JOURNAL OFVIROLOGY,Apr. 1992,p. 1971-1976 0022-538X/92/041971-06$02.00/0
Copyright © 1992,American SocietyforMicrobiology
Gliotoxin: Inhibitor of Poliovirus
RNA
Synthesis
That
Blocks
the Viral RNA Polymerase
3DPo1
PEDRO L.RODRIGUEZAND LUIS CARRASCO*
Centro deBiologiaMolecular, Universidad Aut6noma, CantoBlanco, 28049Madrid, Spain Received23 September1991/Accepted3 December1991
Themodeof action of gliotoxinagainstpoliovirushas beenanalyzedindetail. Thisfungalmetabolite inhibits theappearanceofpoliovirus proteinswhen present from thebeginningofinfection but hasnoeffectonviral
translation when added atlate times. In agreementwith previous findings, this toxin potently inhibited the incorporation of[3Hluridineintopoliovirus RNAsoonafteritsadditiontothe culture medium.Analysisof the synthesis of poliovirus plus-orminus-strandedRNA inthepresenceofgliotoxinsuggests that thiscompound effectively hampered bothprocesses.This resultcontrastswith the mode of action of other inhibitors ofpoliovirus RNAsynthesis, suchasguanidineorflavones, thatselectivelyblockplus-strandedRNAsynthesisand suggests that the target ofgliotoxindiffers from the target ofguanidine, i.e., poliovirus protein2C.Indeed, gliotoxinwas found to be a potent inhibitor ofpoliovirus RNA synthesis in cell-free systems, using membranous crude replication complexes,areaction that isnotblockedby guanidineorRo 09-0179.Moreover,invitroactivityof
thepurified polioviruspolymerase3D°"'wasefficientlyinhibitedby gliotoxin.These resultsindicate that this toxin
actsonthe poliovirus polymerase3D1"',providingthe firstdescriptionofaninhibitor of this viralenzyme.
Picornaviruses are among the simplest and best-known animalviruses in molecularterms(10). Despitetheir simplic-ity,severalaspectsof theirreplicative cycleremainobscure (3, 11, 16, 25). Hopefully, detailed studies of the mode of action of selective inhibitors ofpicornavirus replicationwill provideabetterunderstandingof thoseprocesses. Although the main steps in the replication of poliovirus RNA have been delineated, there are still some reactions that remain puzzling (25). Thus, it is known thatoncethegenomicRNA is free in thecytoplasm, it is first translatedtogiverisetoan array of viral proteins (3, 11, 16). An unknown number of
these viralpolypeptides usetheplus-stranded genomicRNA as a template to make a complementary minus-stranded
RNA(19, 25). Thisnegative-stranded RNA isused, inturn,
as atemplatetosynthesizemorecopiesofpositive-stranded RNA molecules that can fulfill several requirements, i.e., they can be used as mRNAs to make viral proteins or as templates to make more copiesofnegative-stranded RNA, ortheycanbeencapsidatedtogenerate maturevirions(19, 25).
The virus-encoded RNA-dependent RNA polymerase, knownasprotein 3DPo1,is ableto elongate already initiated plus- or minus-stranded RNA molecules (24, 25). It was suggested that thegenome-linkedprotein VPg (viral protein 3B),wasusedas aprimertoinitiatenewchainsof viral RNA (19, 20). However, recent evidence indicates that this
pro-tein iscovalentlylinkedtothe RNAwell after the initiation event (22). Therefore, the exact mechanism of initiation of picornavirus RNA synthesis still remains controversial. Moreover, viral RNA replication complexes are tightly
bound to membranes, and the inhibition of membrane for-mation leadstoimmediatearrestofpoliovirus RNA synthe-sis(8).Perhapsviralprotein2Coritsprecursor2BC playsa
part in thecoupling between the RNAreplication complex and the membranous vesicles that proliferate throughout infection (1). A role for protein 2C in poliovirus RNA replication was suspected on the basis that the selective
*Correspondingauthor.
inhibitor ofpoliovirusRNA synthesis, guanidine, acted on protein 2C (17, 25). Apartfromthatfinding,theexactroleof protein 2C in RNAsynthesisis obscure. It isevenpossible
that other viralproteins,suchas2Bor3A,arealsoinvolved
in the replication of viral genomes (19, 25). Therefore, a
better understanding of the mode ofaction of inhibitors of poliovirus RNAreplicationcanhelptounravel thisprocess.
