0022-538X/82/110530-08$02.00/0
Copyright C 1982,American SocietyforMicrobiology
Conserved Sequences
at
the
Origin of Adenovirus
DNA
Replication
BRUCEW.STILLMAN,* WILLIAMC.TOPP,ANDJEFFREY A.ENGLER Cold Spring Harbor Laboratory, ColdSpringHarbor, New York 11724
Received 23 April 1982/Accepted 13 July 1982
The origin ofadenovirus DNA replication lies within an inverted sequence
repetition at either end ofthe linear, double-stranded viral DNA. Initiation of DNA replication is primed by adeoxynucleoside thatis covalentlylinked to a protein, which remains bound tothe newly synthesizedDNA. We demonstrate
thatvirion-derivedDNA-protein complexes fromfive human adenovirus
serologi-cal subgroups (A to E) can act as a template for both the initiation and the
elongation of DNA replication invitro, using nuclear extracts from adenovirus
type 2 (Ad2)-infected HeLa cells. The heterologous template DNA-protein
complexeswere not asactiveasthehomologousAd2DNA,mostprobablydue to
inefficientinitiation by Ad2replicationfactors. In an attempt toidentify common
features which may permit this replication, wehave alsosequenced the inverted
terminal repeated DNAfrom human adenovirus serotypes Ad4 (group E), Ad9
and AdlO (group D), and Ad31 (group A), and we have compared these to
previously determined sequences fromAd2 and Ad5 (group C), Ad7 (group B),
and Adl2 and Adl8(group A) DNA. In all cases, the sequence around theorigin
ofDNAreplicationcanbe divided intotwostructural domains: aproximalA*
T-richregion which ispartially conservedamongtheseserotypes,andadistalG* C-rich region which isless wellconserved.TheG*C-richregion containssequences
similartosequences presentinpapovavirus replication origins.The twodomains
may reflect a dual mechanism for initiation of DNA replication:
adenovirus-specific proteinpriming of replication,andsubsequentutilizationofthisprimer by
hostreplication factors forcompletion ofDNAsynthesis.
Adenovirus DNA replicates by astrand
dis-placement mechanism from origins of
replica-tion that are located withinaninvertedrepeated
sequence at each end of the linear
double-stranded DNA (see the review by Winnacker
[37]). Replicationisinitiatedateitherorigin,and
elongation ofDNAchains proceedstoward the
opposite end of the molecule, displacing the
nontemplate DNA strand (type I replication [16]).Thedisplaced strand is thenreplicatedas a
result of a separate but analogous initiation
event (type IIreplication).
The viral DNA has a 55,000-dalton protein
(terminal protein, TP) covalentlylinked toeach
5' terminus (21-23) via aphosphodiester bond
between the
13-OH
of a serine residue in theprotein andthe5'-OHof the terminal
deoxycyti-dine residue (2, 9). The terminal protein is
synthesized as an 80,000- to87,000-dalton
pre-cursorprotein (pTP) (2, 31) which iscleavedto
TP late in infection (6). ThepTP is encoded by
early region E2b on the adenovirus genome,
which forms part ofacomplextranscriptionunit
encoding otherreplication proteins (31).
Intra-cellular DNA (6, 8, 14, 29, 30, 36) and nascent
DNA strands replicated in vitro (2, 28) also
contain protein linked to each 5' terminus of
DNA, and this protein has been identified as
pTP (2).
Recentdevelopments have demonstrated that
pTPfunctions in the initiation of DNA
replica-tionby covalently binding the first
deoxynucleo-tideresidue,which then provides a 3'-hydroxyl
groupforsubsequentchainelongation (7, 11, 17,
20, 33). This mechanism is broadly consistent
witha model originally proposed by Rekosh et
al. (21). A DNApolymerase activity is
associat-edwith aprotein complex consisting of pTP and
a 140,000-dalton protein (11). The parental TP
thatis covalently attached to the templateDNA
is not required for the formation of the
pTP-dCMP complex; rather, a specific DNA
se-quence has to be located at the end ofa linear
molecule (33).
The inverted terminal repetitions (ITRs) ofa
number of adenovirus serotype DNAs have
been sequenced and include human
adenovir-uses type 2 (Ad2) (1, 25) and AdS (27), both
group C viruses; Adl2 and Adl8 (26, 32, 35;
C. F.Garon,R. P.Parr,R. K.Padmanabhan, I.
530
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 44, 1982
Roninson, J. W. Garrison, and J. A. Rose,
Vi-rology, in press), bothgroupAviruses; and Ad3
and Ad7 (10, 35), both group B viruses. The
sequenceofthe ITRs ofsimianadenovirus SA7
(35), mouse adenovirus FL (34), and the avian
CELO virus (P. Alestr6m, A. Stenlund, P. Li,
and U. Pettersson, Gene, in press) have also
beendetermined.
