JOURNALOFVIROLOGY,Mar.1970,p.368-380
Copyright( 1970 AmericanSociety forMicrobiology
Vol.5,No. 3 Printed inU.S.A.
Molecular
Recombination
in
T4
Bacteriophage
Deoxyribonucleic
Acid
III.
Formation of Long Single
Strands
During
Recombinationl
ROBERT C. MILLER, JR.,2 ANDRZEJ W. KOZINSKI, AND SAMUEL LITWIN
Department of Medical Genetics, University of Pennsylvania, Philadelphia, Pennsylvania 19104
Received for publication3November 1969
Evidence was presented to support thehypothesis thatlong single strands
ap-pearingatlatetimes(15 min afterinfection) areproducedasaresult of
recombina-tionand not asa continuous elongation during thereplication process. The
pro-duction oflong strands does notdepend onthemultiplicity ofinfection, and the
first long strandsappearatthetime when20to50phage equivalent units of
deoxy-ribonucleic (DNA)aresynthesized,and notearlier.Theaddition ofchloramphenicol
at5 min, which preventsmolecular recombinationbutallowsreplicationofDNA,
prevents the formation oflong, single strands. Chloramphenicol added between 8
and10minafterinfection,atimeatwhichmolecularrecombinationisfully expressed
andcovalentrepair of recombinant molecules isallowed, doesnotpreventformation
of long single strands. Cutting of single-strand DNA with a limited amount of
endonuclease I allows confirmation that the fast-sedimenting characteristic of
intracellular denatured DNA is caused primarily bythelength of thestrands, and
not by the formation of aggregates.The computer simulation of two
recombina-tion models indicates the feasibility ofrandom breakage and rejoining of
mole-cules in generating long concatenates.
Thedeoxyribonucleic acid
(DNA)
moleculesofT4 bacteriophage are
circularly
permuted andterminally
redundant(8,
12).
Circularpermuta-tion can be created by at least two different
mechanisms.Onewouldinvolveforminga
physi-cal circle of the DNA during the replicative
process. Thesecondprocesswouldrequire, prior
tomaturation,theformation of molecules much
longerthan onephageequivalentunit.This would beconcludedbyexcision of moleculesof aboutone
phageequivalent unitinlength
during
thematu-rationprocess. The
long
molecules could becre-atedbysome mechanismof
replication
inwhichthe DNA iscontinuously elongated,orbya
mech-anism of recombination, which we favor. Since
neutral sucrose gradient analysis doesnotreliably
measure the molecularweight ofreplicative DNA
(6),
thepresence oflong single strandsinanalka-line sucrose gradient would be a desirable
indica-tion that double-stranded DNAlongerthan one
phage equivalentunit does exist in the
intracellu-lar DNApool.
1Submitted byR. C. Miller,Jr., to the University of
Penn-sylvania inpartial fulfillment of the requirements for the Ph.D degree.
2Present address: InstituteforEnzymeResearch, University ofWisconsin, Madison, Wis.
When newly synthesized DNA is labeled with
3H-thymidine during later stages ofinfection of
Escherichia coli B bybacteriophageT4, some of
the label is found in material which sediments
faster than reference DNA in analkaline sucrose
gradient (1). It has been postulated that this
material representssingle-strandedDNAwhich is
longer than the single strands which can be
iso-lated from maturephage.Experiments testingthis
hypothesis are reported here. Net synthesis of
DNA was measured simultaneously with the
production of long single strands to determine
whether thelongsinglestrandswereformed
dur-ing the initial stages of replication or arose at
later times, concomitantly with the onset of
re-combination. Furthermore, it will be
demon-strated that one caninhibitlongsinglestrand
for-mationwithout inhibitingDNAsynthesis.
Studies tobe reported here support the
conten-tion that the fast-sedimenting DNA, in alkaline
sucrose gradients, is indeed composed of single
strands longer than those isolated from mature
phage. Estimatesof net DNAsynthesis
concomi-tant with long single strand production indicate
that approximately 20 phage equivalent units of
DNA are synthesized before a measurable
frac-tion of intracellular DNA enters single strands
368
on November 11, 2019 by guest
http://jvi.asm.org/
exceeding in size the reference mature DNA.
Addition of chloramphenicol (CM) to infected
cells at a time which leads to the inhibition of
re-combination inhibits the formation of longsingle
strands even though DNA synthesis is not
im-paired. Furthermore, an insignificant extent of
long singlestrand production canbe detected in
cells grown in 5-bromodeoxyuridine (5-BUdR) medium even though this condition allows
pro-ductionof viablephageof normalburst sizes. Two
modelsof recombinationwill be simulated in the
computer, and the results will be evaluated in
respect to thefeasibilityof the formation oflong
molecules asa result ofrecombination.
MATERIALS AND METHODS
Most of the procedures for density or isotopic
labelingandsucrose ordensity gradient analysishave
beendescribed elsewhere (6).Some sucrosegradients
wereunderlayeredwithapad ofsaturatedsucrose to
guarantee quantitative recovery ofthe input
radio-activity; these gradients were prepared by
under-layering3.5 mlof 5to20%sucrosegradientwith 1.0
ml of saturated sucrose with a canulating needle
attached to a syringe. The lysozyme-Triton X-100
lysis (LTL) method wasperformed in the following
manner. Cellswere suspendedinamixture of 0.05 M
tris(hydroxymethyl)aminomethane (Tris), 0.05 M
NaCl, 0.05 M ethylenediaminetetraacetate (EDTA),
andlysozyme (100,ug/ml), pH 8.0,andchilledat4C
for 10 min. Triton X-100 wasaddedtoa final
con-centration of1%,and themixtureswerechilledforat
least 10minat4 Cbeforethe addition ofalkali. The
programmingapproachesfor computersimulationwill
beoutlinedin thetext.
RESULTS
Net DNA synthesisversus long strand. Iflong
single strandsare
produced simply
as a functionof
replication,
theyshould be detected beforetwoor three phage
equivalent
units of DNA perbacterium are
synthesized.
