Copyright© 1978 AmericanSocietyforMicrobiology Printed inU.S.A.
Biochemical Studies
onBovine Adenovirus
Type
3
III. Cleavage Maps
of
Viral DNA
by Restriction Endoncleases
EcoRI,
BamHI, and
HindIII
T.KUROKAWA,* K. IGARASHI,ANDY.SUGINO
BiologicalResearchLaboratories, CentralResearchDivision, Takeda ChemicalIndustries, Ltd.,
Osaka,Japan
Received forpublication 2 March1978
Cleavageof bovine adenovirustype3 (BAV3) DNAbyrestriction
endonucle-asesEcoRI, BamHI,and HindIIIyielded 7 (AtoG),5 (AtoE), and 12 (A to L) fragments, respectively.Theorderof thesefragmentshas been determined to be GDACBFEfor EcoRIfragments, AEBDCfor BamHIfragments, and JEBKAC-DHFGIL forHindIlI fragments, and cleavage sites of these enzymeshavebeen
mapped on the genome of BAV3. BAV3 preparation contains incomplete virus whosegenomehasadeletion of about13% ofcompletevirusgenome.Restriction
endonuclease digestionoftheincomplete virus DNA revealedthat EcoRI E and F, BamHI C and HindIll G, I, and Lfragments were deleted. Therefore, the deletedregion of incompletevirus DNAislocatedneartheright-handend ofthe BAV3DNAmolecule,aresult consistentwithourprevious electron-microscopic observationsonheteroduplexmoleculesformed betweencompleteandincomplete BAV3 DNA.
Genomecleavagemaps, constructedby
order-ingDNAfragmentsproduced by digestionwith
restriction endonucleases, are an essential tool
for thedetailedanalysisof the genomefunctions of various DNA tumor viruses and cellular trans-formation by their genomes. Cleavage maps have already been constructed in many DNA tumorviruses (2, 8, 11) andutilizedasthebasis for further studies.Cleavagemaps of adenovirus genomeshave also been producedin many hu-man(15, 17, 19, 19a,21) andmouse (14)
adeno-viruses.Accumulation of these data might give
some insight into the evolutional relationships
amongthese viruses.
Wehavepreviously reportedthebiochemical
properties of bovine adenovirus type3 (BAV3) (9, 16).The genome of BAV3 isalinearduplex
DNAmolecule withamolecularweightof 24 x
106. To provide specificDNAfragmentsforuse in further analysisof BAV3functions,we have
attemptedtoconstructcleavagemaps ofBAV3 DNA by the use of restriction endonucleases
EcoRI, BamHI, and HindIII. In this paper,
cleavage sites of these enzymes have been
mappedmainly by two methods: (i) analysis of
overlappingsetsofspecific fragments produced
by different endonucleases and (ii) cleavage of DNApreviouslyend-labeledbyT4 DNA
polym-erase.
Original stocks of BAV3 contain a unique population ofincomplete virus (16), whose
ge-nomehas a deletion whichoccupies about 13%
of the total genome of complete virus, as
re-vealed by electron-microscopic observation in our previous study (16). This observation was further confirmed by analysis of the deleted
fragmentsonrestriction endonucleasedigestion
of theincompletevirus DNA.
MATERIALS AND METHODS Virus andDNA. Two clonalviruses, BAV3-0 (16) and BAV3-1 (16), originating from BAV3 prototype
(WBR-1) (3) were growninconfluentmonolayers of
CKT-1 cells(12) (an establishedcelllinederived from
calf kidney) at an input multiplicity of about 10
PFU/cell.
The procedures for purification of complete and
incomplete BAV3 and for extraction of viral DNA
have been describedpreviously(16).
Enzymes. Restriction endonuclease EcoRI was
purifiedfromEscherichiacoliRY 13 by the method
of Greene et al. (7). Restriction endonuclease BamHI
from Bacillus amyloliquefaciens (22) and HindIII
fromHaemophilus influenzae(13) were gifts from H.
