Copyright 0 1974 AmericanSocietyforMicrobiology Printed inU.S.A.
Molecular
Weight Determination
of
Sendai and Newcastle
Disease Virus RNA
DANIEL KOLAKOFSKY, EDWARD BOY DE iATOUR, AND HAJO DELIUS
Departement de Biologie Moleculaire, Universite deGeneve, Geneve, Switzerland,and ColdSpring Harbor Laboratory, Cold Spring Harbor, New York 11724
Received forpublication21September 1973
The molecular weights of Sendai and Newcastle disease virus RNA were
estimatedby sedimentation insucrosegradients andby lengthmeasurementsin
the electron microscope under both denaturing and nondenaturing conditions.
Sedimentation analyses under denaturing conditions yielded molecular weight
estimates of 2.3 x 106 to 2.6 x 106, whereas length measurements yielded
estimates of 5.2 x 106 to5.6 x 106for both denatured and nondenatured viral
RNA. It would appearthat theconditions ofdenaturation used (99% dimethyl
sulfoxide at26 C, and reaction with 1.1 M formaldehyde for 10 minat60C) do
notequally denature parainfluenza virus RNA andother RNAs, suchascellular
rRNA, 45S rRNA precursor, and R17 RNA.
The molecular
weight
of Sendai RNA esti-matedby
sedimentation in sucrose gradients containing 99%dimethyl
sulfoxide (DMSO)was found to be approximately half of that estimated undernondenaturing conditions and
accountsforonly half the RNA estimatedtobe
present in the viral nucleocapsid (13). Three
possible explanations of this difference have been investigated.
(i) The Sendai viralgenome, like thatofthe
RNAtumorviruses, iscomposed ofacomplex of
more than oneRNA chain which disaggregates
upondenaturation (8, 10, 14).
(ii) The apparentmolecular
weight
obtained by sedimentation analysis undernondenaturing
conditions isartificially high duetothesecond-ary structure of
Sendai
RNA, and the estimateobtained under
denaturing
conditions is morereliable. In this case, the viral nucleocapsid would contain two separate RNA chains of similarlength, thoughnotnecessarily of
identi-calsequence.
(iii) Conversely, sedimentation analysis in 99% DMSO is unreliable due to different
de-grees ofdenaturation of Sendai and the marker
RNAs we have used for
calibration,
and thevalue obtainedundernondenaturing conditions
istherefore morereliable. Inthiscase,the viral nucleocapsid would contain one continuous RNA chain ofapproximately 5 x 10'daltons.
By usingmixing experiments in 99%
DMSO,
sedimentationanalysis
of formaldehyde-reacted RNAs in sucrosegradients
with and without 1.1 M formaldehyde,polyacrylamide
gel electrophoresis offormaldehyde-reacted
RNAs, and
length
measurements of DMSO-treated,formaldehyde-treated,and heatedSen-dai and Newcastle disease virus
(NDV)
RNA inthe electron microscope, weconclude thatonly the third
possibility
is correct; that is, Sendai RNA is asingle,
continuous RNA chain ofap-proximately5 x 106daltons.
MATERIALS ANDMETHODS
Preparation of32P-48SSendai RNA.
Three-day-old confluent cultures of MDBK cells (obtainedfrom
P. Choppin) in 75-cm2 Falcon tissue culture flasks
wereinfected with10 meanegginfectivedoses per cell
ofSendaivirus (Harris strain).After45minat30C,
theinfectingmediumwasremoved andreplacedwith 8ml of mediumcomposedof 9 parts ofphosphate-free
medium 199 (Wellcome Laboratories) and 1 part of DulbeccomodifiedEagle mediumcontaining50
ACi
of carrier-free 32po4 perml. At 18h post-infection, the medium was removed (and discarded) and replacedwith 8 ml of the above medium containing 10uCi of
32PO4 perml.At36hpostinfection, the mediumwas
removed andcentrifuged for10minat 5,000 x gto remove cellular debris, and the virus was pelleted throughacushion of25%glycerolinTNE(13) for2h at 36,000 rpmin theInternational A-170 rotor (8 C). The virus pellet was dissolved in TNE containing 0.5% sodium dodecyl sulfate (SDS) and 0.2 mg of Proteinase K (Merck) per ml at an approximate concentration of2mg/mlandincubated for15minat
25C, and
150-Aliter
amounts were then centrifugedon5 to 23% sucrosegradientsaspreviouslydescribed (13). Fractions containing the 46 to50S RNA were pooled, recovered byethanol precipitation, and dis-solved in 1 mM sodium acetate (pH 5.3), 1 mM EDTA, and0.05%SDS(RNAbuffer).RNAprepared
as above had a specific activity of approximately
261
on November 10, 2019 by guest
http://jvi.asm.org/
400,000 counts per min per lAgon isolation.