Thefollowing
experi-ment was conductedto determinetheamountof
DNA synthesized before
long
single
strands can bedetected.E. coli B23 was grownto 3 X 108 cells/ml in
low phosphate TCG medium. The culture was
splitinto two
samples;
thefirst one(part 1)
wasinfected with T4
bacteriophage
[multiplicity
of infection(MOI)
=5.0]
labeled with32p
at aspecific activity of 5.0 mc/mg of P. The second
sample was
(part
2)
infected with coldphage
(MOI = 5.0).P2p
at aspecific
activity
of 0.5mc/mgofP was added tothe second
sample
at2 min afterinfection. DNA
synthesis
wasmeas-ured by uptake of 82P to trichloroacetic
acid-precipitable, alkali-resistant material
(9).
Atvarioustimes after
infection, samples
from part 1 weretransferredtoKCNandlysed
withlysozyme
and sodium lauryl sarcosinate (1) or with lyso-zyme and Triton X-100. At the same time, sam-ples from part 2 were precipitated with
trichloro-acetic acid and the amount of DNA synthesized was determined after alkali digestion of the sam-ples by procedure of Schmidt et al. (9). The num-ber of phage equivalent units of DNA
synthe-sizedwas calculated on the basis that 1 ,ug of P =
5 X 10'° phage equivalent units. 8H reference
phage were added to the lysates from part 1; themixtureswere treated with alkali and layered
on 5to 20% sucrose gradients which were
under-layed with 1 ml of saturated sucrose. The satu-rated sucrose pad provided quantitative recovery
of the material from the gradients.
Alternatively, if progeny strands were labeled
with 32p incorporated after infection (part 2),3H
reference phage was added, similarly to part 1, butfractionswere collected into conical tubes and werealkali-digested (9) prior to being counted in a scintillation counter at various times after in-fection (Fig. 1). Figure 2 shows the results of
sucrose gradient analysis of lysates obtained at
corresponding timesafter infection. This result is
independent of the MOI up to at least 15 phages per bacterium. Figure 2 represents the fate of
parental 32p label. Progeny label was analyzed at
the same times, and the patterns obtained in
sucrose gradient analysis were virtually
indistin-guishablefrom thoseofparental DNA; therefore,
they are not documented inthis paper.
Inhibition of long strand formation. Kozinski
has shown that addition ofCM at about 5 min
after infection inhibits recombination without im-pairingDNAreplication (6). If long singlestrands
are formed as a result of recombination, the
addition ofCM to the infected bacteria at this
time shouldinhibit their assembly. Indeed, if CM
isaddedat 5 minafter infection andthecellsare
incubatedfor 45minutes, eventhough50 to 100
phageequivalent units of DNA weresynthesized
(a figureatleasttwiceaslargeasthat whenlong single strands arefirst formedduring normal
in-fection),nolongsingle strandsarefound(Fig. 3).
This indicates thatsynthesiscanproceed without
necessarily forming long single strands. In
con-trast, addition ofCM at 8 to 10min allows the
appearance of
fast-sedimenting
DNA.When the infected bacteria were pregrown in
5-BUdR, an unmeasureable amount of long
singlestrands was formed in the absence of CM.
When CM was added to bacteria infected in
5-BUdR medium, the amount of long single
strands formed was much less than in
light
medium (Fig. 4). When one notesthat
5-BUdR-labeledDNA sediments faster thanan
unsubsti-tuted DNA molecule of the same size
(10),
theactualamountoflong
single
strands which wereon November 11, 2019 by guest
http://jvi.asm.org/
MILLER, KOZINSKI, AND LITWIN
Z 200
LLC
0t
/
unZ/
z/ ow
z
Z
100->Sz/
X. 50
w
0 5 lo 15 20 25
MINUTES AFTER INFECTION
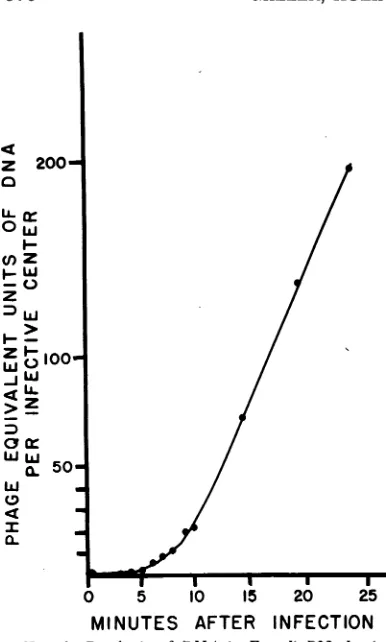
FIG. 1. Synthesis ofDNA in E. coil B23, by
in-fected bacteriophage T4B01r. E. coli B23 was grown
to 3 X 108 cells/ml in low-phosphate TCG medium. The culture was divided into two parts: (i) infected
with T4 bacteriophagelabeledwith 32p (see Fig.2for
sucrosegradient analysis ofthispart); (ii) infectedwith
coldphage. To the latter 32pwasaddedat 2 minafter
infection,and DNAsynthesiswasmeasuredby uptake
of 32ptotrichloroacetic acid-precipitable,
alkali-resis-tantmaterial.
synthesizedinthepresenceof CMdecreases
fur-ther. However, bacteria infected in 5-BUdR
medium liberates5-BUdR-labeled, viableprogeny
in normal quantities. It appears, therefore, that
the formation of long single strands is not an
obligatory intermediate of DNA replication in
T4-infected cells.
Inallcases,theprocedure yielding thegreatest amountoflong singlestrandswasto add CMat
10to 12 min afterinfection, a time whenall the
enzymes necessary forrecombination and repair
of recombinantmoleculeswerepresentandyeta
timewhen maturationwasinhibited.