Shimojo and H. Ariga and from K. Fujinaga and K.
Sekikawa, respectively. T4 DNA polymerase was
kindly providedby M. Takanami.
Digestion ofDNA with restriction
endonucle-ase.Digestion ofBAV3 DNAwith restriction
endo-nucleasesEcoRI,BamHI,andHindIII wascarriedout
accordingtoPetterssonetal. (18), Wilson and Young (22), and Danna et al. (2), respectively. The reaction was stopped by the addition of 0.1 volume of 60%
glycerol containing0.2 M EDTA and 0.05% bromo-212
on November 10, 2019 by guest
http://jvi.asm.org/
phenol blue, and products ofthe reaction were sub-jectedtoanalysis byelectrophoresisongels.
Gelelectrophoresis.Twotypesofcylindrical gel (0.6 by 26 cm) were used: (i) 0.9 or 1.2% agarose (Seakem, BioProducts,Rockland, Maine) gel (20); (ii) 0.7% agarose-2.4% acrylamide composite gel (18). ElectrophoresiswasconductedwithEbuffer (40 mM Tris-hydrochloride, 5 mM sodium acetate, 1 mM
EDTA, pH 7.8)at 1.6V/cm for20h for both types of gel. TheDNAbandswerestained with ethidium bro-mide contained in thegel and E bufferatthe concen-tration of0.5 ,ug/ml and were visualized under UV light and photographed. Theradioactivity of gel slices was counted withtoluene-based scintillation cocktail aftersolubilization of gel slices with0.4 ml ofNCS (Amersham/Searle,Arlington Heights, Ill.).
For the extraction of DNA from agarose gel, gel slicesweremashed in buffer(10mM Tris-hydrochlo-ride, 1 mMEDTA, 0.1% sodium dodecyl sulfate, pH 8.0), and theresultingslurrywasincubatedovernight
at45°C. Thesupernatantwascollected and extracted threetimes withphenol, and then DNA was precipi-tatedby addition of2volumes of ethanol and dissolved inbuffer (10 mM Tris-hydrochloride, 1mM EDTA, pH7.8).
End-labeling of DNA by T4 DNA polymerase. Linear duplexDNAmoleculescanbe labeled witha
3H-labeleddeoxyribonucleosidetriphosphateattheir 3'terminiby T4 DNApolymerase(5). This method is basedonthe fact that T4 DNApolymerase contains
apolymerizingaswellasa3'-5'exonucleolytic activity. For theterminallabelingofBAV3DNA, the follow-ing reaction mixture (0.1ml)wasincubated for30min
at28°C:70mMTris-hydrochloride (pH 8.0),7mM
2-mercaptoethanol, 10,Lgof BAV3 DNA,and 0.2Uof
T4 DNApolymerase. Thena
10-pi
sample ofamixture of5 mMdeoxyribonucleotide triphosphates (dATP, dGTP,anddCTP)and100uCi of[3H]dTTP(2nmol, Daiichi Pure ChemicalCo.,Japan)wereaddedtothe*mixture,and incubationwascarried out foranother1
h at28°C. DNA thus labeledwaspurified from the mixture by phenol extraction followed by passage throughaSephadex G-75 column (0.5by10cm). In thiscondition, 3H labelwasintroduced into about20
nucleotides from both 3' ends of the linear duplex moleculeof BAV3DNA.
RESULTS
Restriction endonuclease digestion of BAV3 DNA. BAV3 DNA was digested with restriction endonucleases EcoRI, BamHI, and
HindIII,
and eachdigestion product
wasana-lyzed by electrophoresis on0.9 or 1.2%agarose
gels (Fig. 1). Digestion of BAV3 DNA with
re-striction endonuclease EcoRIyieldedseven
frag-ments, which were
designated
in order ofde-creasing electrophoretic
mobility
asAtoGac-cording to Danna et al. (2). BamHI digestion yielded five (AtoE)
fragments.