In addition to guanidine, which has been the classic inhibitor ofpoliovirusRNAsynthesis (21),otheragentshave been reported to selectively block the step of viral RNA replication. Thus, some natural flavones, such as 3-meth-ylquercetin (4, 5),orRo 09-0179 (7, 9),selectively block the synthesis of plus-stranded RNA without inhibiting minus-strandedRNAsynthesis at concentrations 1,000-fold lower than those forguanidine (7). 2-(a-Hydroxybenzyl)-benzimid-azole (HBB) (6), penicillamine (14), and gliotoxin (15) also block viral RNAreplication, but the detailedmechanism of action of thoseagentsis stillunknown. High concentrations ofpenicillamine (above 7 mM)orHBB (above 0.1 mM)are requiredtoblockpoliovirus (6, 14). Gliotoxin andanumber
ofrelated compounds,either from naturalsources or chem-icallysynthesized, arethemostpotent agentsthat interfere with poliovirus RNA synthesis (15, 23). We undertook an
analysis of the mode of action ofthis toxin, andthe results presented suggest that it differs from the effect of known poliovirus RNA inhibitors, such as guanidine and the fla-vones.
MATERIALS ANDMETHODS
Cells and viruses. HeLa cells were grown in Dulbecco
modified Eagle's medium supplemented with 10% newborn calfserum (GIBCO, Grand Island, N.Y.) andincubated at
37°C ina5% CO2 atmosphere. Poliovirus type 1 (Mahoney strain) (American Type Culture Collection) was grown in
HeLa cells. Cell culture and virus titration were as
previ-ouslydescribed(7).
Compounds. 4'-5-Dihydroxy-3-3',7-trimethoxyflavone
(Ro-1971
Vol.66, No. 4
on November 9, 2019 by guest
http://jvi.asm.org/
1972 RODRIGUEZ AND CARRASCO
+
POLIOVIRUS
1 2345 6 7
-.M-- --2C
-.
1 2 345 6 7
B
+ POLIOVIRUS MocKinf ected cells
0 h.p.i. 4 h.p.i. 0 h.pi
addption
00.3061.5 3 0 030.61.53 00.30.61.53 GTX (pM)
Pi-
IIrn=_
4- 44
iim1 g.t"_
1-0
*0_ s t a"IC=_as
C am*"_am
" At NM
li~ ~~, _ _
-Act'in
1C-_a
FIG. 1. Effect of gliotoxinontranslation. (A) Timecourseof the proteins synthesized in poliovirus-infected cells minus (-)orplus(+)
1.5 ,uM gliotoxin (GTX) fromzerotime. HeLa cellsgrownin 24-well plateswereinfected with poliovirusatanMOIof50PFUpercell. After
a60-min adsorption period, cellswerelabeledeveryhour with 20 ,uCi of [35S]methionineperml for 1 h. Proteinswereanalyzedon a15%
polyacrylamide-SDS gel asindicated in Materials and Methods. (B) Analysis of the proteins synthesized in poliovirus-infected cells or
mock-infectedcells between4to5hp.i. Different concentrations of GTXwereaddedat0or4h p.i.
09-0179)waskindly provided byH. Ishitsuka and K.Yokose, Nippon Roche Research Center (Kamamura, Japan). Glio-toxin, guanidine, and dactinomycin were purchased from Sigma Chemical Co., St. Louis, Mo.
Virus infection ofcell monolayers. HeLa cells were
in-fected with poliovirus (-1 h) in Dulbecco modified Eagle's medium supplemented with 2% newborn calfserum. After incubation at 37°C for 1 h, unattached viruswas removed and fresh mediumwas added(zero hours).