Length of denatured DNAasafactorprimarily
responsible for fast sedimentation. Incubation of
the infected bacteriawith lysozymepriorto
alka-linelysis isimperative; withoutlysozyme present
in the lysing solution, material which sediments
much faster than reference DNA in alkaline
sucrose
gradients
canbe detectedeven if CM isadded at the moment of infection. When this material is incubated with lysozyme, the parental label is found to sediment
identically
with thereference DNA. The
alkali-resistant,
lysozyme-sensitivecomponentpresumably
isanassociation oftheparentalDNAwithcell wallmaterial. This association takes placeevenifCMisaddedatthemomentofinfection,eveniftheparental phage is
inactivated with ultraviolet light, and it takes place with ambermutants, deficient inDNA
syn-thesis.
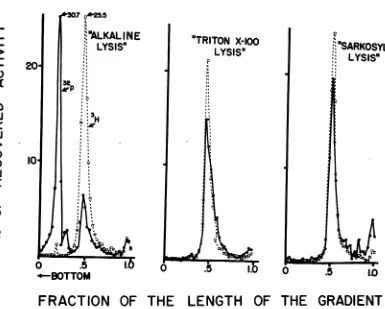
Figure
5 illustrates the results of alkalinesucrose
gradient
analysis
ofonelysate treatedinthreeways:
(i) lysed
in 0.25MKOH,(ii)
lysed by thesarkosyl method,
and(iii) lysed
by the Triton method. The lysate was E. coli Binfected withT4BO1r
inthepresenceof CMandwasincubated
for30minbeforesampling.
Oneexplanation ofthelysozyme-resistant, fast-sedimenting material could bethatthereplicating pool ofphage DNA is such a tangled mass of
nucleicacid that during alkali denaturation indi-vidual single strands become trapped in a
"chicken wire-like" mesh which would sediment
as an aggregate in alkaline sucrose gradient. A
secondexplanation might bethat theassociation with cellwallmaterial which is sensitiveto
lyso-zyme when thetrapped DNA is of conservative
nature might be resistant when the DNA is in
replicative form, since replicativeDNA may have
a structurewhich facilitates the association with
cell wall material. Recombinant DNA is known
to have
"puffs,"
"gaps," and a highly tangledstructure(3,
6);
thisstructuremight maintainanassociation withcell wallmaterial which couldbe more intimate and resistant to lysozyme. This would
impart
afast-sedimenting
characteristicto the DNA. Two experiments argue against this hypothesis.First,fast-sedimenting, denaturedDNAcan be
isolated from an
alkaline
sucrose gradient,dialyzed, and heated for2min at95 Cin standard saline-citrate
containing
1%
formaldehyde, andthematerialwillstill sedimentas a
fast-sediment-ingcomponent. A partially renatured, mesh-like
structure might be expected to decompose at
meltingtemperatures in thepresence of
formalde-hyde.
Second, if the fast-sedimenting, denatured
DNAisincubated with
endonuclease
I so that theDNA receives, on the average, one to two single
strandbreaks perphage equivalent unitof length,
the fast-sedimenting DNA is cut to short
frag-ments as predicted for a
linear
molecule. Themolecular-weight distributionofthe fragments of
a moleculewhich has been randomly cut can be
370 J.VIROL.
on November 11, 2019 by guest
http://jvi.asm.org/
[image:3.497.56.249.52.373.2]C]
LLI
H
H 0
C)
0
1-0 H
z
Lli
w-'.'V28.3%
:L
.1.
9 min.
-BOTTOM
FRACTION
OF THE
LENGTH
OF THE
GRADIENT
FIG. 2. Alkaline sucrosegradient analysis of intracellular parental 32P-labeled DNA. Samples from part iof
the experiment (describedinFig.1)weretransferredatvarious timesafter
infectiont
toKCN,chilled,andlysed withlysozymeandTriton X-100.3Hreference phagewereadded to the lysates and the mixtures were treated with
alkali andwerelayeredonS to20% alkalinesucrosegradients whichwereuniderlayeredwith1.0ml of saturated
sucrose. 3H = reference, integralDNA.
predicted; furthermore,thesedimentationpattern
of thesefragmentsina sucrose
gradient
has beensimulated in a computer
(7).
Figure
6 illustratesthepredicted distribution of
fragments produced
by cutting molecules of differentlength (L
=phageequivalentsof
length)
with various averagenumbersofcuts(x = number of
cuts)
perphage
equivalent unit of DNA. When one to two cuts
per phage equivalent unit have degraded the
target molecules, the products all have
approxi-mately the same distribution regardless of the
originallength of the molecule. Afteroneto two
cuts per phage equivalent unit of DNA, the
products of size 1 molecules will sediment in a
sucrosegradient similarlytothoseof sizes3, 5,or
15, for example. However, ifthetargetmolecules
11
..o.
on November 11, 2019 by guest
http://jvi.asm.org/
[image:4.497.98.393.63.507.2]MILLER, KOZINSKI, AND LITWIN
I-C)
H
llJ
0
llJ
0
11
0
A
5'cm
0
.5
-BOTTOM
B
8'cm
C
lO'cm
FRACTION OF THE
LENGTH
OF THE
GRADIENT
FIG. 3. Effect ofCMonformation oflong single strands. Inanexperimentsimilartothat describedin Fig.
1, CMwas addedat the indicated timesafter infection,andthecellswere incubatedforanadditional 45 min.
3H= reference,integralDNA.
are notlinear single-stranded DNA, theproducts
ofcuttingmaynotbeexpected tosediment simi-larly to reference DNA treated in an identical
manner. Figure7 illustrates theprinciple of this
experiment. If the fast-sedimenting material
ac-quires its sedimentation characteristics from a
structureresemblinga "chicken wire-like" mesh,
it would notbeexpected tofallcompletely apart
afterone to two cutsperphageequivalent unitof
length. Also, ifthelabeledDNAwereattachedto somelysozyme-resistant, alkali-resistantmaterial, a portion ofthe label would still sediment fast
after cutting. With these ideas under
considera-tion, thefollowing experiment wasconducted to testthehypothesis that thefast-sedimenting
ma-terialwascomposedofsinglestrandslongerthan
those isolated from mature phage.