HindIII
diges-tion product was separated into 11 bands by electrophoresison acolumn ofagarosegel.
Fur-theranalysis
showed that one of these bandswascomposedof two DNAfragmentswith
sim-(a)
(b)
(C)
A
A
B C
D
E
F
G
A
B
C
D
B C
D
E E
F
G
H I
J
K
L
FIG. 1. Analysis of BAV3 DNA digested with re-striction endonucleaseEcoRI, BamHI, and HindIII. BAV3 DNA(about1
jig)
wasdigested with the three restrictionendonucleases, and the products were ap-plied togels. Electrophoresis wasconductedat 1.6V/cm for 20h andphotographed under UV light. DNAfragment bands were designatedasA, B, C, andsoon, inorderof their size. (a) EcoRI digestion products on 0.9% agarosegel. (b) BamHIdigestion products on 1.2%agarosegel. (c)HindIII digestion products on 1.2% agarosegel.
ilar sizes (H and I
fragments). Therefore,
HindIII
digestion
gave 12fragments (A
toL).
HindIII
fragments
H andI could beseparated
onagarose-acrylamide
composite
gel (data
notshown).
Estimation of molecular
weight
of eachfragment.
The size of eachfragment
as a per-centageof the whole DNAwasestimatedonthe basis of therelativeradioactivity
recovered in each fragment fromuniformly
labeled BAV3[32P]DNA.
In Table 1, relative molecular weights ofEcoRI,
BamHI,
and HindIIIon November 10, 2019 by guest
http://jvi.asm.org/
[image:2.500.250.443.74.425.2]mentsareshown. For somefragmentsthat were
difficult to separate from each other on gels (EcoRIAandBfragments andHindIII Hand
Ifragments), the sumof the values oftwo
frag-mentsis shown.
The molecular
weight
of eachfragment
wasestimated from its relative
mobility
on agarosegel electrophoresis. The
relationship
between mobility and molecularweight
wasplotted (Fig.
2) for some marker DNA
fragments,
and fromthisplot the molecularweightof eachfragment
wasdetermined(Table 1).
Reciprocal digestion of
specific
frag-ments
with
different restriction [image:3.500.261.450.67.224.2]endonucle-ases.
(i)
Combination ofEcoRI and Hindm. The orderoffragments
on the DNA molecule couldbedeterminedby
overlapping
thespecificTABLE 1. Molecularweights ofBAV3DNA fragments produced by restriction endonuclease
digestion
Molwt(% of BAV3
DNA) Mol wt(x
Fragment 105)
Radioac- Moilityb
tiVitya
Mobity
EcoRIfragment
A 46.9 24.9 57.2
B ) 46.9 2315 53.4
C 18.6 18.4 42.3
D 17.0 15.8 37.0
E 10.5 9.7 22.3
F 3.8 4.1 9.4
G 3.4 3.6 8.3
BamHIfragment
A 55.4 54.0 124.3
B 16.6 18.2 41.9
C 13.9 13.4 31.7
D 8.6 8.5 19.5
E 5.6 5.9 13.5
HindIIIfragment
A 22.2 22.4 51.5
B 15.1 15.0 34.5
C 11.4 11.3 26.0
D 10.3 9.4 21.6
E 6.5 6.9 15.9
F 6.4 6.1 14.0
G 5.6 5.7 13.1
H 10.5 5.2 12.0
I J5.2 12.0
J 5.0 5.0 11.5
K 4.7 4.5 10.4
L 2.4 3.4 7.8
aDetermined from the distributionof3Pradioactivityon
eachfragment.Gelsweresliced into1-mmslices, and
radio-activitywascounted. EcoRIAandBfragmentsandHindIII HandIfragmentswere notcompletely resolvedon electro-pherograms, and the numberof counts in bothfragmentswere addedtogether.