Challberg and Kelly (5)have described a
cell-free system that is capable of initiation and
completion ofoneround oftypeIreplicationon
anexogenously added templateDNA. Since this
system faithfully mimicsthe mechanism oftype
I replication in vivo, it provides a convenient
means for studying the sequence requirements
forthe initiationof DNA replication. We
com-pared the ability of the DNA-protein complex
fromrepresentative adenovirus serotype groups
A to E to initiate andelongate DNAreplication
in heterologous adenovirus type 2-infected
HeLacell nuclear extracts, andwefound thatall
DNA-protein complexes tested did replicate to
some extent. The replication of Ad7
DNA-pro-tein complex in an Ad2 nuclear extract was
reported previously (28). As a first attempt to
determine the DNA sequencesrequiredfor the
initiation of adenovirus DNA replication, we
extended the analysis ofITR sequences to
in-clude representatives of group D (Ad9 and
AdlO)and group E (Ad4) humanadenoviruses,
as well as another memberof group A(Ad3l).
Thesecombineddatahave enabledustoidentify
the DNA sequences that are common to
differ-ent human adenovirus serotype DNAs which
mayallow DNAreplication in vitro.
MATERIALSANDMETHODS
Cellsandvirus. HeLa S3 cells weregrown in
sus-pension in Joklik modified Eagle Spinner medium
(GIBCO Laboratories) containing 5% calf serum
(FlowLaboratories, Inc.). A549cells,derived from a human lung carcinoma, were obtained from Walter A. Nelson-Rees (Naval Biosciences Laboratory, Naval
Supply Center, Oakland, Calif.)and grown as
mono-layer cultures in Dulbecco modified Eagle medium
(GIBCO) containing10ocalfserum.Ad2 was grown in both HeLa and A549 cells. Adenovirus serotypes
Ad4, Ad7, Ad9,AdlO, andAd3l wereobtained from
the American Type Culture Collection, plaque
puri-fied,andgrown in A549cells.
PurifictionofvirusandDNA. Infectedcells
show-ingextensivecytopathic effect werecollectedby low-speed centrifugation, frozen and thawed, and suspend-ed in cell lysis buffer (0.5% Nonidet P-40-10 mM
MgCl2-10mM Tris [pH7.9]) at about 5x107cells per
ml.After 10min on ice, the cell debris was removed by centrifugation, and virus was purified by two succes-sivebandings in CsCl as described previously (18). Viral DNA was purified by digestion of purified virus
by previously self-digested pronase (1 mg/ml) in the presence of 0.5% sodium dodecyl sulfate (SDS), fol-lowed by phenol and CH3Cl-isoamyl alcohol (24:1)
ADENOVIRUS DNA REPLICATION 531
extractions and ethanol precipitation. DNA-protein complex was prepared as describedpreviously (28).
Labeling and DNA sequencing of adenovirustermini.
The 3' labeling of thetermini of Ad4, Ad9,AdlO,and
Ad3l DNA was performed as described byChallberg
and Englund(3), except that the Klenowfragmentof DNApolymerase I (NewEngland Biolabs)was used instead of T4 DNA polymerase. DNA (30 to 50 1Lg) isolated from phenol-extracted virions was used in eachlabelingexperiment. [a-32P]dGTP(400Ci/mmol, Radiochemical Centre, Amersham,England)wasused forlabeling. Inaddition, each reaction contained0.3 mMdATP, 0.3 mMTTP, and 1 ,uM dGTP. Labeling
was at 12°C for 3 h. Afterlabeling, DNA samples were digested with restriction endonuclease RsaI (New EnglandBiolabs) before being loaded on an 8%
poly-acrylamide gel (40:1, acrylamide-bisacrylamide) in TBE(135 mM Tris OH-45 mM boricacid-2.5 mM
disodium EDTA). Purification oflabeled DNA from unincorporated nucleotidetriphosphates, extractionof DNA from polyacrylamide gels, and nucleotide se-quencing have beendescribedby Maxamand Gilbert
(19).
DNA repUcation in vitro. Nuclear extracts were prepared from Ad2-infected HeLa S3 cells as de-scribedpreviously (5). Reactionconditionshavebeen described previously (28); each 50-,ul reaction con-tained 80 ng ofundigestedorHindlll(NewEngland Biolabs)-digestedDNA-protein complexand was incu-bated for 1 h at30°C.Thereaction wasstopped bythe additionof EDTA to 15 mM andofpronase to 1mg/ml
and further incubated for 1 h at 37°C. The DNA
products were then analyzed by either neutral or
alkalineagarosegelelectrophoresis.
Formation ofpTP-dCMPcomplexes. Apartially puri-fied protein fraction containing active pTP was pre-pared by columnchromatography throughdenatured
DNAcelluloseasdescribedpreviously (33). Reaction
conditions have also beendescribedpreviously;each
reaction,which contained 40 ,ug ofDNA-protein
com-plex,wasincubated for 1 h at30°C.Theproductswere analyzed on 10%o SDS-polyacrylamide gels as de-scribedpreviously(33).