E. coli B23 was grown to 3 X 108cells/ml in
high phosphateTCG medium and infected with
T4BOir
(MOI = 5.0) labeled with 32p at aspecific activity of 3.0. Chloramphenicol (100
Ag/ml)
was added to the infected cells 10 minafter infection, and the cells were incubated for
45min at 37 C. Thecells werelysedbythe
lyso-zyme-Triton method;
38H
reference DNA wasadded to the lysates, and themixture was incu-batedfor 20minwith 0.25 M KOH. Themixture
was analyzed in an alkaline sucrose gradient
which had been underlayered with a pad of
saturated sucrose. 32P-labeled DNA reachingthe
pad was isolated, dialyzed against 0.07 M Tris
(pH 7.6) and supplementedwith single-stranded
3I reference DNA. This 32P-labeled DNA was
considered to be apparently long single strands.
Partof themixturewasreanalyzedin an
alkaline
sucrosegradient (Fig. 8A);this servedasacontrol
which showed that fast sedimenting strands
re-tained their characteristics after the isolation procedure.Partof the mixturewasheated to 95 C
for 2 min in standardsaline-citratecontaining 1%
formaldehyde; this part was analyzed on a 5 to
20% sucrose gradient containing 1%
formalde-hyde(Fig. 8B). Onecanseethat suchheat
treat-mentdoesnotdrastically affect thesedimentation
characteristic of the 32P-labeled DNA. Another
part of the fast-sedimenting moiety was
incu-372 J. VIROL.
on November 11, 2019 by guest
http://jvi.asm.org/
[image:5.497.60.448.63.362.2]F-C)
LLI
llJ
0
Q
LLJ
bi
11
0
0 5
0-BOTTOM
9'CM
1.0
(D
0
12'
CM
. .
::1 ::1 : :,
: :, . I
: :,
I,
.,.,
::~
.5
1.0
FRACTION
OF THE
LENGTH OF THE GRADIENT
FIG. 4. EffectofCMonformationoflong single strands producedinbacteria infectedin5-BUdRmedium. Cells
weregrownfortwogenerationsin5-BUdRmedium andwereinfectedwith 32P-labeledT4BO1r. CMwasadded at
various timesafterinfection,andthe cultureswereincubatedfor45min.Thecellswerelysed,andalkalinesucrose
gradientanalysis oftheintracellular, 32P-labeledDNAwasperformedwith the 3H integral DNA asa
referenice
bated with a very low concentration of
endo-nuclease I. As an internal
control,
3H-labeledmaturephageDNAindenatured formwasadded
prior to
digestion.
After treatment with endo-nuclease I, the mixture was incubated with 0.25M KOH-0.02 M EDTA for 20 min and then
analyzed by
alkalinesucrosegradient
sedimenta-tion(Fig.
8D).
Itisobviousfrom thisgraph
that the32P-
and 3H-labeled DNA sedimented in asimilar fashionaftertreatmentwithendonuclease
I. The
apparently long
single
strands werecut tothesize
fragments
predicted
for linear molecules. To test the size ofresulting
fragments,
fresh 32p referenceDNAwasadded inexcesstothemixturetreated with endonuclease Iand themixturewas
analyzed in an alkaline sucrose
gradient (Fig.
8C).By
comparing
thedistance sedimentedby
the32p reference DNA (D1) with the distance
sedi-mentedbythe3H DNA
(D2),
one candeterminethenumber of
single-strand
scissionsreceivedby
the DNA per
phage
equivalent
unitof DNA.Thiscanbe done
by
comparing Fig.
8CwithFig.
7orby
using
thegraph
of number of scissions(X)
versusD2/D1ofLitwin,Shahn,and Kozinski (7).
Thenumberofscissions whichthe DNA received
from the endonuclease I was between one and two perphage equivalent unitof DNA. This
ex-periment supports the contention that the
fast-sedimentingDNAis actually composed of single
strandslonger thanonephageequivalentunit and
argues against thepossibilityoftheartefacts
dis-cussedbefore.
Simultaneous maturationof progeny and paren-tal phage DNA. Some theories of the in vivo replicationof T4bacteriophageDNApredict that
labeled parental DNA should appear in mature
progeny phage at a rate different from that of
labeled progeny DNA. Forexample, if T4 phage
DNAreplicatedinacontinuouslyelongating
con-catinate (11), 50% of theoriginal parental label
would be extendedout at oneend. IfphageDNA
then is encapsulated at this end, 50% of the
parental label would beencapsulatedintheinitial
stagesof maturation. The
"rolling
circle" modelpredicts, on the other hand, that some of the
parentalDNAresides ina sanctuary attached to
on November 11, 2019 by guest
http://jvi.asm.org/
[image:6.497.52.438.65.366.2]MILLER, KOZINSKI, AND LITWIN
L-a
w
w
0
w
0L
0
0 .5
*-BOTTOM
0
"TRITON
X-1OO
LYSIS"
p:..
N
.5
lb
[image:7.497.63.448.57.366.2]FRACTION OF THE
LENGTH OF
THE
GRADIENT
FIG. 5. Effect oflysing procedureonsedimentationcharacteristicsofparentalDNAin alkalinesucrosegradients.
E. coli B23 cellswereinfectedwith32P-labeledT4BOi',andCMwasadded at themomentof infection. The
cul-tureswere incubatedfor30minandchilled in NaCI-EDTA. Thesuspensionsweredivided into threeparts;each
partwaslysedbyadifferentmethod:A,lysedin0.25mKOH;B, lysed bythesarkosylmethod(I);andC,lysed
by the Triton method. 3H referenceDNA was addedtothe lysates, andthe mixtures were treated withalkali. The alkali-treatedmaterialwas layered on S to 20% alkaline sucrose gradients underlayered with a pad of
saturatedsucroseandanalyzedasusual. 3H = reference, integralDNA.