bDetermined from mobility on agarose gel. Molecular
weightwasestimated fromrelativemobilityofeachfragment on agarose gelelectrophoresis, andthe percentage ofviral genome was calculated fromthe molecular weightof each fragment, using2.4x107forthatof whole BAV3 DNA.
x10 6
20-10
6 a
4
LM
~~~~c
,012_ C D
E
:
11XI.*o : 0.2 0.6 1.0 1.2
0.6 Mobility
FIG. 2. Relationship between mobility and molec-ularweight. In this experiment, 1.29o agarose gel was used. EcoRIfragments (A toF) and HindIII
frag-ments(a-d)ofhuman adenovirus type 2 DNA were
usedasmolecular weight markers.Molecular weights of these markers were adoptedfrom the values re-ported by Petterssonetal. (18) andDunn andHassell
(4).
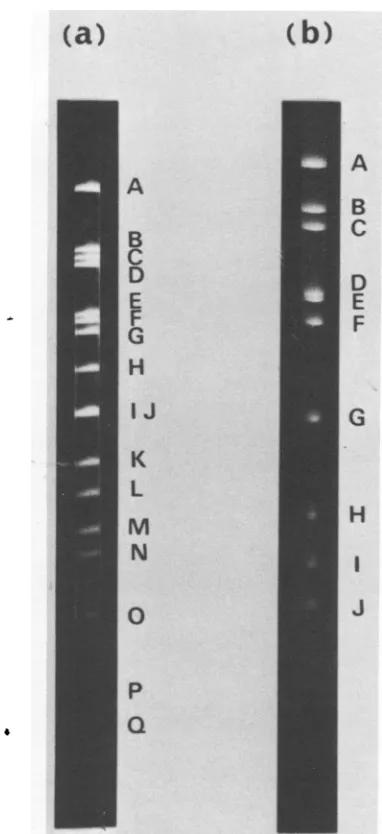
fragments produced bytwodifferent restriction endonucleases. We applied thismethod to the combinationofEcoRIandHindIII. Successive digestion of BAV3 DNA with EcoRI and HindIII gave 17 fragment bands (A to Q) on
agarose gel (Fig. 3). Since the cleavage sitesof
both enzymesontheDNAmoleculewere differ-ent, 18fragments might be producedby double digestion with both enzymes. In our
experi-ments, one very small fragment, produced by closely located cleavage sites ofboth enzymes,
could be missed on thegel.
BAV3 DNA was cleaved with EcoRI or
HindIII, and each resulting fragment was
iso-lated from agarose gels. These fragments were
redigested with another endonuclease, i.e., EcoRIfragment with HindIII andHindIII frag-mentwith EcoRI, and the products were
ana-lyzed on agarose gels. From these results, we
examinedtherelation betweenthespecific frag-ments of EcoRI or HindIII and the
double-digestion fragments produced by additional digestion with reciprocalendonucleases (Table 2). The molecular weight of each specific frag-mentobtainedby thefirstenzyme digestionwas identical to or very close to the sum of the
molecular weightsof theproducts derivedfrom it by the second enzyme digestion (data not
shown). Accordingto thedata shown in Table 2, EcoRI and HindIII fragments could be
over-lapped throughdouble-digestion fragments, ex-cept at one site where one lost fragment of
double-digestion products mightbelinked. The orders of EcoRI andHindIIIfragments,deduced from their overlapping, were GDACB, EF and
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.500.56.244.268.570.2]enzymes; one fragment could not bedetected, as in the case of the combination of EcoRI and HindIII (Fig. 3). The relation between specific fragments of EcoRI and BamHI and the double-digestion fragments produced by further diges-tion with reciprocal enzymes is presented in Table 3. The order of BamHI fragments was
A
deduced to beAEBD,
C,
and that ofEcoRIfragments
wasascertainedtobe(GD)ACB, EF,
B thesame asinthe combination with HindIII.