Fragmentation of 48S Sendai RNA. A 0.4-ml amount of 0.2 M sodium carbonate was added to 1.2 ml of water containing 0.65 mg ofSendai 48S RNA (both solutions having been prewarmed to 50 C), and the solution was incubated for 15 min at 50C. After addition ofsodium acetate(pH 5.3)toafinal concen-tration of 0.2 M, the RNA was precipitated with 2 volumes ofethanol, recoveredbycentrifugation, dis-solved in 0.3 ml of RNA buffer, and sedimented on two 5 to 23% sucrose gradients (a third gradient contained "C-labeled cell RNA markers) for 3 h at 52,000 rpm(80)intheSW56 rotor (13). By monitoring absorbance at 260 nm, the RNA was found to sedi-ment as abroad band (3 to 27S)with apeak at 16S. The RNA was divided into 3 to 12S and 12 to 27S portions, and the 12 to 27S RNA was recovered by ethanol precipitation and again treated withsodium carbonate as aboveforafurther 7 minat50C. Upon centrifugation in sucrose gradients, the RNA was found to sediment as a broad band (3 to20S)with a peak at10S. The 3 to 12S RNA from both gradients was pooled, precipitated with 2 volumes of ethanol, recovered by centrifugation, and dissolved in RNA buffer. Sendai RNA fragments prepared as above represented approximately 65% ofthe starting RNA. Polyacrylamide gel electrophoresis. Gels of 2.2% acrylamide, 0.11% bis-methylene acrylamide, and 0.6%agarose werepolymerized in 9-cm (0.5-cm inter-nal diameter) quartztubes in a buffer containing 40 mM triethanolamine (pH 7.8), 20 mM sodium ace-tate,2mMEDTA,and2.5%glycerol. Electrophoresis
was carried out for 2 h at 10 V/cm and 4 C (until bromophenol blue marker dye had just run off the gel) in the abovebuffer plus0.2%SDS.Afterscanning at 260 nm in aGilford spectrophotometer, the gels were frozenand cut into 2-mm slices.Slices were incubated with 0.4 ml of 0.5%SDS for 6 h at 50 C and were then countedby liquidscintillation after the addition of3.6 ml ofAquasol (New England Nuclear Corp.).
Other methods and materials. 9H-labeled 48S Sendai RNA, non-radioactive Sendai and NDV 48S
RNA, and "4C-labeled RNA from uninfected mouse kidney cells were prepared as previously described (13).Formaldehydetreatment ofRNA andits centrif-ugation insucrosegradients containing1.1M formal-dehyde in SPB buffer wereperformed essentially as described byBoedtker (5).
RESULTS
Does Sendai RNA disaggregate upon
denaturation? Aspreviously shown (13),
Sen-dai RNA which sediments at 48S in ordinary
sucrose gradients sediments at 32SDMSO in
su-crosegradients containing 99% DMSO (relative
to 18 and 28SrRNA markers arbitrarily set at
18 and
28SDMSO).
Calibration of these DMSOgradientswith18SrRNA, R17 RNA,28S rRNA,
and 45S rRNA precursor yielded an estimated
molecular weight of2.3 x 106 for Sendai RNA
under these conditions, whereas calibration of ourordinary sucrose gradients with 18 and28S
rRNA yielded a molecular weight estimate of
5.0 x 106. Anestimationbasedonthe chemical
composition and lengthofthe viral
nucleocap-sid relative to tobacco mosaic virus yielded a
value of 5.0 x 106to5.6 x 106 daltons of RNA
pernucleocapsid. The possibility that the
Sen-dai genome like that of avian myeloblastosis
virus(AMV)wassegmented (8, 10, 14) and that
the approximate 5.0 x 106 daltonsofRNAthat
each Sendai nucleocapsid contained was
pres-ent as anoncovalentcomplexof morethanone
RNA chainwasalso examined. RNA which had
sedimented at 32SDMSO wasrecovered from the
DMSO gradient by ethanol precipitation and
was then centrifuged on an ordinary sucrose
gradient with untreated Sendai RNA. The
re-sults of this experiment (13) showed that, in
contrast to AMV RNA, the DMSO-treated
Sendai RNAwas indistinguishable in its sedi-mentation properties from untreated Sendai
RNA.This suggested that, if Sendai RNAwas
disaggregatingin99%DMSO, it wascapable of
rapid and quantitative reassociation upon
re-moval of the RNA from DMSO (presumably
during its concentration
by
ethanol precipita-tionand solutioninwater). Totestthispossibil-ity further, the following experiments were carried out.