thecell membrane or resides as the core of the
rolling circle (2). Parental label would, in this
case, beexpectedtomature atarateslower than
that ofprogenyphage.Incontrast,thepossibility
that recombination disperses all parental
ma-terialuniformly throughout the pool of
replica-tive DNA should guarantee that both parental
andprogenyDNAin thepoolshould haveequal
chance ofbecoming incorporated into maturing
phages. To test this hypothesis, parental phage
DNA was labeled with 82p and progeny phage
DNA was labeled with tritiated thymidine, and
then the percentage of each label which became
resistanttodeoxyribonucleasewasdetermined as
afunction of time afterinfection.Theexperiment
wasconducted in thefollowingmanner.Aculture
of E. coli B23 was grown at 37 C to 3 X 108
cells/mlinhigh phosphateTCGmedium
supple-mented with 5 Mg ofthymidine per ml, 5 ,ug of
5-fluorodeoxyuridineperml,and 25,gof uridine
perml. The culture was infected at an MOI of
5.0 with T4 bacteriophage labeled with 32p at a
specific activity of 0.3 mc/mg ofP. At 11 min
after infection, 3H-thymidine was added to the
cultureat a final specific activity of 2 mc/mg of
thymidine. At 12minafterinfection,the infected
cells were chilled, centrifuged, washed with and
suspended in high phosphate TCGmedium
sup-plemented with 100 Mgof cold thymidineperml.
Theincubation of the cellsat37 Cwascontinued,
andatintervalsthereafter samplesoftheinfected
bacteriawerechilled; oneportionof each sample
was tested for the presence of mature progeny
phage, andoneportionwaslysed by the
lysozyme-Triton method. The lysates were incubated with
pancreatic deoxyribonuclease; before and after
treatmentwith deoxyribonuclease, aportionwas
precipitated with trichloroacetic acid, transferred
toaglass-fiber filter, dried, and countedfor
radio-activity. (Aseparate testshowedthat inamixture
v
.
SARKOSYL
LYSISN
.:1
.,
_J
0
w
a
374
J. VIROL.6 a
on November 11, 2019 by guest
http://jvi.asm.org/
D
A B
Reference DNA Reference
DNA
ENDO I +ENDO I
[image:8.497.50.462.61.658.2]*
r_
FIG. 7. Hypothetical results of cutting long single
strandswith limited amount ofendonuclease I. A, true
Ll long strand; B, fast-sedimenting artefact.
of 3H-labeledDNA and 32P-labeled phage which
was treated in a reconstruction
experiment,
the3H was rendered completely soluble in 0.3 M
trichloroacetic acid, whereas the 82p was
com-pletely
insoluble;
i.e.,
theprocedure truly
dis-criminated betweenDNA outside and insidethephage
head.)
InFig.
9, the percentage of eachlabel which became resistantto
deoxyribonuclease
is plotted, as well as theproduction
of matureprogeny phage. It is apparent from this graph
that the SP and 3H become resistant to
deoxy-ribonuclease at almost identicalrates; thisindi-catesthat the
parental
andprogeny DNAformedpriortomaturation become enclosed inthephage
L-l headatthesamerate.
L.2
Computersimulation
ofrecombination
and theL*3
Lz15 FIG. 6. Predicted size distributions of DNA
frag-mentsresultingfromrandom breaks. The graphs
pre-sentedin thisfigure were produced by the methodof
Litwin,Shahn,andKozinski (7).The graphsrepresent
computer-simulated sedimentation patternsoffragments
of long molecules. Thefragments were produced by
0 cuttingdifferent-length "molecules" (L= phage
equiv-alentunitsoflength) withvariousaverage nwnbersof
12/D TOP cutsperphageequivalentunitofDNA (X = cuts per
phage equivalentunitofDNA).
A
=
0.5
Ls2
Lw2
A=2.0
2
Botom
on November 11, 2019 by guest
http://jvi.asm.org/
[image:8.497.236.440.69.339.2]MILLER, KOZINSKI, AND LITWIN
20-c
3ap
I-10
-0
.S 1.0 0 .5 1.0
LGBA D
20
LL_ 0
Z 10
0~
0 .5 0 .5
-i--BOTTOM
FRACTION OF THE LENGTH OF THE
[image:9.497.258.457.46.315.2]GRADIENT
FIG. 8. Sedimentation of long single strands after
heatingat95 CoraftertreatmentwithendonucleaseI.
Long single strands labeled with parental 32p were
isolatedfrom thepad ofanalkalinesucrose gradient,
supplemented with 3H integral DNA, and dialyzed
against0.07MTrisbuffer (pH 7.6).This material was
treated in thefollowing manner. (A) Part was
reana-lyzedinanalkalinesucrosegradientunderlayeredwith
apad. (B)Partwasheatedto95Cfor2minin standard
saline citrate containing 1% formaldehyde and
ana-lyzedinasucrosegradientcontaining1%formaldehyde.
(CandD)Partwasincubated with endonuclease Iand
split into twoportions; one (D) was analyzed on an
alkaline sucrose gradient and one (C) was
supple-mented with a 10-foldincrease of "2P-labeled,
single-strandedreferenceDNA.
production oflong molecules. Evidencepresented
in this paper supports thehypothesis previously
expressed (6) that recombination between
frag-mentsof DNAmayleadtotheformation of long
molecules and ultimately to the production of
circularly permuted, mature DNA molecules. It
remainstobeproven,however, howfeasiblethis
mechanismis,froma stochasticpoint of view.
Thisquestioncanbeapproached by the method
cn
4-c
0 0
30
a
a-20
0
I-c
a)
0
0 101
I..
C
0~
-10 aX
C
-8 0
80
16 t, a}
C
'4
12 CL
Minutes after Infection
FIG. 9. Maturation ofprogeny ('H) andparental
(32p) phageDNA. A cultureofE. coliB23wasgrown
to 3 X 108 cells/ml in high-phosphate TCG medium
and was infected with "2P-labeled T4BO,r
bacterio-phage. 3H-thymidine was pulsed from 11 min after
infectionito12min,and thenwasfollowedbya"chase"
as described in the text. At intervals thereafter,
in-fectedcellswerechilled andlysed bythe LTLprocedure.