Polyacrylamide gel analysisofproteins.At the times indi-cated,cellswereincubated in methionine-free medium in the
presence of 20 ,uCi of [35SJmethionine (1,450 Ci/mmol; Amersham) perml. One hour later the radioactive medium
was removed, the cell monolayer was washed with phos-phate-buffered saline, and cellswere collected in 200
RI
of buffer containing 62 mM Tris-HCI (pH 6.8), 2% sodium dodecyl sulfate (SDS), 0.1 M dithiothreitol, 17% glycerol, and 0.0024%bromophenolblue. Eachsamplewassonicatedto reduce viscosity and heated at 90°C for 5 min. Five microliterswasappliedtoa15%polyacrylamide gelandrun overnightat 100 V. Fluorographyof thegelwascarriedout with 2,5-diphenyloxazole-dimethyl sulfoxide (20% wt/vol).
Estimation of uridine incorporation in RNA. HeLa cells
grown in 24-well plates were infected with poliovirus at a multiplicityof infection (MOI)of 50 PFUpercell. Dactino-mycin (5
jig/ml)
wasadded atzero hours. Everyhour after infection, 10 ,uCi of [3H]uridine (25 to 30 Ci/mmol; Amer-sham)permlwasadded. Thelabelingmediumwasremovedafter 1h, and the cellmonolayerwastreated with 0.5 ml of
5% trichloroacetic acid, washed twice with ethanol, dried under an infrared lamp, and dissolved in 200 ,u of 0.1 N NaOH-1% SDS. Samplesof 150plwerecounted inaliquid
scintillation spectrophotometer.
Synthesis of labelled RNA probe. The poliovirus
strand-specificRNAprobewasgenerated byinvitrotranscription ofafragmentofpoliovirus cDNA encompassing nucleotides 2099to4600, subcloned inaBluescriptvector (Stratagene). The poliovirus cDNAsequence was flanked by T7 and T3
promoters. Using T7 (New England Biolabs) or T3 RNA polymerase (Stratagene), weobtained RNAprobes specific
tothe viral minus-orplus-stranded poliovirus RNA,
respec-tively.Theprobeswerelabeled with[32P]CTP (400 Ci/mmol; Amersham).
62~~~~~~~~~2
A a B
0
so 480
.20-ol . . ...__. L.
0 1 2 3 4 5 6 7 8 910 0 0.5 1 1.5
TIME (h.p.l.) GilotoxinILN
FIG. 2. Inhibition ofpoliovirus and cellular RNAsynthesis by
gliotoxin. (A)TimecourseofpoliovirusRNAsynthesis.Conditions
of infection and analysis of RNA synthesiswere as described in
Materials and Methods. Poliovirus-infected cells in the absence of
inhibitor(O)orin thepresenceof 1.5p,M gliotoxinfromzerotime
(0) or at 1.5 (Cl), 3.5 (-), or 5.5 (A) h p.i. Pulse-labelingwith [3H]uridinewascarried outevery hourp.i. (B)Effect of different
concentrations ofgliotoxinonRNAsynthesisin mock-infected cells
in the absence ofdactinomycin (0)orinpoliovirus-infectedcells in
thepresenceofdactinomycin (-). RNAsynthesiswasdetermined
between 4to5 hp.i.Gliotoxinwasaddedat1.5 hp.i.
A
GTX 1.5 pM
TIME (h.p.i.)
Actin
J. VIROL.
Asz6 AA m
on November 9, 2019 by guest
http://jvi.asm.org/
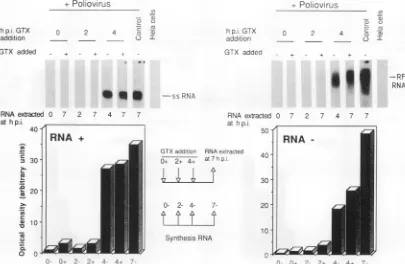
[image:2.612.115.516.82.318.2] [image:2.612.322.550.503.627.2]GLIOTOXIN BLOCKS POLIOVIRUS 3DPo' 1973 + Polhovirus
hp.i.GTX 0 2 4
addition
- Poiiovirus
0
GTX added
-5
hp.i. GTX 0 2 4 o
addition __
a,
v
0
-a:
GTX added _- +
RNAexrcted 0 7 2 7 4 7 7 at hp.i.