(i) Since briefperiods ofheating have been
shown to disaggregate AMV RNA (8, 10, 14),
32P-48S
Sendai RNA in 1 mM EDTA, pH 7.0(pretreated by passage through a column of
Chelex-100) was heated in
boiling
water for 2 min, quick cooled, and then sedimented inordinary sucrose gradients. Although some
(minimal) thermal degradation did take place, the sedimentationrateofthe bulk of theSendai RNA was unaltered (data not shown). There-fore, if the Sendai genome were segmented, heating for 2 min at 98C was not sufficient to
effect thedisaggregation.
(ii) To avoidstepswhich tendtoconcentrate
RNA and therefore enhance its
possible
reas-sociation,
32P-48S
Sendai RNA was placed in99% DMSO at a concentration of 1
ug/ml,
diluted 10-fold with water, and directly
cen-trifuged in ordinary sucrose gradients. Sendai
RNA so treated was found to sediment
unal-tered as a homogenous band at 48S (datanot
shown). Although unlikely, thepossibility
can-notbeexcluded thatSendai RNA, even atsuch
low concentrations (0.1
,ug/ml),
was capableofreassociation.
(iii)
If 48S Sendai RNA is composed of acomplexof twoRNA chains whichdisaggregate
in99% DMSOto form two32SRNAs and then
reaggregate upon removal of the RNA from the
denaturing solvent, mixing of the 32S RNAs
should take place during this treatment.
Fur-thermore, if while disaggregated, RNA frag-262
on November 10, 2019 by guest
http://jvi.asm.org/
mentsderivedfrom48S SendaiRNA are added
in excess, 32S Sendai RNA should
preferen-tially reassociate with these fragments rather
than with themselves to form a complex which
would sediment between 32 and 48S, depending onthe size of the fragment. Non-radioactive 48S
Sendai
RNAwas therefore fragmented by lim-ited base hydrolysis to a size of approximately20,000 to 350,000 daltons (3 to 12S) (see
Materials and Methods). 32P-48S Sendai RNA
wasfirstsedimented in a99% DMSO gradient, and the four peak fractions sedimenting at
approximately
33SDMSO
(relative to 'IC-labeledcell RNA markers sedimented in a separate
tube) were pooled. Two samples were then
mixed with a 200- and a 400-fold excess by weight of Sendai RNA fragments (in 90%
DMSO), respectively, while a third sample
receivednoaddition. Afterheatingfor 5 min at 37
C,
the RNA wasrecovered from theDMSO solution by ethanol precipitation and dissolvedin water, and
"C-labeled
cell RNA was addedas sedimentation markers and resedimented in
ordinary sucrose gradients. The results(Fig. 1)
demonstrate that even a 400-fold excess of
fragments does notsignificantly affect the
sedi-mentation properties of the 32P-Sendai RNA.
This suggests that 48S Sendai RNA does not
disaggregate
into more thanone RNAchain in99%
DMSO,
butrepresents asingle,continuousRNA chain.
Thereis, however, apossible objectiontothe interpretation of this last experiment. The above model assumes that very limited
se-quences of RNA in a much larger RNA are
involved in holding RNA chains together. It is
possible that,
forthe association totake place,the sequences involved must exist in a given
structure whichis somaintainedby therestof
the RNA chain and that fragmentation ofthe RNA
destroys
thisstructure.If thiswereso,the non-radioactive fragments added in the above experimentmightnotcompete inthe reassocia-tion of 32P-Sendai RNA even at a 400-foldexcess of RNA chains ranging in size from
20,000to350,000 daltons. Although each of the above experiments does not unequivocally
ex-clude the
possibility
that 48S Sendai RNAdisaggregates
upondenaturation,
takento-gether they strongly suggest that 48S Sendai
RNA represents a single, continuous RNA
chain.
Sedimentation of Sendai RNA under
con-ditions offormaldehydedenaturation. In view of the different sedimentation properties of
Sendai RNA relative to rRNAs in water and
99% DMSO, we next examined the possibility
that the sedimentation value obtained in 99%
DMSO waspeculiartotheuseof the
nonaque-8
---(c)
6
-4-
12I,
I0
20 30E
8-
(b)
a-6 6 6g/\ fragments
10 20 30
48
-(a)
a-FIG.