RESULTS
DNAsequencing of terminal repetitions of Ad4,
Ad9, AdlO, and Ad3l. The sequences of the
termini, determinedby thechemical degradation
procedures of Maxam and Gilbert (19), are
shown inFig. 1B. Weinferthat thefirst
nucleo-tide of each ITR isdC, since preliminary
experi-mentsshowed that
[a-32P]dGTP
gavethe highestincorporation of radioactivity into adenovirus
DNA incubated with the Klenow fragment of
DNA polymerase I (data not shown). Where
determined, every other human adenovirus
DNA had adC residue at the 5' end.
The ITRs ofAd4 (group E), Ad9 (group D),
and Ad31 (group A) are 116, 158, and 148
nucleotideslong,respectively. The ITR of AdlO
is identical to that of Ad9, at least to nucleotide
135.The ITRsof the group C (Ad2 andAdS)and
groupB adenoviruses (Ad3 andAd7) have been
shown to be 103 and 137 nucleotides long,
on November 10, 2019 by guest
http://jvi.asm.org/
532 STILLMAN, TOPP, AND ENGLER J. VIROL.
U
'C 'C 'CH 'C
I-C3)- U 'C 'C 'C
<: ~ ~~~~~~~~~~U U 'C 'C< O <:~ ~ ~ ~ - H HC 'C U'Cb
UH 0~~~~~~~~~~~~~~~~~u L
H w<: *'e *e 'C *'; *U *C
'C 'C 'C 'C H H 'C H
H H H 'C 'C 'C 'C
0. H H H 'C 'C U 'C
'C U U C) C.
eC 'C 'C 'CU U -) C.)
> ~ ~~~'C 'C 'C H U 'C
'C H
3 *)()H e * C.) C. 'C H
Hn 'C 'C ' 'C U 'C
= ~ ~~~H H C- C- C-) U
C~~T --.r 'C'~ C) U
<5) ~
~~
Ce
. H~C.) UO H HF H HF
(_)~ ~ ~ ~' 'C 'C 'C 'C 'C 'C
F
~~~*H
-H -H - *H -H *H 'C 'la 'C ' ' 'C/i Hr. 'eC' H
Na<e vHU;UOOH eH 'CH 'CH 'CH HUE 'C
U H HU U
U;
H UH: U 'C H H H UHz
4) U
N 0's m - - N
NM t
rC, tici
m~a~ ~ 40 S-*= .
o a v o Sw4. C
4CC)~~~~~~~~~~~~~~~~~~~.
O00*.) -U (
7*O* o * 2Cr< <r. Qs. 5o
C-) H~~~~~~~~~4.
U e - E
X U U -0 =
--U
-C|
v *UOcn3
=
o
OU. e * UV C- C
e
U
U
u,
r-D4)
o<;
*-o
zce < < v 3; ~~~~~~~~~~o*5);^O N *3=.;&=
Uo'C.e.e . so .A. 4r)
U C- UW . .
f-. H e Xb$M~
U - 'C
-0~
'CQ
;
o > om > e >
>_D P
ir<4
U C) C-)
oHUL LiU C U H 4) 1y °
cc O W .=e |
C-, C C U
C-)) C-) U C)'
4)ato'C 'C
C-4\Jb )H H C- U '0W 4. .
iVivcc 'C U en]IU
U U U .>
-) H H 4)--> ,
H'C-~~~~~~~C-) 'C 'C 04)~~~~m .
4-H C- C- UWV'
-UUC-)C-) U H U ~~~~4
W.
C- u U H U -0 'CJ W '
U C) C- L)H C- Ut
'C Hc H >CUU: t.U= 0
U0E H H 'C C)C)CI0''
-UU UH U U
-U HLC)C)~4
~~~L ) ~~~~~]~C.) 4-4; 4.
-U-H U
-'-O CU U
-'C Uc U Hc Hc Uc H4
UUU H ~~~~HU U
UUU U 'C~~~~~-U U- 4W)
H- H H LI) U 'C H
U 'C 'C H H U
'CHHH H 'C H~~~~~~~~~~~C E-- -cc -Uc -C -4
on November 10, 2019 by guest
http://jvi.asm.org/
ADENOVIRUS DNA REPLICATION 533
respectively. Two other group A adenovirus DNAterminihavebeensequenced; the ITRsof
Adl2 and Adl8 are both 165 nucleotides long
and arequite homologous. The ITR of Ad3l, on
the other hand, isshorter and exhibitsimperfect
homology with Adl2 and Adl8 in only two
sections: nucleotides 1 through 68 and 120
through 148oftheAd31 sequence. For
compari-son,the DNAsequences of the ITRs fromAdM2,
Ad7, and Ad2 are shown in Fig. 1B. It is
noteworthy that whereas Adl2 and Adl8 grow
theleast well of the 20 virus serotypes wehave
propagatedon A549cells,Ad31 growsasrapidly
astheviruses ofsubgroup B,thoughnot aswell
asthe groups C, D,and E(datanotshown).
