,thesephenomenaareconsistent with our observation of
virus-specific proteins inAcNPVOT2-infected and AcNPV OT2-BmNPV-coinfected BmN cells at 5 and 12 h p.i. (at
very lowlevels). On the basis of these data, it is difficult to
conclude at present whether the difference in the abilities of
that the host range of baculoviruses may be determined by
the ability or lack of ability of host cells to combat
virus-induced cytotoxicity. Virally encoded proteins with helicase activity (e.g., the putative RNA helicase of the turnip crinkle
virus [3]) and cell-specific host factors (e.g., those induced
by the parvovirus minute virus of mice [1]) have been
implicated in inducing
cytotoxicity
in several host systems(20).
Aclear understanding of the mechanisms of host range
determination of baculoviruses is essential for their safe use
and acceptance as insecticides and expression vectors. At
present, the exact mechanism by which the putative AcNPV
DNA helicase inhibits viral replication in BmN cells is
unknown; however, our results indicate that the AcNPV
DNAhelicase itself may inhibit either directly or indirectly
thetranslational machinery of BmN cells by inducing
cyto-toxicity.
REFERENCES
1. Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone ofparvoviralgenes encodingnon-structuralproteins. EMBO J.9:2989-2995. 2. Clem, R. J., M.Fechheimer,and L. K.Miller. 1991.Prevention
of apoptosis by abaculovirus gene during infection ofinsect cells.Science 254:1388-1390.
3. Colimer, C. W., L.Steinzier,X. Chen, N.Fay, and D. Hacker. 1992. Single amino acid change in the helicase domain of the putativeRNAreplicaseofturnipcrinkle virus alters symptom intensificationbyvirulentsatellites.Proc. Natl. Acad. Sci. USA 89:309-313.
4. Friesen, P. D., and L. K. Miller. 1987. Divergenttranscriptionof early 35- and 94-kilodalton protein genes encoded by the HindIII K genome fragment of the baculovirusAutographa califomica nuclear polyhedrosisvirus. J.Virol. 61:2264-2272. 5. Granados,R. R., and B. A.Federici (ed.). 1986. The biology of
baculoviruses,vol. I andII.CRC Press, Boca Raton, Fla. 6. Guarino, L. A., and M. D. Summers. 1986. Functional mapping
of atrans-activatinggenerequired for expression of a baculo-virusdelayed-early gene. J. Virol. 57:563-571.
7. Guzo, D., H.Rathburn, K. Guthrie, and E. Dougherty. 1992. Viral and hostcellular transcriptioninAutographa califomica nuclear polyhedrosis virus-infected gypsy moth cell lines. J. Virol.66:2966-2972.
8. Hershberger, P. A., J. A. Dickson, and P. D. Friesen. 1992. Site-specific mutagenesisof the35-kilodalton protein gene en-coded byAutographa califomica nuclear polyhedrosis virus: cell line-specificeffects on virus replication. J. Virol. 66:5525-5533.
9. Huh, N. E., and R. F. Weaver. 1990. Identifying the RNA polymerases that synthesize specific transcripts of the Au-tographa californica nuclear polyhedrosis virus. J. Gen. Virol. 71:195-201.
on November 9, 2019 by guest
10. Iatrou, K., K. Ito, and H. Witkiewicz. 1985.Polyhedringeneof Bombyx mon nuclearpolyhedrosisvirus.J. Virol.54:436-445. 11. Kamita, S. G., K. Majima, and S. Maeda.1993. Identification and characterization of thep35 gene ofBombyxmon nuclear polyhedrosis virus that prevents virus-induced apoptosis. J. Virol. 67:455-463.
12. Kondo, A., and S. Maeda. 1991. Host range expansion by recombinationof thebaculovirusesBombyxmon nuclear poly-hedrosis virus andAutographacalifornicanuclearpolyhedrosis virus.J.Virol.65:3625-3632.
13. Laemmli, U. K. 1970.Cleavage of structuralproteinsduringthe assembly of the head ofbacteriophage T4. Nature (London) 227:680-685.