[image:4.500.32.223.65.482.2]c Determination of
terminal
fragments.
TABLE 2. Relationbetween BAV3 DNA EcoRIor D HindIIIfragments and the fragmentsdouble
E digested with both restriction endonucleases
F Double-digested
Specific fragment
fjagmentEcoRIfragment
A.A,F,K
B.B,H,I,O
.C,D
D.G, E,Q
E.J, N,P F .L G
H
HindIIIfragment
A
B
C.
J D
E F
G ...
H ...
I...
J ...
K ...
L ...
A, C E,F
D,0
B G H L, P I
J
M, Q K
N
FIG. 3. Analysis of BA V3 DNA digested succes-sivelywithtworestrictionendonucleases. Conditions ofelectrophoresis were the same as in Fig. 1. (a)
EcoRIandHindIIIdigestion productson1.2%
aga-rosegel. (b) EcoRI and BamHI digestionproductson
1.2%agarosegel.
JEBKAC(DHF), GIL, respectively. Cleavage sites ofHindIIIthatyielded D,H, and F frag-mentsalllay in the EcoRIBfragment,andthe order of these three fragments could not be determined in thisexperiment.
(ii) Combination of EcoRI and BamHI. Thesameoverlapping procedurewasappliedto
the combination of EcoRI and BamHI. Ten fragment bands (A toJ) wereobtainedon
aga-rose gels by digestionof BAV3 DNAwith both
TABLE 3. Relation between BA V3 DNA EcoRI and BamHIfragments and thefragments double digested with both restriction endonucleases
Specificfrag-inent Double-digested
Specific fragment
fragnent EcoRIfragmentA ... A
B ... C,F C ... E, G,J D ... B E ... D F ... H G ... I BamHIfragment
A ... A,B, E,I B ... C,J C ... D,H
D ... F E ... G
(a)
(b)
A
B
C
D
E
F
G
H
IJ
K
L
M
N
0
p
Q
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.500.244.437.202.508.2] [image:4.500.243.437.458.651.2]BAV3 DNA waslabeled with
[3H]dTTP
in its 3'-terminalregionsbyT4DNApolymerase.
The end-labeled BAV3 DNAwasdigested
withre-striction endonucleases
EcoRI, BamHI,
and HindIII, and radioactivities of theresulting
frag-ments were determined. The
radioactivity
of each fragment, expressed in terms of percentdistribution, is shown in Table 4. The radioac-tivitywasconcentratedat twoof thefragments produced by each restriction endonuclease in about equalproportions. From the data in Table
4,it isclear that the
fragments, EcoRI
E andG,
BamHI A and C, and HindIII J and
L,
werepreferentially labeled and that these
fragments
[image:5.500.57.246.250.657.2]werelocalizedatthetermini of theBAV3 DNA molecule.
TABLE 4. Distributionof
radioactivity
on the restriction endonucleasefragmentsofterminal-labeled BAV3 and DNAa
Fragment
~~Distribution
ofra-Fragment dioactivity(%)
EcoRIfragment
A .,
B .... ...
5.5
C ... 2.6
D ... 3.6
E ... 47.8
F.. .... 3.1
G.. .. 37.4
BamHIfragment
A ... 38.2
B .... 8.0
C .... 37.4
D . ... 8.5
E ... 7.8
HindIIIfragment
A .. ... 4.3
B ... 6.7
C ... 6.1
D.. .. 5.1
E ... 5.6
F ... 4.9
G ...) 5.8
H 8.0
J... 23.1
K ... 3.7
L ... .. 26.7
'Terminal-labeled
BAV3
DNA wasdigested
with restriction endonucleases EcoRI, BamHI, and HindlIl,andthedigestion products of each weresep-arated by agarose gel electrophoresis. Gels were
stained with ethidiumbromide, the gel slices contain-ingDNAbandswere cut out under UV light, and the radioactivities were counted. EcoRI A and B
frag-mentsandHindIII H and Ifragments were not
sepa-ratedongels, and for these fragments the sum of the values oftwo fragments is shown.