1.6
Eff
\ no fragments4 / \
0
~~~0
20 30FRACTION NUMBER
FIG. 1. Effect ofSendai RNAfragmentson renatu-ration of Sendai RNA from 99%o DMSO. "P-48S
Sendai RNA (0.16 Mg, 24,000 counts/min) and
14C-labeled cell RNA markers were sedimentedseparately
in 99%o DMSO-sucrose gradients as previously
de-scribed(13). The four peakfractionsof the 32P-Sendai
RNA, which sedimented at approximately 33SDMSo
(13), were pooled and divided into three 170-jiliter
samples containing 0.034MUgof32P-Sendai RNA (5,200
counts/mmn)each.Non-radioactive SendaiRNA
frag-ments (1.36 mg/mlin 90%DMSO;seeMethods and
Materials) were added to two of the samples as
indicated (6.8,ugof fragments,representinga200-fold
excessby weight, panelb,and13.6Mgof fragments,a
400-foldexcess,panel c),and the RNAwasrecovered
from theDMSOsolutionbyethanolprecipitationand dissolved in0.1mlof RNA buffer. Marker"4C-labeled
cell RNAwas addedtoeachsample, whichwasthen sedimented in 5 to 23% sucrosegradientsaspreviously described (13).
ous solvent. Reaction of RNA with aqueous
formaldehyde (HCHO)
is also known todena-ture RNA (4) and, althoughformaldehyde does
notdestroysingle-stranded stacking (15) and is
therefore not as strong a
denaturing
agent forRNA as DMSO, Boedtker has shown that this
method destroys secondary structure
suffi-cientlytoallow the resolution of RNAsby chain length (5, 6). 3H-labeled Sendai RNA and
"4C-labeled
cell RNAwere therefore heated(10
min at 60C) inthe presence of1.1M
formalde-hyde and
centrifuged
inaqueous sucrosegradi-263
on November 10, 2019 by guest
http://jvi.asm.org/
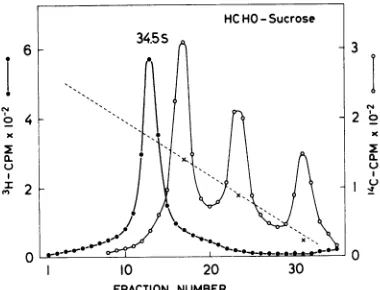
[image:3.493.248.440.70.351.2]ents containing 1.1 M formaldehyde. The
re-sults (Fig. 2) show that Sendai RNA sediments
as ahomogeneous band at 34.5SHCHO relative to
18 and 28S rRNA markers, a result similar to
that found with sucrose gradients containing 99%DMSO. By using Escherichia coli4S, 16S, and
23S
RNA as calibration markers,Sendai
RNA isfoundtosedimentat37SHCHO
(datanotshown). Since reaction with formaldehyde is
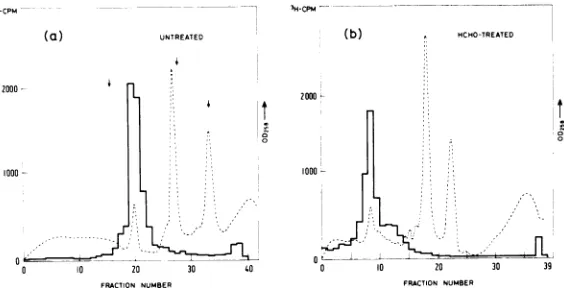
onlyslowlyreversible (4), the absence of formal-dehyde inthesucrosegradient doesnot signifi-cantly affect the result. Figure 3 shows the result of asimilar experiment in which
dupli-cate samples of 3H-labeled Sendai RNA and
"4C-labeled
cell RNA were both placed in 1 Mformaldehyde,
but only one sample (panel b)was heated in
formaldehyde
and thencen-trifuged
on sucrosegradients
not containing formaldehyde. Formaldehyde-treated (i.e., heated in formaldehyde) rRNA sediments onlyslightly
slower thanuntreated rRNA (comparepanels a andb), whereas formaldehyde-treated
Sendai
RNA sediments considerably moreslowly then untreatedSendai RNA. Calibration of these
gradients
withmouse 18and28S rRNA and E. coli 16 and23S
rRNAyield molecular weight estimates of2.6 x 106 to 2.8 x 106forSendai
RNA. The difference in sedimentation properties ofSendai RNA under denaturingand nondenaturing conditions previously noted withDMSO
is therefore not peculiar to the use ofthis nonaqueous
solvent.
!-
4x
I
0
I 4
x
LI
a.
10 20 30
[image:4.493.265.451.77.377.2]FRACTION NUMBER
FIG. 2. Sedimentation of HCHO-treated Sendai RNA in sucrose gradients containing 1 M HCHO.
Elevenmicroliters of28% HCHO in SPBbufferwas
addedto3,500counts/min of 3H-labeled SendaiRNA and4,000counts/min of 14C-labeled cell RNA in 90
gliters
of SPB buffer, and the samplewasheated for10minat60C. Thesamplewasthencentrifugedon5 to23%sucrosegradientsinSPBbuffer containing1.1
MHCHO (5) for190minat50,000rpm (80) inthe
SW56rotor.Fractionswerecollectedandprocessedas
for ordinarysucrosegradients(13).