The lysates were treated with pancreatic
deoxyribo-nuclease, andaportionwasassayed fortrichloroacetic
acid-precipitable material.
of computer simulation. The T4 DNA molecule
can be represented by a series of numbers
(2 X 105) arranged in a circularly permuted
manner and endowed with a 5% terminal
re-dundancy (7). Two basic hypothetical modes of
recombination can be considered: (model 1, cut
and strip model) one which would demand
double-strand cutting and enzymatic removal of
opposite strands, resulting, in effect, in the two
original neighbors being unabletorecombine with
each other [a model similartothat postulated by
Thomas(11)]; and (model2,diagonalcutmodel)
onewhichwillgeneraterecombining fragmentsby
a diagonal cut between two nicks located in the
trans position. Nicks or cuts can be introduced
intothesimulatedmolecules byarandom-number
generator by a procedure previously described
(7).Theprograminvolved in simulating
recombi-nation ina simulated cell is as follows. A
popu-lation ofsimulated bacteria are infected with a
poissonly distributed MOI of12 phages, 2 light
and 10 heavy. (The difference in the density in
376 J. VIROL.
on November 11, 2019 by guest
http://jvi.asm.org/
[image:9.497.58.252.58.393.2]thecontext of this paperplaysno role, butwillbe
used foranalysis ofdensity classes of simulated
recombinants presented elsewhere.) Each of the
infecting phage DNA receives a randomly
dis-tributed number of nicks or cutsassigned bythe
random-number generator. After completion of
thisstep, theprogram calls forseparationof each
infecting molecule intosubunits eitherby (model
1) introduction ofa cut on the opposite strand, which will be followed by the removal of 400
"nucleotides" on twoopposite strands (let'ssay, fromsimulated 5' end ofstrands) or(model2) by
performing a diagonal cut between two closest
translocated nicks. The resulting fragments are
characterized
according
totheirsize, number,andsequence of numbers within the open
comple-mentary area, a sequence, which being unique,
offers no ambiguity. The
listing
ofthe resulting subunitsand their terminalcomposition isstoredin thememory ofthemachine. Thenext stepisa
simulated recombination which proceeds as
follows. Thecomputerretrievesatrandomoneof thefragmentsand matchesit
against
arandomly retrieved additionalfragment out ofthepool offragments in the simulated bacterium. Ifthereis
no homology between the two fragments ofat
least 12bases, thesecond
fragment
istransferredbacktothepool.Themachineproceedsto
pick
atrandom a third subunit, checking it for possible homology. If
homology
isindeedfound,
adimer(recombinant)
is formed which is returnedtothepool in which further random
matching
opera-tions are performed until all such possibilities
have been exhausted. After
completion
of therecombinational process, recombinant molecules
are characterized
according
to theirlengths,
densities, possible
unmatched areas, and sizes of gapsand"whiskers."The results ofthesecharac-terizations are stored in the memory of the
machine. After
completion
of thismatching
processwithinthe
bacterium,
thecomputerkeeps
tallyof the
resulting population
offragments
andrecombinants and proceeds to evaluate another
bacterium in a similar manner. The simulation usually involves
103
to 10 simulated bacteria. Uponcompletion
oftheexperiment,
the resultsare retrieved from the memory of the machine
and canbe
represented
inavariety
ofways. Themachinemaybe instructedtowithdrawthe
mole-cules fromonebacterium and drawthemolecular
configurations
of the recombinants(Fig. 10).
Alternatively, the machine may be instructed tosubdivide the
resulting fragments
in the entireexperiment
according
to their molecular lengthsand draw the results as
simulated,
sucrosegradient distribution
(7).
In this case, the
dispersion
of r = 0.0323 hasbeen allowed asthis
corresponds
to the bestob-served dispersion of intact molecules in sucrose gradient in our laboratory. The simulation of both
models ofrecombination reveals that whereas the
model invoking cutting and denuding the ends, "cut and strip model" proposed by Thomas (11), is not compatible with the formation of long molecules and, for that matter, produces
recombi-nants of short size, the model ofdiagnonal cut
produces a sizeable proportion of recombinant
molecules which are larger than size one. Signifi-cantly, the ends of the participating molecules
were not allowed to be engaged in
recombina-tional events, i.e., to be partially denuded. Even
without this option, the distributionof observed
sizes of recombinants, for a limited number of
nicks and limitednumber ofinfectingmolecules,
resembles pretty well the distributions observed
in an in vivo experiment.
Wedo want toemphasizethatcomputer
simu-lation proves the stohastical feasibility of the
diagonal cut-typerecombination as a mechanism
generating long molecules. Scrutiny of the draw-ing represented in Fig. 10 shows that recombinant
molecules obtained by this method contain
nu-merous single-stranded gaps and "whiskers."
Those could quite feasibily be filled or excised,
respectively, by the proper enzymes. Indeed,
electron microscopy of recombinant molecules
isolated from CsCl reveals a sizeable amount of
"whiskers" and branches (3).
DISCUSSION
T4bacteriophage DNA iscircularly permuted
and terminally redundant (8, 12). Circular
per-mutation could be achieved by a number of
different mechanisms. One mechanism could
in-volve theformation of acircularDNA molecule
during thereplication process. A second
mecha-nismcouldinvolve alongmolecule of intracellular
phage DNA which would becut into units ap-proximately one phageequivalent unit in length
during the maturation process. Thelongmolecule
could be created either through replication of
progeny DNA or through recombination.
Ob-servation of
fast-sedimenting
DNAinanalkalinesucrose gradient indicated that long molecules
mayindeed exist atlatertimes after infection.