From these results, we concluded that the orders ofEcoRI, BamHI,andHindIII
fragments
were GDACBFE,
AEBDC,
and JEBKAC-(DHF)GIL, respectively. The cleavage mapsofBAV3 DNA with restriction endonucleases
EcoRI and BamHI weretentativelyconstructed asshown inFig.4.
DeletedregionofincompleteBAV3 DNA. The BAV3preparation contains aunique
pop-ulation of incomplete virus (16). The DNA of this virus has a deletion of 13% of the genome of
completevirus. Incomplete virus DNA was
di-gestedwith restriction endonucleases and
ana-lyzed by agarose gel electrophoresis (Fig. 5). Compared withthe digestionproducts of
com-pletevirusDNA,those ofincomplete virusDNA
lacked EcoRI E and F fragments, BamHI C
fragment, and HindIII G, I, and L
fragments.
Therefore, the deleted region ofincompletevirus DNA was positioned at the right-hand end of
completevirusDNA, asdepicted inFig.4.
On gel electrophoresis, EcoRI B, BamHI D, and HindIII F fragments of incomplete virus
DNA were shownto beaccompanied by some
minor bands adjacent to them (Fig. 5). This
implied that thesefragments had minor hetero-geneities in their sizes, that is, the incomplete virus preparation containedseveral minor pop-ulations with differentdeletion
lengths,
and the deletedregions extended into the corresponding fragments.The deletion of incomplete virus DNA was
thought tobe inone site; the presence of
par-tially deleted HindIIIFfragment in incomplete
virus DNA indicated that it was a neighborto thedeleted HindIII G fragment.
Moreover,from the analysis of partial diges-tion products of HindIII, it was resolved that HindIII C and D fragments were neighbors (data notshown).
Therefore,the order ofHindIIIfragmentswas concluded to be JEBKACDHFGIL, and from thisresult the cleavage map with HindIII was constructed as showninFig. 4.
DISCUSSION
In this paper, we showed cleavage maps of
BAV3DNAconstructedby use of three restric-tion endonucleases, EcoRI, BamHI, and
HindIII.
Ourexperiments showed that the gene nec-essary for cell transformation was located in EcoRI D orHindIII J-Efragments (10), and so we have tentatively placed these fragments at
theleft-handend ofthe maps shown in Fig. 4, as proposed in humanadenovirus(1).
Fragment orderswere determined mainlyby
overlapping ofspecific fragments produced by
on November 10, 2019 by guest
http://jvi.asm.org/
EcoRI [GI D BaffHI I
A C
A
B ,F, E
E B D C
Hindlil t1JI E I B IKI A C I D HIF G I IL 0 10 20 30 40 50 60 70 80 90 100
FIG. 4. Cleavage maps of BA V3 DNA. Maps are divided into 100 units. The deletion of incomplete BA V3
DNA is indicatedby (+-*).
differentrestrictionendonucleases. The order of
EcoRIfragments, asdeterminedbytwo differ-ent combinations of enzymes, EcoRI-HindlIl andEcoRI-BamHI,wasthesame.Three restric-tion endonucleases all cleaved BAV3 DNA at near 86 map units (Fig. 4). For this, one very
shortfragmentwasproduced,which wasmissing
on the analyses ofdouble-digestion fragments,
forboth endonucleasecombinations, making it
difficulttodeterminethefull order offragments by the overlapping method. The ordering of EcoRI andBamHIfragmentswasaccomplished bytheanalysisof thecleavage productsof end-labeled BAV3 DNA. Thefull order ofHindIII fragmentswasobtainedbytheaid of additional
observations: the analysis of partial digestion products and the examination of the deleted region ofincompletevirusDNA.