,_ AI
40-3
30- L-W
20--1
ce
, 10 cU ID
-ssRNA
RNA extracted 0 7 2 7
ath pi.
GTX addition RNA extracied
0+ 2+ 4+ at7hpi
L IA
501
40
-
30-20
0- 2- 4-
7-L 7-LJ
f
Synthesis RNA 10
S-I
-RFRNA4 7 7
RNA
-O
O-O- 00- o0 2- 2- 4- 4+ 7- 0- 0- 2- 2- 4- 4+
7-FIG. 3. Effectofgliotoxin(GTX)onthesynthesisofpoliovirusplus-orminus-stranded RNA. HeLacellsgrownin 60-mm-diameter dishes wereinfectedwith poliovirus at50PFUpercell. Total RNAwasextractedfromuntreated(-) cellsat0, 2, 4,or7 hp.i.Total RNAfrom cells treated with 1.5p.M gliotoxin (+)wasextractedat7 hp.i. The inhibitorwasaddedatthe indicatedtimes,andincubation continued until the 7thh. TheRNAwasextracted andanalyzedbyNorthernblottingasindicated inMaterials andMethods. Thesameblotwashybridized
topositive-stranded(RNA-)and negative-stranded(RNA+) poliovirus riboprobes.
Northern (RNA) blot
analysis
of poliovirus RNA. HeLa cellsweregrownin60-mm-diameterdishes and infected withpoliovirus at an MOI of 50 PFU per cell. At the indicated timesafterinfection, the cellswerelysedinabuffer
contain-ing10 mMTris-HCl (pH7.8), 1 mMEDTA, 150 mMNaCl,
and 0.65% Nonidet P-40. After removing nuclei by low-speedcentrifugation,supernatantsweremixed withanequal
volume ofabuffercontaining20 mM Tris-HCl (pH7.8),20 mMEDTA, 350 mM NaCl, and 1% SDS. Total RNAwas
extractedwithphenolandprecipitatedwithethanol.
North-ern blot analysis with a radiolabeled probe was performed (7). The quantitation was done with a 3,000-A computing
densitometer (Molecular Dynamics).
In vitro synthesis of poliovirus RNA. HeLa cells were
infected with poliovirus at an MOI of 200 PFU per cell, incubatedat37°C, and harvestedat 7 hpostinfection (p.i.). Preparation of the crude membrane fraction and its treat-ment with DEAE cellulose were performed as previously
described(20, 25).Invitro RNAsynthesiswascarriedoutin
a reaction mixture containing 50 mM N-2-hydroxyethylpi-perazine-N'-2-ethanesulfonic acid (HEPES)-KOH (pH 8),
3.5 mMmagnesiumacetate, 50mM
KCI,
10pg
of dactino-mycin per ml, 2.5 mM phosphoenol pyruvate, 2 ,ug of pyruvate kinase, 10 p,g of DEAE-cellulose-treated mem-branes, and compounds at the concentrations indicated in each experiment. This mixturewasincubated for 30min at 4°C. Then, 50 p,Ci of[3H]ATP
(56 Ci/mmol) per ml-1 mM UTP-1 mMCTP-1 mM GTP-100 pMATPwasadded, and incubation continuedat30°C for various periods of time. The reactionwas stoppedby the addition of 10% trichloroacetic acidat4°C.The RNAwascollectedonWhatmanGF/C fiberfilters, and theradioactivity incorporated in RNAwas deter-mined inascintillation counter.