3
2
0.
_II 10 20 30
a (b) 36s HCHO-treated
2
0
_
10 20 30
FRACTION NUMBER
FIG. 3. EffectofHCHOtreatment onthe
sedimen-tation of Sendai RNA. Duplicate samples for
sedi-mentation analysis contained 3,500 counts/min of
3H-labeled 48S SendaiRNA,and 4,000counts/minof
"4C-labeledcell RNA in20
,gliters
ofSPBbuffer. Two microliters of28% HCHOinSPBwasaddedto eachsample. One sample (panel a) was kept in ice; the
othersample(panel b)washeatedfor10minat60C.
Thesampleswerethen diluted with80glitersofTNE
bufferandcentrifuged on 5 to 23%ordinarysucrose
gradients for 105 min at 50,000 rpm (7.5°) and processed aspreviously described(13).
Molecular weight determination of Sendai and NDV RNA
by
electron microscopy. Sincemolecular weight estimates of Sendai RNA
obtained by sedimentation analysis in sucrose
gradients varied so greatly depending on
whether the sedimentation was
carried
outunder denaturing or nondenaturing conditions
(2.3 x
106
to 2.8 x106
asopposedto 5.0 x 106daltons), and since this difference was
appar-ently not a result ofthe disaggregation ofthe
viral genome upon denaturation, this
discrep-ancy would appear tobethe result of amarked
difference in secondary structure of Sendai RNA relative to the prototype RNAs such as
cellular rRNAs used as calibration markers.
The sedimentation of RNAs in nondenaturing 264
6
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.493.63.253.415.560.2]MOLECULAR WEIGHT ESTIMATION
sucrose gradients at rates inconsistent with their molecular weights is well known. For example, R17 RNA (27.5S), which is barely distinguishable in sedimentation rate in
ordi-nary sucrose gradients from 28S rRNA,
sedi-ments at a rate
(21SDMSO)
consistent with itstrue molecular weight in gradients containing
99%DMSO (13).Itwould thereforeappearthat
themolecular weight estimate of2.3 x 106to2.8
x 106obtained from sedimentationunder
dena-turingconditions ismorereliable, and that the
estimate of 5.0 x 106 obtained under
non-denaturing conditions wasdue tothe factthat the structure of Sendai RNA, like R17 RNA,
was considerably more compact than cellular rRNA.
Totestthishypothesis, the molecular weight of Sendai RNA was estimated by electron
microscopy, amethod independent of
hydrody-namic properties. Electron microscopy
mea-surements werecarriedoutonboth Sendai and
NDV RNA, these parainfluenza RNAs being indistinguishable in sedimentationrate in
ordi-narysucrosegradients (3, 13),sucrosegradients
containing 99% DMSO (13), andsucrose
gradi-ents containing 1 M formaldehyde (data not
shown). By using T4 gene 32 protein toaid in
unfolding the RNAs and PM2 DNA as ah
internal calibration marker, both Sendai and
NDVRNAweremeasured at5.6 x 106 daltons
(Table 1). Moreover, neither heating the
par-ainfluenzaRNAs for 2 min inboilingwaternor
treating Sendai RNA with 99%
DMSO
signifi-cantly altered the lengthmeasurements ofthelongest RNAs (Table 1). An example of these lengthmeasurements is shownin Fig.4.
Electron microscopymeasurementswerealso
carried out on untreated and
formaldehyde-treated NDV RNA without the aid of T4gene32
protein, since itwas found not tobind well to
formaldehyde-treated RNAs in buffer
contain-ing 1.1 M formaldehyde. In these measure-ments, no internal DNA marker was used for
calibration, but R17 RNA, treated identically,
was measured on separate grids. The results
(Table 2) show thatformaldehyde-treated NDV
RNA is the same lengths as untreated NDV
RNA (within the error ofthe measurements),
yielding molecular weight estimates of 5.2 x
106. An example ofthese length measurements
is shown in Fig. 5.
Under all conditions oflength measurement
by electron microscopy, Sendai and NDV RNA, whether untreated, treated with formaldehyde, DMSO, or briefexposure to heat, and spread with and without the aidofT4gene32protein,
have been found to have molecular weights of 5.14 x 10. to 5.67 x 106. Since this molecular
weight estimate is considerably different from
that obtained from sedimentation analysis
under denaturing conditions (2.3 x 106to2.6 x
106), it would appear that Sendai and NDV
RNA do not sediment under these denaturing
conditionsat arateconsistent with their
molec-ularweight.