14. Lu, A., and E. B.Carstens. 1992.Transcription analysisof the EcoRI D region of the baculovirus Autographa califomica nuclear polyhedrosis virus identifies an early 4-kilobaseRNA encodingthe essentialp143gene. J.Virol.66:655-663. 15. Luckow, V. A. 1991. Cloning and expression of heterologous
genesin insect cells withbaculovirusvectors, p. 97-152. In A. Prokop,R. K.Bajpai, andC. S. Ho (ed.), RecombinantDNA technology andapplications. McGraw-Hill, Inc., New York. 16. Maeda, S. 1989. Expression offoreign genes in insects using
baculovirus vectors. Annu. Rev. Entomol. 34:351-372. 17. Maeda, S.1989. Gene transfer vectors of abaculovirus, Bombyx
mon, and their use for expression of foreign genes in insect cells, p. 167-181. In J. Mitsuhashi (ed.), Invertebrate cell systemapplications. CRC Press, Boca Raton, Fla.
18. Maeda, S. Unpublished data.
19. Maeda, S., S. G. Kamita, and H. Kataoka. 1991. The basic DNA-binding protein of Bombyx mon nuclear polyhedrosis virus: theexistence ofan additional arginine repeat. Virology 180:807-810.
20. Maeda, S., S. G. Kamita, and A. Kondo. 1993. Host range expansionofAutographa califomicanuclearpolyhedrosisvirus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating fromBombyx mori NPV. J.Virol. 67:6234-6238.
21. Maeda, S., T. Kawai, M. Obinata, H. Fujiwara, T.Horiuchi, Y. Saeki, Y. Sato, and M. Furusawa. 1985. Production of human alpha-interferonin silkwormusing abaculovirusvector.Nature (London)315:592-594.
22. Maeda, S., and K.Majima. 1990.Molecular cloning and phys-icalmappingof the genome ofBombyxmorinuclear
polyhedro-sis virus. J. Gen. Virol. 71:1851-1855.
23. Maeda, S., Y. Mukohara, and A. Kondo. 1990. Characteristi-cally distinct isolates of the nuclear polyhedrosis virus from Spodopteralitura. J. Gen. Virol.71:2631-2639.
24. Maeda, S., S. L. Voirath, T. N. Hanzlik, S. A. Harper, K.
Majima,D.W. Maddox, B. D.Hammock, and E. Fowler. 1991.
Insecticidal effects of aninsect-specificneurotoxinexpressed by arecombinant baculovirus. Virology184:777-780.
25. Miller, L. K. 1988. Baculovirusesas geneexpressionvectors. Annu. Rev.Microbiol.42:177-199.
26. Morris, T. D., and L. K. Miller. 1992. Promoterinfluence on baculovirus-mediated gene expression in permissive and non-permissiveinsect celllines. J. Virol.66:7397-7405.
27. Oellig,C.,B.Happ, T.Muller, and W. Doerfler. 1987. Overlap-pingsetsof viralRNAsreflect thearrayof polypeptides inthe EcoRI J and N fragments (map positions 81.2 to 85.0) of the Autographa californica nuclear polyhedrosisvirus genome. J.
Virol. 61:3048-3057.
28. O'Reilly,D.R.,L. K.Miller,and V. A. Luckow. 1992.
Baculo-virusexpression vectors: alaboratorymanual. W. H.Freeman &Co.,New York.
29. Pham, D.Q., and N.Sivasubramanian. 1992. In vivo transcrip-tionalanalysisof three baculovirus genes: evidenceof homol-ogybetween viral andhosttranscripts. Virology190:288-297. 30. Possee, R. D., T. P. Sun, S. C. Howard, M. D. Ayres, M.
Hill-Perkins, and K. L.Gearing. 1991. Nucleotide sequence of the Autographa californica nuclear polyhedrosis 9.4 kbp EcoRI-I and-R(polyhedrin gene) region. Virology185:229-241. 31. Stewart, L.M., M. Hirst, M. L. Ferber, A. T. Merryweather, P. J. Cayley, and R. D. Possee. 1991. Construction of an
improved baculovirus insecticide containingan insect-specific toxingene.Nature (London) 352:85-88.
32. Sugimori, H., T. Nagamine, and M. Kobayashi. 1991. Protein synthesis in BM-N cells infected with Bombyx mori nuclear polyhedrosis virus.J. Invertebr. Pathol. 58:257-268.
33. Tomalski, M. D., and L. K. Miller. 1991. Insect paralysis by baculovirus-mediated expression of a mite neurotoxin gene. Nature(London)352:82-85.
34. Whitford, M., S. Stewart, J. Kuzio, and P. Faulkner. 1989. Identification and sequenceanalysisofageneencoding gp67,an
abundantenvelope glycoproteinof the baculovirusAutographa californica nuclearpolyhedrosis virus. J. Virol. 63:1393-1399.