DNA replication in vitro. Since adenovirus
DNAreplicationstarts atthe endsof the DNA,
only the terminal fragments of restriction
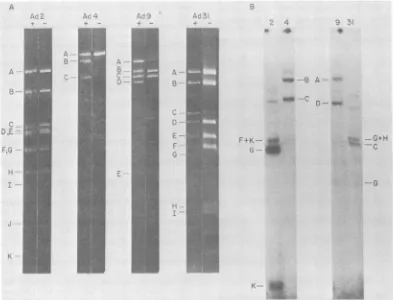
en-zyme-digested adenovirus DNA-protein com-plexreplicatespecificallyin vitro(13).We have
used this assay to determine whether
DNA-protein complex fromAd4, Ad9, and Ad3l will
replicate in vitro, using nuclear extract from Ad2-infected HeLa cells. First, the terminal
restriction fragments were identified by their
inability to enter agarose gels without prior
digestion withpronase. DNA-protein complexes
were digested with HindIII, and samples were
subjected to electrophoresis in an agarose gel
before and after digestion with pronase (Fig.
2A). The terminal fragments bound to TP were
retarded in the gel and only migrated in the
correct position after pronase digestion, thus
identifying the terminal HindlIl fragments as B
and C of Ad4, A and Dof Ad9, and C and G of
Ad3l. The terminal HindIlI fragments of Ad2
are G and K. HindlIl-digested DNA-protein
complex from either Ad2, Ad4, Ad9, or Ad31
was incubated with Ad2 nuclear extract under
standard reaction conditions, digested with
pronase, andsubjected toelectrophoresis on an
agarose gel(Fig. 2B). In each case, the terminal
-G+H
b--1 ? :.
[image:4.491.45.439.310.610.2].~~~~~~~~~~~--G
FIG. 2. Replication ofheterologous DNA-protein complexes in vitro. (A) DNA-protein complex derived from eitherAd2, Ad4,Ad9,orAd3l virionswasisolated,digestedwithHindIlI, andsubjected toagarosegel electrophoresis either before (-) or after (+) digestion with pronase (1 mg/ml). Ad9 fragment F and Ad3l
fragmentsJ toS(Y.Sawada,T.Yamashita,and K.Fujinaga, personal communication)werenotvisible. Thegel
wasstained withethidium bromide andphotographed. (B) DNA-protein complexfrom eitherAd2, Ad4, Ad9,or
Ad3lwasincubated withextractfrom Ad2-infectedHeLacells. Theproductswerethendigestedwithpronase
and subjected to electrophoresis through a 0.8%agarose gel. Thegel was dried and autoradiographed. The
positionsof the terminalfragmnentsareshown;theHindlIll digestfor both Ad2 andAd3l waspartial.
VOL. 44,1982
on November 10, 2019 by guest
http://jvi.asm.org/
restriction fragments were preferentially la-beled, indicating that each DNA-protein
com-plex had the capacity to act as template for replication in vitro. TheHindIIIdigests of Ad2
andAd3l were both partial, and replication of
theterminal and subterminalfragments is
indi-cated (Fig. 2B). Ad7DNA-proteincomplexwas
previously shown to replicate by using an Ad2
nuclear extract (28).
Todetermine whetherreplication in vitro
re-sulted in genome-length DNA molecules, uncut
DNA protein complex from either Ad2, Ad4, Ad7, Ad9, or Ad31 was used as template in an
Ad2-infected HeLa cell nuclear extract. The
products were digested with pronase and
ana-lyzed by electrophoresis on an alkaline agarose
gel (Fig. 3). In eachcase,full-lengthproduct was
observed, but the amount of product of each
heterologous template was reduced relative to
the homologous Ad2 template. In a number of
experiments with different preparations of
DNA-protein complexfrom each serotype,
dif-ferentamounts ofproduct were observed. This
has also been observed with various
prepara-tions ofAd2complexandreflectsthe amount of
proteolyticdegradation and the concentrationof
thestockof each DNA-proteincomplex
(unpub-FIG. 3. Alkaline agarosegelelectrophoresisofthe
replication reaction products. Undigested DNA-pro-tein complex from either Ad2, Ad4, Ad7, Ad9, or
Ad31 wasincubated inareaction mixturecontaining
nuclearextractfromAd2-infected HeLa cells for1hat
37°C. Thereactionproductswere digestedwith
pro-naseandsubjectedtoelectrophoresis througha0.7% alkaline agarosegel.Thegelwasdried and
autoradio-graphed.