Experiments were undertaken to demonstrate that this
fast-sedimenting
DNA was actually composedofsinglestrandslongerthanonephage equivalent unit. Because neutralsucrosegradient analysis had been shown to be an unreliableindicator of molecularweightofreplicativeDNA
(6), it was feared that analysis atpH 12.5might
also be unreliable. For
example,
itwasconceivablethat DNAsedimentedfast in thesucrosegradient
atpH12.5notbecause of itslengthbut because of
its association with some
lysozyme-resistant,
on November 11, 2019 by guest
http://jvi.asm.org/
MILLER, KOZINSKI; AND LITWIN
I
I.0EL
.4.0k %,_312 -5112
i.\ I9"9b .6633
4
5
6 SI
7
13 W
14 -\_._ :_ \
15
16
[image:11.497.62.451.61.382.2]N
FIG. 10. Computerdrawing of the contentsofa "single cell" infectedwith 10 heavy (thick line) and2light
(thin line) phage. After completion ofthe recombinationprocess, performed according to the "diagonal cut"
model, 16fragmentsresulted. Thosearedrawn bythe computeras twoparallel linesrepresentinga bistranded
structure. Note thediversity of length (some sizably largerthan onephage equivalent unit) andthe presence of
single-strandedgapsand"whiskers." Note also that thediagonalcutmodel allows reunionoftwooriginally
neigh-boringfragments,areunionwhich, exceptforanick (thinslashacrossthesingle strand),isperfect,notendowed
withagap or"whisker,"andshould berepairablein vivoby ligase without the involvementofa "filling"or
ex-cising enzyme. Fragment #2 illustrates thefinedetails ofthematchingprocedureperformed by thecomputer,
namelyby displaying thebase numbers initiating andending each ofthe constituents oftherecombinant. The
drawings wereperformed bythecomputerforarandomlychosen "bacterium."
alkali-resistant material, or because individual
single strands in the pool of replicative DNA
could not, for any number ofreasons, be
dis-associated. Two experiments presented here
argue in favor of the hypothesis that the
fast-sedimentingDNAwasactuallycomposedoflong
single strands. First, the DNA still sedimented
"fast" after reisolation and heating to 95 C.
Second,andmoreimportant,theapparentlylong
singlestrands werecut tothe sizefragments
pre-dictedbycomputer analysisfor linear molecules
exposedtoalow concentration of endonuclease I
(aconcentration which gaveonetotwo cutsper
phage equivalent unit oflength). Onthe basis of
any of the proposed artefacts, a residual,
fast-sedimenting core of this DNA moiety should
have remained after only one to two cuts per
phage equivalentunit ofsingle strandedDNA.
Having supportedthe contentionthat the
fast-sedimenting DNA, atpH 12.5, was indeed long
singlestrands, the nextquestioninvestigatedwas
whether the long single strands were produced
bya mechanism ofreplication orby
recombina-tion. A computer simulation had in fact shown
that long molecules could be produced by a
mechanism of random breakage and reunion.
Thisshould be followed by efficient repairof the
single-strand intersections in the polynucleotide
chain of the recombinant molecules.
If the long singlestrandswerebeing produced
asa result ofrecombination, one should expect,
first ofall, to seethe appearance oflong single
378
.02*'
.s18" .95r1.50t.05'3
3
J. VIROL.
8
9
10
II z
12 ' _ _
/'
-- 2 i! 56
ALS).94r .112.% -N.-.-,
a- x
A
N
.01
.@" .S453VW .-YN.25
,/
-.Z
".7
on November 11, 2019 by guest
http://jvi.asm.org/
DIAGONAL CUTMODEL
SI. t.00 6.00 0.60o .es LENGTH (PEU) a
DIAGeNAL CUT MODEL SIGMA = 0.0323
CUTANDSTRIP MODEL SIGMA= 0.0323 DIAGONALCUT MODEL SIGMA =0.0323
Ws S _8-NICKS
D2/0l '?oo I00D2/D o
I~~~~~~~~~~~~~~~~
FIG. 1 1. Simulatedsucrose gradient distributions ofthleproducts of recombination in bacteria infected with 12 phages (no replication allowed). The infecting phages were distributed poissonly and received 4, 6, or 8 nicks (also distributed poissonly). For 6-nicks class, two models ofrecombination were simulated, diagonal cut model and cut and strip model. ineach panel, a broken sigmoid curve represents the summed percentage of recovery (integral graph) ofunnicked, integral DNA. Notethiatfor any given number of nicks in the diagonal cut model, recombina-tion generates a sizeable fracrecombina-tion of molecules larger than size 1.This can be appreciated while comparing integral graph of per cent recovery of recombinants (solid, continuous sigmoid line) with that of reference DNA (broken line). The cut-and-strip model is notcompetent for production of long molecules, andlrecombinants actually tend to peak at approximately 0.4 phage equivalent unit length (PEU).
strands coincidentallywith theonsetof
recombi-nation, and, second, theaddition of CM at the
propertime would inhibittheformation oflong
single strands while not affecting replication of
DNA. Addition of CM around5min after
infec-tionwasknowntoinhibit recombination without
inhibiting replication (6).Infact,addition of CM
at 5 min inhibited theproduction oflong single
strands, as predicted, even though 45 phage
equivalent unitsof DNAweresynthesizedinthis
particular experiment. (Themaximumfigure,not
reported here,canbeashighas100phage
equiva-lentunits.)
Many of the experiments documented in this
paperhave beenperformedwith aparentallabel;
it should be emphasized here that most of the
results havebeenconfirmedalsoforprogeny DNA
labeledby uptake of 3H-thymidine or32p. The
in-hibition of long single strands by CM without
inhibiting DNA replication is important when
considering another unlikely but possible
ex-planationof theprevious results. Iftherewere20
hypothetical"sites" forreplicationinthecell(as
might be inferred from the fact that 20 phage
equivalent units ofDNA are synthesized before
anylong singlestrandsareformed), and ifallthe
sites hadto beoccupied bya phageDNA
mole-culebeforeanylong singlestrandswereproduced,
then it wouldnotbesurprisingthatnolong single
strandswereproduced duringthe first rounds of
replication. However,sincelong singlestrandsare
not produced when recombination is inhibited
even though 50to 100 phage equivalent units of
DNAaresynthesized, thishypothesis isvery
un-likely.
Theabsenceoflong singlestrandsproduced by
phage-infecting bacteria in 5-BUdR medium
argues against the hypothesis that long single
strands are an obligatory intermediate of DNA
replication.