AreSendaiand rRNA equally denaturable? Since sedimentation under completely dena-turing conditions should provide a reliable
estimate of RNA chain length, we next
ex-amined the possibility that Sendai RNA, under the conditions of denaturation we have used, was not completely denatured but still
pos-sessed some structure which would cause it
tosedimentatarateslower than thatexpected from itsmolecular weight. Structures knownto
behaveaccordingly, suchasextendedregionsof
double strandedness which increase frictional
drag during sedimentation, would also be
ex-pected to decrease theelectrophoretic mobility
of RNA in polyacrylamide gels. However,since
electrophoretic mobility of RNAs in
polyacryl-amide gels isinversely proportionalto
molecu-lar weight, such structures would cause a
de-creaseintheelectrophoretic mobilityand hence
an overestimation of the molecular weight.
Figure 6 shows the results of an experiment
TABLE 1. Molecularweight determination by electron microscopy
RNAinpeak Meanlength Mean Daltons
Sample (AM) PM2length correctiona for (x
10.)
PM2=3.45
NDV 88 6.36 +0.30 3.39 + 0.12 6.47 5.63± 0.27
NDV heated 53 6.22 ± 0.30 3.50 ± 0.12 6.13 5.33 ± 0.26
Sendai 49 6.01±0.18 3.18±0.11 6.52 5.67±0.17
Sendai DMSO 50 6.36±0.38 3.58±0.11 6.12 5.32±0.32
Sendai heated 47 6.46±0.45 3.59±0.15 6.21 5.40 ± 0.39
aPM2DNA, addedas an internal standard,wasused tocorrectfordifferences in
magnification
under theassumption thatitshould havealengthof 3.45um(6.4 x10"daltons) when compared toT7DNA(25.2 x 10' daltons, 13.6 um), which has beenpreviously usedto determine the factor of 870,000 daltons ofRNA/um of RNA-protein complex (7).
on November 10, 2019 by guest
http://jvi.asm.org/
TABLE 2. Molecularweight determination by electron microscopy
RNA Mean length Daltonsa Sample inpeak
(JAm)
(x 106)UntreatedNDV 49 5.14±0.22 5.14±0.22 RNA
HCHO-treated 64 5.24i0.26 5.24±0.26
NDVRNA
_
_
aBacteriophage R17RNA, measured under
identi-calconditionsonseparate grids,wasfoundtohave a mean length of 1.2 gm whether or not it had been reacted with formaldehyde. Molecular weight esti-mates ofR17 RNA varyfrom1.05 x 106(11) to 1.3 x 106 (6). Assuming amolecular weight of 1.2 x 106, 1
,gmofRNAspreadby this techniqueisequivalentto 1.0 x 106daltons. For detailsofspreading technique, see legendtoFig. 5.
FIG. 4. Length measurements of NDV RNA by electron microscopy.NDV RNA wasreacted with T4 gene 32protein,fixedwithglutaraldehyde,andspread
aspreviously described (7). The graph at the top of
thefigure shows thefrequencydistributionoflengths
observed (abscissarepresents length in micrometers; ordinate represents number of molecules). The hori-zontal bar in thegraphdenotes thelengthdistribution of thepeak used to determine the mean length. Bar length in photograph (final magnification, 28,800)
equals1 um.
designedtotestthispossibility. Duplicate
sam-ples of 3H-labeled Sendai RNA and
non-radi-oactivecell RNAwereplacedinSPB bufferand
made 1.1 M in formaldehyde. One sample
(panel b)washeated (10min at 60 C) whilethe
other (panel a) was left in ice, and then both
samples were subjected to electrophoresis in
polyacrylamide gels. The arrows in panel a,
from left to right, show the positions of
formal-dehyde-treated Sendai RNA, 28S rRNA, and 18S rRNA obtained by normalizing their
posi-tions so that untreated and
formaldehyde-treated 18S rRNA coincide. The electrophoretic
mobility of Sendai RNA has been decreased proportionately more than that of 28 and 18S
rRNA by formaldehyde treatment. Assuming
that a linear semilogarithmic relationship of
P. I"-.--I
FIG. 5. Length measurement of HCHO-treated NDV RNA by electron microscopy. RNA was spread without theaid of T4 gene 32 protein essentially as described byInman and Schnos (12). One microgram ofNDV RNA in 50jilitersofSPB buffer was made 1.1 M informaldehydeandheatedfor 10 min at 60 C. Five microliters ofthe RNAsolution was then mixed with 20 jliters of buffer as previously described (12) ex-cept that the pH was adjusted to 6.4, and 20
Aliters
of formamide, 4 jiliters of 0.1% cytochrome and 5
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.493.259.453.78.173.2] [image:6.493.278.439.269.557.2]201
IN
--'H-CPM
(a) UNTREATED (b) ^ HCHO-TREATED 100
2000
3
-~~~~~~~~~~~~~~~~~10
0 10 20 30 40 0 10 00 30 39
FIG. 6. Polyacrylamide gel electrophoresis of untreated and HCHO-treated Sendai RNA. Duplicate samples for electrophoresis contained 12,000 counts/min of 3H-labeled 48S Sendai RNA (8,000 counts per min per 1sg) and 8
Ag
of cell RNA in 20Asliters
of SPB buffer. To one sample (panel B) 2 p liters of 28% HCHO in SPB was added, and the sample was heated for6min at 60 C. Each sample was then diluted with an equal volume of 40% glycerol, subjected to electrophoresis in 2.2%polyacrylamidegels containing 0.6% agarose, and processed as described in Materials and Methods.mobility versus molecular weight holdstrue in
ourgelsystem asinothers (2, 6), the molecular
weight of formaldehyde-treated Sendai RNA
estimated from the data shown in panel b is
approximately 10 x 106, whereas that from
panela (untreated) is 4.7 x 106. Since electron
microscopy measurements of the lengths of
untreated andformaldehyde-treatedNDVRNA
show no significant difference, it seems clear
thattheseparainfluenza RNAs under the
dena-turing conditions we have used still possess
sufficient secondary structure which leads to
unreliable molecular weight estimations using
techniques basedonthehydrodynamic
proper-ties of RNA.