2 4 7 9 31
-fl (20K)
87 K-4 . i-m (85K)
-ma
(66K)
-Z (48K)
FIG. 4. Formation ofapTP-dCMP complex with various DNA-protein complexes as template. Undi-gested DNA-proteincomplex from either Ad2, Ad4,
Ad7, Ad9, orAd31wasincubated in areaction
mix-ture containing [a-32P]dCTP and a partially purified
extractfromAd2-infectedHeLacell nuclei (33)for 1 h
at30°C. Thereactionwasstopped bytheaddition of EDTAto 10 mM and then was subjected to
electro-phoresis through a 10% SDS-polyacrylamide gel as
describedbyLaemmli (15). MarkerproteinswereAd2 virionproteins, and the molecular weightsareshown. The gel was dried and autoradiographed with an
intensifyingscreen.
lished data). However, Ad4, Ad9, and Ad3l
DNA-protein complexes isolated concurrently
with Ad2 complexconsistently incorporated
ap-proximately10-fold fewerpicomolesof
deoxyn-ucleosides than did Ad2DNA-protein complex,
whereasAd7 template showed65% ofthe level
ofincorporation seenin Ad2(28).
InitiationofDNAreplication. Ithas been
pro-posed (21) that the first step in theinitiation of
adenovirus DNAreplication is theformation of complex betweenpTPand dCMP, the5'
termi-nal nucleotide of Ad2 DNA (1, 25). Such a
covalent complex was found when extracts of
adenovirus-infected HeLa cells wereincubated
with
[a-32P]dCTP
andtemplate DNA(7, 17, 33).We detected the formation of a complex
be-tween Ad2 pTP and dCMP whenheterologous
DNA-protein complexes wereusedastemplate.
DNA-protein complexes derived from Ad2,
Ad4, Ad7, Ad9, andAd3l virionswere
incubat-edwiththepartiallypurifiedAd2 nuclearextract
and
[a-32P]dCTP,
and the reaction productswereanalyzedby SDS-polyacrylamide gel
elec-trophoresis (Fig. 4). EachDNA-proteincomplex
facilitated the formation of an 87,000-dalton
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.491.254.449.79.288.2] [image:5.491.118.190.373.595.2]ADENOVIRUS DNA REPLICATION 535
pTP-dCMPcomplex;however, the homologous
Ad2complex wasbyfar themostactive.
Com-plex was not formed when either [a-32P]dATP, dGTP, orTTPwasused (datanot shown). The
relativeefficiency of pTP-dCMP complex
forma-tion for eachheterologous template was
reflect-ed in the relative efficiency of replication as
assayed by specific replication of the terminal
restriction fragments (Fig. 2B) and by the
pro-ductionoffull-lengthproduct(Fig.3),suggesting
that pTP-dCMP complex formation may be a
rate-limitingstepin thereplicationof viral DNA.
DISCUSSION
ChallbergandKelly (5)described the isolation
of a soluble nuclear extract from
adenovirus-infectedHeLa cellsthatwascapable of
replica-tion ofexogenousDNA.Replicationwas
depen-dent upon thetemplateDNAhavingthe terminal
protein bound to each 5' end, since template
DNA that hadbeendigested with protease was
inactive (4). Residual peptides present on
pro-nase-digested DNA template probably inhibit
the initiation reaction (33). It was therefore
suggested thatboththe TP and DNAsequences
at the origin of replication facilitated the
initia-tion of DNA synthesis, with the TPinteracting
with one or more of the proteins in the soluble
nuclear extract. The apparently strict
require-mentforthetemplateDNAtocontain
covalent-ly bound protein made it impractical to
deter-mine DNA sequence requirements attheorigin
byin vitromutagenesis. However,the
availabil-ity ofanumber of adenovirus serotypes, each
having differentDNA sequencesattheoriginof DNAreplication, suggestedaway toovercome
this problem of analyzing DNA sequence
re-quirements forthe initiation ofreplication. We
therefore tested the ability of heterologous DNA-protein complexes from anumber of hu-man adenoviruses, representingfive serological
groups, to act astemplate forboth theinitiation
and theelongation ofreplication in nuclear
ex-tractsderived from Ad2-infectedcells. All
heter-ologous DNA-protein complexeswereactive for
both initiation and elongation of replication in
Ad2 nuclear extracts, but they werealwaysless
active than thehomologoustemplate.Template
DNAsfrom Ad4, Ad9,andAd31formeda
pTP-dCMP complex with low efficiency, but the
complex was sufficient to primespecific
replica-tion from both termini of these DNAs, as
as-sayedby the replication oftheterminalHindIll
restriction enzyme fragments. Thus, we
con-clude that DNA sequences within theITR that
are conserved between these viruses and Ad2
may play an important role in the initiation
reaction. It was not practical to reverse the experiment and use Ad2 DNA-protein complex
as a
template
in aheterologous adenovirusnu-clear extract, since all such viruses we have
used growpoorly inourHeLacells.