The partial digestion methodwas partly
ap-plied to the ordering of EcoRI and HindIII fragments, and the resultswereconsistent with the mapsdescribed
(data
notshown).From the
analysis
ofcleavageproducts
withrestrictionendonucleases,thedeletionof
incom-plete virus DNAcould be
mapped
tothe right-hand end oftheviralgenome. We hadpreviously
observed a
single-stranded
deletionloop
occu-pying 13%ofthewhole genome in electron-mi-croscopic observation of the
heteroduplex
mol-ecule formed betweencomplete andincomplete BAV3 DNA (16).Attheendoftheloop, a shortdouble-strandedstem(about 1% ofthe genome) was observed, and therefore incomplete virus DNApossessed the samesequence is complete
virus DNA for both ends of themolecule. When complete and incompleteBAV3 DNA were denatured and then renatured, many
sin-gle-stranded circular DNAs were seen by the electronmicroscope (16).However, in those fig-ures, no double-stranded "panhandle" struc-tures weredetected; this is not the case in human adenovirus type 18 (6), which hasthe long
in-verted terminal repetitions (about 3% of the
genome)in its DNA. Thesefacts show that both
completeandincompleteBAV3 DNAcontained invertedterminal repetitions and that thelength ofrepetitionis rathershort.
Based on thecleavage maps shown inFig. 4,
(a)
(b)
(C)
A
A B
C
D
G
A B
D
B
C
D
E E
F
H J K
FIG. 5. Analysis of incomplete BAV3 DNA di-gested with restriction endonucleases. Conditionsof electrophoresiswerethesame asinFig.1.(a) EcoRI digestionproducts on 0.9%oagarosegel. (b) BamHI
digestion products on
1.2%o
agarosegel. (c)HindIIIdigestionproductson1.2%agarosegel.
theposition of thegene essential forcell trans-formation on BAV3 DNA was studiedby(i)
Cot
analysis of the viral genome harbored in trans-formedcells (Y. Niiyama,T.Kurokawa,K. Igar-ashi, and Y.Sugino,manuscript inpreparation)
28, 1978
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.500.121.366.71.145.2] [image:6.500.250.442.179.540.2]and (ii) celltransformationby infection of cells with fragmented DNA (10). These results are shown infollowingpapers.
ACKNOWLEDGMENTS
WearegratefultoK.Fujinaga and K. Sekikawa (Cancer Research Institute, Sapporo MedicalCollege, Sapporo,Japan) and H.Shimojo and H. Ariga (Institute of Medical Science, Tokyo University, Tokyo, Japan) for gifts of restriction
en-donucleasesHindIII andBamHI, respectively. Weare also
grateful toM. Takanami (Institute for ChemicalResearch, Kyoto University, Kyoto,Japan) foragift of T4DNA polym-erase.
LITERATURE CITED
1. Bachenheimer, S.,etal. 1977. Adenovirus strand
no-menclature:aproposal.J. Virol. 22:830-831. 2. Danna, K. J., G.H.Sack, Jr.,and D. Nathans. 1973.
Studies of simianvirus 40 DNA. VII. Acleavagemapof theSV40genome.J. Mol. Biol. 78:363-376.
3. Darbyshire,J. H. 1966.Oncogenicityof bovine adeno-virustype3in hamsters.Nature(London)211:102. 4. Dunn, A. R., andJ. A. Hassell. 1977. A novel method
tomaptranscripts:evidence forhomologybetweenan
adenovirusmRNA and discretemultiple regionsof viral
genome.Cell 12:23-36.
5. Englund,P. T. 1971.Analysisof nucleotidesequencesat
3'-terminiofduplexDNA with theuseof the T4 DNA polymerase.J. Biol. Chem. 246:3269-3276.
6. Garon, C. F., K. W. Berry, and J. A. Rose. 1975. Arrangement of sequences in the inverted terminal repetition of adenovirus18 DNA. Proc.Natl. Acad. Sci. U.S.A. 72:3039-3043.