DISCUSSION
Estimates of the molecular weight of RNAs
bysucrose gradient sedimentationor
polyacryl-amide gel electrophoresis with RNAs of known
molecularweight areonly reliable when all the
polyribonucleotide chains used are true
struc-tural homologues, i.e., their Stokes' radius isa
monotonic function of chain length. It is clear
from the data presented in this paperthat the
molecular weights obtained under the
condi-tions of denaturation usedarenot reliable, and
that parainfluenza RNAs and cellular rRNA
therefore do not have the same hydrodynamic
,glitersof this mixture wasthenspreadonwater
proc-essed for electron microscopy as previously
de-scribed (12). Untreated RNA was processed
identi-cally, except that it was not heated. Details of the graphat thetopofthefigurearegivenin thelegend toFig.4.Finalmagnification of photographis33,000.
properties in 99%DMSOorafterheatingin 1.1
Mformaldehyde.
Strauss et al. (16) have shown that 99%
DMSO at room temperature
sufficiently
dena-turesRNAs from 0.55 x 106to2.1 x 106 daltons
to allow their separation
by
sedimentationac-cordingto chain length. It is
unlikely
that this method is inapplicable tolarger RNAs such asNDV RNA or Sendai RNA simply because of
theirsize, since45SrRNA precursor (4.1 x 106
[18]) sediments insucrose gradients containing 99% DMSOat a rateconsistentwithits molecu-lar weight (13). It seems more likely that the assumption that99% DMSO at room tempera-ture equally denatures all RNAs is incorrect.
The finding of Ariv and Faulkner (1) that
Sindbis RNA,
another animalvirusRNA,
sedi-ments in sucrose
gradients containing 99%
DMSO at a rate
considerably
slower than thatexpected from its molecular weight is
note-worthy inthis respect. Furthermore, high
con-centrations offormamide, another nonaqueous
solvent known to destroy
polynucleotide
sec-ondary structure (17), has been foundby some
workers toonlypartially denaturecertain RNAs
at roomtemperature (16).
IntheuseofformaldehydetodenatureRNA,
Boedtker has shown that reactionfor 15 min at
63C is sufficient to permit molecular weight
estimations in sucrose gradients or
polyacryl-amide gels (5, 6). Although intheexperiments
reported here the RNAs had been reacted for
only 10 min at 60C to minimize thermal
degradation, reaction of the RNAsfor 15minat
63C did not
significantly
alter the resultspresented here. Again, since
formaldehyde-e
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.493.106.388.72.216.2]treated Sendai RNAmoves moreslowly relative
tocellular rRNAonbothsucrosegradients and
polyacrylamide gels, it seems clear that these
RNAs donot havesimilarhydrodynamic
prop-erties and that formaldehyde treatment, like
99% DMSO, does not equally denature all RNAs.
The molecular weight estimates from length
measurements in the electron microscope
pre-sented here have not been corrected for
differ-ences in base compositions between R17 RNA
and theparainfluenza RNAs, orforthepossible
varying degree of stretching ofDNA relativeto
the RNA-protein complex where T4 gene 32
protein has been used. For these reasons the
molecular weight estimates have a probable
uncertainty of 10%.Y Nevertheless, in view of
our findings that not all RNAs are equally
denatured under such conditionsas99%DMSO
or reaction with 1.1 M formaldehyde, we feel
that molecular weight estimation methods
which are independent ofhydrodynamic
prop-erties, such as length measurement in the
electron microscope, provide a more reliable
result.