Aproblemposedbythese resultsis the
possi-bleinteraction of the DNA-bound
heterologous
TP with the Ad2 pTP that is present in the
nuclear extracts. We do not know to what
degreethiswillaffect the initiationreaction,and
thuswedrawnoconclusionsas totheefficiency
with which specific sequences within the ITRs
of thedifferent adenovirus DNAs support DNA
replicationin the Ad2 system. We concludeonly
that there is sufficient sequence homology to
correctlyinitiatereplicationatsome
frequency.
However, during the course of these experi-mentsitbecame clear that the TP isnotrequiredforinitiation of replication inapartially
fraction-ated replication extract, since aplasmid DNA
containing the ITR of Ad5was active for
pTP-dCMP complex formation (33). The plasmid
DNA was only active when the origin was
located at the end of the linearized DNA. In
addition, the plasmid DNA was active when it
was heat denatured, again provided that the
origin was located at the end of the molecule.
Thus, interactions between pTP and the TPon
theparental DNA are not obligatory, but they
still may affect theefficiency of initiation.
Thehuman adenovirus serotypes 1 to38 have
been subdivided into five groups on the basis of
DNAhomology thatwasderived from solution
hybridization studies with virion DNA (12).
DNA sequences of the ITRs from group A(Adl2
andAdl8), group B (Ad3 and Ad7), and group C
(Ad2 and AdS) have been determined, and
re-gions ofhomology have been noted (26, 35). The
replication studies suggest that sequences in the
ITR which are important for DNA replication
have been conserved; thus, we have extended
the DNA sequence analysis to include the ITRs
from group D (Ad9 and AdlO) and group E(Ad4)
humanadenoviruses and the ITR of an
addition-al group A virus (Ad3l). Recently, the
se-quences of the ITRs for Ad4 (0. Tokunaga, M.
Shinagawa, and R. Padmanabhan, Gene, in
press) and Adl8 (Garon et al., in press) were
determined. Sequences of the ITRs for some of
the known human adenoviruses are shown in
Fig. 1B.
The ITR for each human adenovirusDNA can
be divided into an A * T-rich region of 50 to 52
base pairs, which occurs at the termini of each
DNA molecule, and a G * C-rich region that
varies in lengthbetween 50 and 110 base pairs.
Indeed, A - T- and G * C-rich sequences are
also found in othereucaryotic origins of
replica-tion, most notably those in the papovaviruses
(reviewed by Seif et al. [24]). Within the ITR,
DNA sequences in the first 50- to 52-base pair
A * T-rich region are themost highly conserved
VOL.44,1982
on November 10, 2019 by guest
http://jvi.asm.org/
among all human adenoviruses (Fig. 1B). This
region is alsopartially conserved in the
adeno-viruses of the lowerprimates (35), rodents (34),
andbirds (Alestrometal., in press). A
consen-sus sequenceforthis regioncanbederived that
conforms with the known sequencesfrom allof
the humanadenoviruses(Fig. 1A).
Within this A T-rich region, there is a
per-fectly conserved sequence of 10 base pairs
(ATAATATACC), and all or part of this
se-quence is alsopresentin the ITR of nonhuman
adenoviruses. In addition, where it has been
determined, all human adenoviruses contain a
5'-terminal dC, which is the base towhich the
TPis covalently attached. Thesequenceof 6to7
bases that lies between the terminal dC and the
conserved10-base sequence isoneoftwotypes:
either relatedtotheAd2sequence orrelatedto
theAd7 sequence. Within this region, all
sero-typeDNAscanbederived from either the Ad2
orAd7 sequencesby base substitutionsor
dele-tion(Fig. 1A). Infact, different isolates of
sero-typesAd2, Ad4, and Ad7 showsequence hetero-geneity within this region (Fig. 1A). At the other
end of the 10-base, perfectly conserved
se-quence is a 9- to 11-base region that has
di-verged, which is followed by a region that is
partially conserved among all human serotype
DNAs.
The G * C-rich region, on the other hand, is
not conserved and is also variable in length.
However, twofeatureswithin this regioncanbe
identified that are common toall ofthe human
adenoviruses. The first is theoccurrenceofone
or morecopies ofasequence verysimilartothe
sequence GGGxGGAG, which ispresent twice
at the origin of replication of human BK virus
(24) and which is alsofoundatthe simian virus
40andpolyoma virus (mouse)origins of
replica-tion (24). All humanadenovirusesand the simian
virus SA7 (35) have the sequence GGGCGGat
leastonce in the ITR. Aspreviously suggested
(24), thispapovavirusconsensussequencemight
play an important function as a recognition
signal foraprotein involved in DNAreplication.