I NEGRAL DNA
S 4-NICKS I;
A6s.60 1.20 1.00 0.00 0.60 0.6 0.20 -'?bo
D2/01
I
on November 11, 2019 by guest
http://jvi.asm.org/
[image:12.497.46.449.67.350.2]MILLER, KOZINSKI, AND LITWIN
Several results presented here and elsewhere
bear directly on various published models of
DNA replication. In any model explaining the
replication of T4 bacteriophage DNA, the
fol-lowing experimental results should be taken into consideration. A pulse of 3H-thymidine
incor-porated in the initialstages ofreplication is not
covalently bonded to the parental DNA; i.e.,
parental 32p and progeny 3H labels separate in
an alkalinesucrose gradient and arerepresented
by both T4 bands (4). An interesting
permuta-tion of this resultisthatapulseof 5H-thymidine
from 3to6.25min after infection is incorporated
intoa piece ofprogenyDNA the samesizeasa
piece labeled from 6to 6.25 min;the size of this
piece is about that of the fragment Kozinski called FSBP (3). The size of the fragment labeled
from 3 to 3.25 min, however, is much smaller.
None of this pulse-labeled progeny DNA was
covalently attached to parental DNA. It should
be noted that these results were obtained by
infection with a 32P-labeled parental phage, so
the integrity of the parental DNA was always
assured in theseexperiments; i.e., the size of the
progeny pieces could not have been caused by
some random nicking by an enzyme such as
endonucleaseI,norcouldthe bulk oftheprogeny
DNA have been separated from parental
ma-terialbysome random-nicking process.
Any model explaining the replication of T4
bacteriophage DNA should be consistent with
theresult that there isnopreferential maturation
of parental DNA into progeny phage. This
proposal is basedonthe result that the increase
in percentage of labeled DNA which becomes
resistant to deoxyribonuclease as maturation
proceeds is the same whether the labeled DNA
is of parental or progeny origin; the progeny
DNA was labeled from 11 to 12 min after
in-fection with a pulse of 3H-thymidine. This
ex-periment discriminates againstamodelbasedon a
continuously elongating concatenate ofprogeny
DNA. This model predicts that when
matura-tionproceeds from the end, 50% of theparental
label should be encapsulated immediately at the
beginning of the maturation process. Even if
parental label were recombined away from the
end of theelongatingconcatenate, someresidual
label would mature preferentially. Predictions
of the rolling circle model (2) of DNA
replica-tion do not agree with this result either; no
sanctuaries such asthe core ofthe rolling circle
or the end attached at a membrane can be
oc-cupied by parental label. In addition, thereare,
apart from physical-chemical controversies,
genetic phenomena which are not included in
the model of the continuously elongating
con-catenate, such as unquestionable fact of
recom-binationand the phenomenon ofclonal
distribu-tion of mutants or recombinants. The best
hypothesisfitting the data is that recombination
distributes parental label randomly through the
replicative pool, so all replicative DNA has an
equal chance of being encapsulated. Thus the
mechanism generating long molecules is a
mo-lecular recombination, a phenomenon
ulti-mately determining circular permutation in T4
DNA.
ACKNOWLEDGMENTS
Thisinvestigationwassupported by NationalScience Founda-tion grant GB 13048 and by Public HealthService grantCA 10055 from the National Cancer Institute. Thecomputer evalua-tion of data was performedin the University of Pennsylvania Medical School Computer Center, supported by Public Health ServicegrantFR15-06.
Oneof the authors (R.C.M.)wassupported by Public Health ServicegrantGM-006-94-09 awardedtotheGraduate Groupon
Molecular Biology, University of Pennsylvania.
LITERATURE CITED
1. Frankel,F. F. 1968. Evidence forlongDNAstrands in the replicating poolafter T4infection. Proc. Nat. Acad. Sci. U.S.A. 59:131-138.
2. Gilbert, W., and D. Dressler, 1968. DNA replication: the rolling circle model. Cold Spring Harbor Symp. Quant. Biol. 33:473-482.
3. Kozinski,A. W. 1968. Molecularrecombinationinthe ligase-negative T4 amber mutant. Cold Spring Harbor Symp. Quant. Biol. 33:375-391.
4. Kozinski, A. W. 1969. Unbiasedparticipation ofT4phage DNA strands in replication. Biochem. Biophys. Res. Commun. 35:294-299.
5. Kozinski, A. W., and P. B. Kozinski, 1963. Fragmentary transfer of82P-labeled parentalDNA toprogenyphage. II. The average size of the transferred parental fragment. Two-cycle transfer. Repair of the polynucleotide chain after fragmentation. Virology 20:213-229.
6. Kozinski,A.W.,P.B.Kozinski,and R.James.1967. Molecu-lar recombination in T4 bacteriophage deoxyribonucleic acid:I.Tertiarystructureofearly replicativeand
recom-bining deoxyribonucleicacid.J. Virol. 1:758-770. 7.Litwin, S. E.,S.Shahn,andA.W.Kozinski, 1969.
Interpreta-tion ofsucrose gradient sedimentationpattern of DNA fragmentsresultingfrom randombreaks. J. Virol.4:24-30. 8. MacHattie, L.A.,D. A.Ritchie,and C. A.Thomas. 1967. Terminalrepetition inpermutedT2 bacteriophage DNA molecules. J. Mol. Biol. 23:355-363.
9. Schmidt, G., B. Hershman, and S. J. Tannhauser. 1945. The isolation oflambda-glyceryl-phosphorylcholine from incubated beefpancreas; its significance for the inter-mediary metabolismof lecithin. J. Biol.Chem. 161:523. 10. Shahn, E., and A.W. Kozinski. 1966.Fragmentarytransfer
of32P-labeled parental DNA toprogeny phage. III. In-sertion ofa single parental fragmrnt to the progeny
molecule.Virology30:455-470.
11.Thomas, C. A. 1966. Recombination of DNA molecules. Progr. Nucl. AcidRes.Mol. Biol. 5:315-335.
12. Thomas, C. A.,andI.Rubenstein. 1964. The arrangements
of nucleotidesequencesinT2 and T5bacteriophageDNA molecules. Biophys.J. 4:93-106.