Enzymatic assay of poliovirus RNA polymerase. Purified poliovirus RNApolymerase was kindly provided by S. J. Plotch (Lederle Laboratories, Pearl River, N.Y.). Poly(A)-dependentoligo(U)-primed poly(U) polymerase activitywas
measured at 30°C for 60 min as previously described (18),
with the modifications that dithiothreitol (DTT) was not added and[3H]UTPwas used asthe labeled nucleotide (45 Ci/mmol;Amersham).
RESULTS
Effect of gliotoxin on poliovirus protein synthesis. Glio-toxin, a fungal metabolite, inhibits the growth of several animal viruses, including picornaviruses. Earlier evidence indicated that it was a selective inhibitor of picomavirus
RNAsynthesisand had lesseffectonprotein synthesiswhen added laterduringinfection(15, 23). However,thosestudies
onlyrelied upon theincorporationofradioactiveprecursors into trichloroacetic acid-precipitable material. Thus, itwas ofinteresttoexamine the effects ofgliotoxinoncellular and viral protein synthesis and to analyze the proteins synthe-sizedbypolyacrylamide gel electrophoresis. Concentrations ofgliotoxin above 0.6 ,uM are effective in the inhibition of viralprotein synthesis (Fig. 1B)if presentfrom thebeginning
ofinfection, butno effectwasobservedonviral translation when gliotoxinwas added 4hp.i. Gliotoxin concentrations of 3 ,uM had almostno effecton cellularprotein synthesis. On theotherhand,although gliotoxin effectivelyreducedthe
synthesis of viral proteins, itwas ineffective inpreventing VOL. 66,1992
on November 9, 2019 by guest
http://jvi.asm.org/
[image:3.612.101.506.82.346.2]1974 RODRIGUEZ AND CARRASCO
+Poliovirus -Pohiovirus
.Ej C)
h p.i. compound 0 2 4 0 a,
addition - C} I
Compound - +G±R - +G+R - +G +R
-added
* _M -ssRNA
RNAextracted 0 7 7 2 7 7 4 7 7 7
at hp.f 30-"
- RNA+
vw a
E!
20-CA 10 a
U,_
0.2
o o
0-OGOR2-2G22R4-4G4R
7-h p.i. compound 0 2 4 o
c-addition
Compound +G+R - GGRRG -R
added
,A&
_ _RNA extracted at hp.i. 50'
40
30
20
10
0
..v...
0 7 7 2 7 7 4 7 7 7
RNA
-O-OGOR2-2G2R 4-4G4R
7-- RF RNA
FIG. 4. Effect ofguanidineand Ro 09-0179 onthesynthesis ofpoliovirus plus- or minus-stranded RNA. The experiment was carried out asdescribed in thelegendtoFig. 3, exceptthat3mMguanidine (+G)or3,uM Ro 09-0179(+R)wasadded instead ofgliotoxin.
the shutoff of host translation induced by the infection of
poliovirus athigh multiplicities (Fig. 1A). These results are
similartothosefound for other inhibitors ofpoliovirusRNA replication (5, 7).
Actionofgliotoxin on viralRNAsynthesis.Figure 2 shows the ability ofgliotoxin to block poliovirus RNA synthesis
when addedat different timesp.i. When gliotoxin (1.5 ,uM) wasadded at zero time or 1.5 hafter infection, viral RNA
synthesis was completelyblocked. Addition of thedrug at 3.5 h p.i. significantly reduced the peak of viral RNA
synthesis observed at 5 h (Fig. 2A). A concentration of
gliotoxin of 0.6 ,uMinhibited poliovirus RNA synthesis in infected cellsby80%,whereas cellular RNAsynthesiswas only slightly affected by 1.5 ,uM gliotoxin. These results agreewell with previous
findings
ongliotoxin activity.
Ina similar assay, the flavone Ro 09-0179 was twofold lessefficacious,whereasconcentrations of 200 ,ug of HBB per ml or1.5 mMguanidinewererequiredtoblockpoliovirusRNA
synthesis by 80% (results notshown).