A second feature of the G * C-rich region is the occurrence of the sequenceTGACG, which
occurs ator nearthe endof allhuman
adenovi-rus ITRs; related sequences occur in murine
(FL) andavian(CELO)adenoviruses (35;
Ales-trom etal., in press). It is noteworthy thatthis
sequenceisnotalwaysatthe endoftheITR,as
previously observed (26); itcanbefound within
theITR (Ad9, AdlO, andAd3l) orjust outside
the ITR(Ad4).Afunctionhas not beenascribed
tothis conserved sequence.
Itislikelythat theseconserved features
with-intheITRplayseparaterolesin theinitiationof
DNAreplication. Onepossible role for the
high-ly conservedA * T-richregionwouldbetoserve
as an origin for the adenovirus-specific protein
priming of DNA synthesis. The complex
be-tween pTP and the 140-kilodalton protein (11) maybindto DNAsequenceswithin thisregion.
The conserved sequences within the G * C-rich
regionaresimilarto thosefound in other
eucary-otic origins and may serve as cellular DNA
polymerase recognition sites. Thus, the two
structural domains may reflect two functional
sites that initiate DNA replication. The recent
demonstration that a linearized plasmid DNA
containing the ITR atthe end of the molecule
canfunction for initiation of DNA replication in
vitro (33) will enableus todefinemoreprecisely
the adenovirus origin of replication by site-directed mutagenesis.
ACKNOWLEDGMENTS
We thankJohn L. Hierholzer for verifying the serotypes of viruses used in this study and J. A. Rose and R. Padmanabhan forcommunicating sequence data before publication. We also thankMichael Mathews and Fuyuhiko Tamanoi for discus-sions and critical reading of the manuscript and Elizabeth Woodruff,DianeSmith, and Mark Hoppe for technical assist-ance.
Thisresearch was supported by grants from the National CancerInstitute(CA-13106) and from the AmericanCancer Society(RD-126). J.A.E.is a Special FellowoftheLeukemia Society ofAmerica.
LITERATURE CITED
1. Arrand,J. R., and R.J. Roberts. 1979. The nucleotide sequences at the termini of Ad2 DNA. J. Mol. Biol. 128:577-594.
2. Challberg, M.D., S. V. Desiderio, and T.J. Kelly, Jr. 1980. Adenovirus DNAreplicationinvitro: characteriza-tion of a protein covalently linked to nascent DNA strands. Proc. Natl. Acad. Sci. U.S.A. 77:5105-5109. 3.Challberg, M. D., and P. T. Englund. 1980. Specific
labelingof 3'terminiwith T4 DNApolymerase. Methods Enzymol.65:39-43.
4. Challberg,M.D.,and T.J.Kelly,Jr. 1979.Adenovirus DNAreplicationinvitro:originanddirectionofdaughter strandsynthesis.J. Mol.Biol. 135:999-1012.
5. Challberg,M.D.,andT.J.Kelly, Jr.1979. Adenovirus DNAreplicationinvitro.Proc.Natl. Acad. Sci. U.S.A. 76:655-659.
6. Challberg,M.D.,andT. J.Kelly,Jr.1981.Processingof theadenovirus terminalprotein.J.Virol.38:272-277. 7. ChaHberg,M.D., J. M.Ostrove,andT.J. Kelly, Jr.1982.
Initiation of adenovirus DNA replication: detection of covalentcomplexes between nucleotide and the 80-kilo-daltonterminalprotein.J.Virol. 41:265-270.
8. Coombs, D. H., A. J. Robinson, J.W. Bodnar, C.J. Jones, andG.D.Pearson.1978. Detection of DNA-pro-teincomplexes:theadenovirus DNA-terminalproteinand HelaDNA-protein complexes. ColdSpringHarborSymp. Quant.Biol. 43:741-753.
9. Desiderio, S. V.,andT.J. Kelly,Jr. 1980. Structure of the linkage between the 55,000 molecular weight terminal protein.J. Mol. Biol.145:319-337.
10. Dijkema,R., andB. M. M. Dekker. 1979. The inverted
terminal repetition of the DNA ofweakly oncogenic adenovirus type 7. Gene 8:7-15.
11. Enomoto, T., J.H. Llchy,J. E.Ikeda,andJ. Hurwitz. 1981.AdenovirusDNAreplicationin vitro:purificationof the terminal protein in a functional form. Proc. Natl. Acad.Sci.U.S.A.78:6779-6783.
12. Green, M., J.K.Mackey,W. S. M.Wold,andP.Rigden.
on November 10, 2019 by guest
http://jvi.asm.org/
ADENOVIRUS DNA REPLICATION 537
1979. Thirty-one human adenovirus serotypes (Adl-Ad3l) form five groups (A-E) based upon DNA genome homologies.Virology93:481-492.
13. Horwltz, M. S., andH.Ariga. 1981. Multiplerounds of adenovirus DNA replication in vitro. Proc. Natl. Acad. Sci.U.S.A. 78:1476-1477.