7.Greene,P.J.,M.C.Betlach,and H. W.Boyer.1974. The EcoRIrestrictionendonuclease,p.87-105. In R. B. Wicker(ed.),Methods in molecularbiology,vol. 7. MarcelDekker,New York.
8. Griffin, B. E.,M.Fried,and A. Cowie. 1974.Polyoma DNA: aphysicalmap. Proc. Natl. Acad. Sci. U.S.A. 71:2077-2081.
9.Igarashi, K., Y.Niiyama,K. Tsukamoto, T. Kuro-kawa, and Y. Sugino. 1975. Biochemicalstudieson
bovine adenovirustype 3.II. Incompletevirus. J. Virol. 16:634-641.
10.Igarashi, K., R. Sasada,T.Kurokawa,Y.Niiyama, K. Tsukamoto, and Y. Sugino. 1978. Biochemical studiesonbovine adenovirustype3. IV.Transformation
by viral DNA and DNA fragments. J. Virol. 28:219-226.
11.Kelly,T.J., Jr., and D. Nathans. 1977. The genome of
simian virus 40. Adv. Virus Res. 21:86-173.
12. Kimura, S., T.Kamogashira, F. Ota, K. Fukui, and Y. Sagara. Growth of bovine adenovirus type 3 incells cloned from a cell line of calfkidney. Tokushima J. Exp. Med. 18:33-37.
13.Lai,C.-J.,and D.Nathans. 1974. Deletion mutants of
simian virus 40generated by enzymatic excision of DNA segments from the viral genome. J. Mol. Biol. 89:179-193.
14.Larsen,S.H.,and D.Nathans. 1977. Mouse adenovirus:
growth ofplaque-purified FL virus in cell lines and characterization of viral DNA. Virology 82:182-195. 15.Mulder, C., J. R.Arrand,H. Delius,W.Keller, U.
Pettersson, R. J.Roberts, and P. A. Sharp. 1974. Cleavage maps of DNA fromadenovirus types 2 and 5 by restriction endonucleases EcoRI and HpaI. Cold Spring HarborSymp.Quant. Biol. 39:397-400.
16.Niiyama, Y., K. Igarashi,K.Tsukamoto, T.
Kuro-kawa, and Y.Sugino. 1975. Biochemical studies on bovineadenovirus type 3. I. Purification and properties. J. Virol. 16:621-633.
17.Ortin, J., K.-H., Scheidtmann, R. Greenberg, M.
Westphal, and W.Doerfler. 1976. Transcription of the genome of adenovirus type 12. III. Maps of stable RNA fromproductively infected humancellsand abor-tively infected and transformed hamstercells.J. Virol. 20:355-372.
18. Pettersson, U.,C.Mulder, H. Delius, and P. A. Sharp. 1973. Cleavage of adenovirus type 2 DNA into six unique fragmentsby endonuclease R.RI. Proc. Natl. Acad. Sci. U.S.A. 70:200-204.
19.Sambrook,J., J.Williams,P. A.Sharp, and T.
Grod-zicker. 1975.Physical mapping of temperature-sensi-tive mutation ofadenoviruses. J. Mol. Biol. 97:369-390.
l9a.Sekikawa, K., and K.Fujinaga. 1977.Cleavage maps
of human adenovirus type 7 DNA by restriction endo-nuclease HindIII andEcoRI. Virology 82:509-512. 20. Sharp, P. A., B. Sugden, and J. Sambrook. 1973.
Detection of two restrictionendonuclease activities in Haemophilusparainfluenzae using analytical agarose ethidium bromide electrophoresis. Biochemistry 12:3055-3063.
21. Tibbetts, C. 1977.Physicalorganization of subgroup B
human adenovirus genomes. J. Virol. 24:564-579.
22. Wilson, G.A.,and F. E.Young. 1975.Isolation of a
sequence-specific endonuclease (BamI) from Bacillus
amyloliquefaciensH.J. Mol. Biol. 97:123-125.