We foundrecentlythatflavones, suchas
3-methylquerce-tin or Ro 09-0179, were selective inhibitors of poliovirus
plus-stranded RNA synthesis, whereas the formation of minus-strandedRNA was evenstimulated in the presence of Ro 09-0179 (7). To test the action ofgliotoxin on plus- or minus-stranded RNA synthesis, specific riboprobes were made and the formation ofplus-orminus-stranded poliovi-rusRNA wasmeasuredthroughout infection.Inparallel,the inhibitorgliotoxinwas added at different timesp.i. and the RNAwas extracted at the end ofpoliovirus infection (7 h
p.i.). Figure3 shows thatgliotoxinefficientlyinterfereswith thesynthesisof bothplus-and minus-stranded RNAchains when present at any timep.i. It is ofparticular interestto focusonthe inhibition inducedbygliotoxinatthe 4th h
p.i.
Atthat time mostof the
plus-stranded
RNAs havealready
been synthesized, but the increase in plus-stranded RNA from 4 to7 h ishampered bygliotoxin. On the otherhand,
mostviral minus-stranded RNAismade from the 4th tothe
7th hp.i. This increase innegativeRNAis also blocked by the toxin. In a similar assay, guanidine had no effect on negative RNAsynthesis, whereas Ro 09-0179increased its formation by 200%(Fig. 4).
Inhibition of in vitro poliovirus RNAsynthesis. Noneof the
available inhibitors ofpoliovirusRNAsynthesisis effective
in cell-free systems. A system that faithfully synthesizes
both poliovirus plus- and minus-stranded RNA in vitro has been developed by purifying the replicative complexes
bound to membranes, the so-called crude replication com-plex (CRC) (25). Curiously enough, gliotoxinwas apotent inhibitor of this reaction (Fig.
5A).
Gliotoxin (750p.M)
inhibited RNAsynthesisin CRCby60%,whereasguanidine
and Ro 09-0179 had littleor no effect
(Fig. SA).
The RNA made in these systems by crude replicative complexes mainlycomesfromelongationcarriedoutbythepolymerase
3DP"°,
although initiation ofnewchains in such systems has beendescribedpreviously (19, 25). Thus, althoughgliotoxin
is the first compound shown to inhibit
poliovirus
RNA synthesisin theCRC, itcannotbeconcluded that itactson polymerase3DP"°.
In orderto testthispossibility,
theeffect of gliotoxin was assayed in cell-free systems that usedpurified 3D polymerase
(18). Figure
6A, shows that theincorporation of [3H]UTPdirected bypoly(A) in the pres-enceofoligo(U)is alsoblockedbygliotoxin.Theinhibition of thisreactionbygliotoxinismorepotentthan theblockade ofthe CRC. Thus, 150 ,uMgliotoxinblocked the
incorpora-tion of
[3H]UTP
in this assayby 80%.On the otherhand,
this inhibitionwasspecificforgliotoxin,since otherinhibitors ofpoliovirus RNAsynthesis, such asguanidineor R09-0179,
had no effect (Table 1). The presence of a reducing agent J. VIROL.
on November 9, 2019 by guest
http://jvi.asm.org/
[image:4.612.115.511.76.352.2]GLIOTOXIN BLOCKS POLIOVIRUS 3DP°' 1975 E
0)
i
E 120
0
1004
0
4-
20-!
n-0 20 40 60 0 150300400600750900
inme nln Glktoxln pi
FIG. 5. Effect of gliotoxin, guanidine,and Ro 09-0179oninvitro
synthesis of poliovirus RNA. (A) Preparation ofthe crude
mem-brane fraction and in vitro RNA synthesis were carried out as
described in Materials and Methods. The DEAE-cellulose-treated membraneswereincubatedat30°Cfor variousperiodsof time inthe absence of inhibitor (0)orinthepresenceof750 p.Mgliotoxin(0), 30 mM guanidine (E), or 750 puM Ro 09-0179 (-). (B) Effect of different concentrations of gliotoxinoninvitro RNAsynthesis. such as DTT prevents the inhibition of poliovirus RNA synthesisinintact cells(23), suggestingthat the blockadeby gliotoxin involvestheformation of disulfidebridgesbetween gliotoxin and essential sulfhydryl groupson theviral poly-merase. The data of Table 1 indicate that the presence of DTT in the cell-free system partially reduced itsinhibitory potential, in agreement with the idea that reducing the sulfhydrylgroups abolishes theactivityofgliotoxin (23).