14. Kelly, T. J., Jr., and R. L. Lechner. 1978. The structure of replicatingadenovirusDNA molecules: characterization of DNA-protein complexes from infected cells. Cold SpringHarbor Symp. Quant. Biol. 43:721-728. 15. Laemmli, U. K. 1970. Cleavage of structural proteins
during the assembly of the head of bacteriophage T4. Nature(London)227:680-85.
16. Lechner, R. L., and T. J.Kelly,Jr. 1977. Thestructure of replicating adenovirus 2 DNA molecules. Cell 12:1007-1020.
17. Lchy, J. H., M. S. Horwitz, and J. Hurwitz. 1981. Formation of a covalent complex between the 80,000 daltonadenovirusterminalprotein and5'-dCMPin vitro. Proc.Natl.Acad. Sci.U.S.A.78:2678-2682.
18. Lonberg-Holm,K., and L.Philipson. 1969.Early eventsof virus-cellinterctionin anadenovirus system. J. Virol. 4:323-338.
19. Maxam, A.,andW.Gilbert.1980.Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol.65:499-560.
20. Plncus, S., W. Robertson, and D.M. K. Rekosh. 1981. Characterizationof theeffectof aphidicolinonadenovirus DNAreplication:evidenceinsupportofaproteinprimer modelofinitiation. Nucleic Acids Res. 9:4919-4938. 21. Rekosh, D. M. K., W. C. Russell,A.J. D. Belett, and
A. J.RobIson.1977.Identification ofa protein linked to the endsof adenovirus DNA. Cell 11:283-295. 22. Robinson, A. J., and A. J. D. BeUett. 1974. A circular
DNA-proteincomplexfrom adenoviruses and its possible role in DNA replication. Cold Spring Harbor Symp. Quant.Biol. 39:523-531.
23. Robinson,A.J., H. B. Younghusband, and A. J. D. Bel-kett.1973. Acircular DNA-proteincomplexfrom adeno-viruses. Virology56:54-69.
24. Self,I., G. Khoury, and R. Dhar. 1979. The genome of humanpapovavirusBKV.Cell18:963-977.
25. Shinapwa,M., and R. Padmanabhan.1979. Nucleotide sequences at the inverted terminal repetition of Ad2
DNA.Biochem. Biophys.Res.Commun.87:671-678. 26. Shinagawa, M., and R.Padmanabhan. 1980. Comparative
sequence analysis of the inverted terminal repetitions from different adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 77:3831-3835.
27. Steenbergh, P. H.,J. Maat, H. vanOrmondt,and J. S. Sussenbach. 1977. The nucleotide sequence at the termini of adenovirus type 5 DNA. Nucleic Acids Res. 4:4371-4389.
28. Stillman, B. W. 1981. Adenovirus DNA replication in vitro: a protein linked to the 5' end of nascent DNA strands. J. Virol.37:139-147.
29. Stilman, B. W., and A. J. D. Bellett. 1978. Replication of DNA in adenovirus-infected cells. Cold Spring Harbor Symp. Quant. Biol. 43:729-739.
30. Stillman, B. W., and A. J. D. Bellett. 1979. An adenovirus protein associated with the ends of replicating DNA molecules. Virology93:69-79.
31. StUlman,B. W., J. B.Lewis, L. T. Chow, M. B. Mathews, and J. E. Smart. 1981. Identification of the gene and mRNA for the adenovirus terminalproteinprecursor. Cell 23:497-508.
32. Suglsaki, H., K. Sugimoto, M. Takanami, K.Shiroli, I. Salto, Y. Shlmojo, Y.Sawada, Y. Uemizu,S.-I. Uesugi, and K.Fujinaga. 1980.Structure and geneorganizationof the transforming HindIII-G fragment of Adl2. Cell 20:777-786.
33. Tamanol, F., and B. W.StUlman. 1982. Function of the adenovirus terminal protein in the initiation of DNA replication. Proc. Natl. Acad. Sci. U.S.A. 79:2221-2225. 34. Temple, M., G. Antoine, H. Delius, S. Stahl, and E.-L. Wlnnacker. 1981.Replicationof mouse adenovirus strain FL DNA.Virology109:1-12.
35. Tolun, A., P.Alesrm,andU. Pettersson. 1979.Sequence of inverted terminal repetitions fromdifferent adenovi-ruses:demonstration of conserved sequences and homol-ogybetween SA7terminiandSV40 DNA. Cell 17:705-713.
36. VanWeilink, P. S., N. Naaktgeboren, and J. S. Sussen-bach.1979. Presence ofproteinatthe termini of intracellu-lar adenovirus type S DNA. Biochim. Biophys. Acta 563:89-99.
37. Winnacker, E. L. 1978. AdenovirusDNA: structure and functionof a novelreplicon.Cell 14:761-773.
VOL.44, 1982
on November 10, 2019 by guest
http://jvi.asm.org/