ACKNOWLEDGMENTS
We thank Pierre-Francsois Spahr for generous advice, encouragement, and support. Andree Bruschi and Edith Gallayfor excellent technicalassistance, andPeterBromley forhelpful discussion.
This workwassupported by research grantno. 3.816.72 and 3.810.72fromthe FondsNational Suisse de la Recherche Scientifique.
ADDENDUM
Duesberg and Vogt have recently estimated the molecularweightofSendaiRNAby electrophoresisin formamide-polyacrylamide gels with 18S rRNA, 28S rRNA, andtobaccomosaic virus(TMV) RNAas
cali-bration standards (9). By using either 18 and 28S rRNAsor18SrRNA and TMV RNAto constructtwo linear calibrationcurves,they haveobtainedby extrap-olation molecular weight estimates for Sendai RNA ranging between 2.5 x 106 and 3.8 x 106. However,
since they did not observe a linear relationship be-tween the mobilities of the three marker RNAs and thelogarithmoftheir molecularweights,itseems
un-likelythat linearextrapolationwillyieldanaccurate
molecular weight estimate ofSendai RNA. As
dis-cussed above, the non-linearity they have observed
maybeduetounequaldenaturation of their marker RNAs.
LITERATURECITED
1. Arif, B. M., and P. Faulkner. 1972.Genome of Sindbis virus. J. Virol. 9:102-109.
2. Bishop, D. H. L., J. R. Claybrook, and S. Spiegelman. 1967.Electrophoretic separation of viral nucleic acids
onpolyacrylamide gels. J. Mol. Biol. 26:373-387.
3. Blair, C. D., and W. S. Robinson. 1968. Replication of Sendai virus, comparison of viral RNA and virus specificRNA synthesis with Newcastle diseasevirus. Virology 35:537-549.
4. Boedtker, H. 1967. The reaction of RNA with formalde-hyde. I. Optical absorbance studies. Biochemistry 6:2718-2727.
5. Boedtker, H.1968. Dependence of sedimentation coeffi-cientonmolecularweight of RNA after reaction with
formaldehyde. J. Mol. Biol. 35:61-70.
6. Boedtker, H.1971.Conformation independent molecular weight determinations of RNA by gel electrophoresis. Biochem. Biophys. Acta 240:448-453.
7. Delius, H., H. Westphal, and N. Axelrod.1973.Length
measurementsofRNAsynthesized in vitro by
Esche-richia coli RNA polymerase. J. Mol. Biol.74:677-687. 8. Duesberg, P. H. 1968. Physical properties ofRous
sar-coma virus RNA. Proc. Nat. Acad. Sci. U.S.A.
60:1511-1518.
9. Duesberg, P. H., and P. K. Vogt.1973.Gel electrophore-sisof avian leukosisandsarcomaviral RNA in
formam-ide: comparison with other viral and cellular RNA species. J. Virol. 12:594-599.
10. Erikson, R. L. 1969. Studies on the RNA from avian
myeloblastosis virus. Virology37:124-131.
11. Gesteland, R. F., and H. Boedtker.1964.Some physical properties of bacteriophage R17 and its ribonucleic acid. J. Mol. Biol.8:496-507.
12. Inman,R. B., and M. Schnos.1970.Partial denaturation ofthymine- and 5-bromouracid-containing ADNAin
alkali.J. Mol. Biol. 49:93-98.
13. Kolakofsky, D., and A. Bruschi.1973. Molecular weight determination of Sendai RNA by dimethyl sulfoxide gradient sedimentation. J. Virol. 11:615-620. 14. Montagnier, L., A.Golde,and P. Vigier.1969.A possible
subunitstructureofRoussarcomavirusRNA. J. Gen.
Virol. 4:449-452.
15. Stevens, C. L., and A. Rosenfeld. 1966. Thesecondary structure of polyadenylic acid. Inferences from its reaction with formaldehyde. Biochemistry 5:2714-2721.
16. Strauss, J.H., Jr., R. B. Kelly, and R. L. Sinsheimer. 1968. Denaturation of RNA with dimethyl sulfoxide. Biopolymers6:793-807.
17. Ts'o, P. 0.P., G. K. Helmkamp, and C. Sander. 1962. Secondary structuresofnucleic acids inorganic
sol-vents.II.Opticalproperties ofnucleotides and nucleic
acids.Biochem. Biophys.Acta 55:584-600.
18. Weinberg, R. A., and S. Penman.1970.Processing of45S nucleolar RNA. J.Mol.Biol.47:169-178.