DISCUSSION
Elucidating the exact steps involved in replication of poliovirus RNA is an important goal. Although the major
biochemical events are known, there are still a number of
stepsthat remain obscure(3, 19,25). Thus, theexactmode of initiation of viral plus-andminus-stranded RNA synthe-sis, the coupling between membranes and replication com-plexes, and the identification of host proteins, if any, in-volved in this process need to be clarified (8, 19, 22). Elucidation ofsomeof theseprocessesatthemolecularlevel will be facilitatedbythe use ofselective inhibitors of viral RNAsynthesis. Inordertousethese inhibitorsproperlywe
need first to carry out detailed studies of their mode of action.
Few inhibitors ofpicornavirus RNA synthesis are
avail-I
0 20 40
Tlm min.
60 0 150 300 450 600
Glktoxinpm
FIG. 6. Effect ofgliotoxinoninvitro poly(U) polymerase
activ-ity.(A) Enzymaticassayofpoliovirus RNApolymerasewascarried
out as described in Materials and Methods. Poly(U) polymerase
activitywasmeasured forvariousperiods of time inthe absence (0)
orpresence(-)of600,uM gliotoxin. (B) Effect of different
[image:5.612.64.298.78.203.2]concen-trationsofgliotoxinonpoly(U) polymerase activity.
TABLE 1. Effect ofinhibitors of poliovirus RNAsynthesis oninvitro
3DPo0
activity'[3H]UTP incorporated
Incubationconditions
cpm %Control
Time, 0 min 373 0
Time,60 min + 5 mM DTT 20,063 95
Time, 60 min
Minus DTT 21,075 100
Minus
3DPO1
524 0.7Minuspoly(A) 423 0.3
Minus oligo(U) 2,134 9
Plus 2% dimethyl sulfoxide 20,468 97
Plus 30 mM guanidine 16,973 80
Plus 750 ,uM Ro 09-0179 22,676 107
Plus 600 ,uM gliotoxin 1,697 6
Plus 600 ,uM gliotoxin + 5 mM 7,291 33 DTT
a Poly(U) polymerase activitywasmeasured at30'Cfor60minasdescribed
in thelegendtoFig. 6. Inhibitorswereaddedat zerotime. Dataarethe means
ofduplicate samples.
able at present(2). Only the modes of action of guanidine and some flavones arepartially known (7, 21). These com-pounds inhibit polioviruspositive-stranded RNA synthesis (7). Flavones stimulate the synthesis of minus-stranded RNAininfected cells byastill unknownmechanism(7). The poliovirus protein target of flavone's action still remains to beelucidated. In fact, theonlyknown targetforaninhibitor ofpoliovirus RNA synthesis is protein 2C, which is the site of action of guanidine (17). This constitutes the major evidence implicating this protein in viral RNA synthesis.
Otherwise, the exactrole ofprotein 2C in poliovirus RNA replication is still puzzling (13, 25). This protein may be involved in thephysicalconnection between membranes and the viralreplicationcomplexes(1). Of interest in this respect is the recent discovery of helicase activity in a potyvirus
protein similartothatofpoliovirus2C(12).
Gliotoxin iscertainlythemostpowerfulinhibitor of polio-virus RNA synthesis in cultured cells(15, 23). The action of gliotoxin differs from the effect of guanidine and flavones, since thesetwoagents preferentiallyinhibit thesynthesisof
poliovirus positive-stranded RNA (7). On the contrary,
gliotoxin does notshow this selectivity and interferes with thesynthesisofbothminus-andplus-stranded RNA synthe-sis.Our studies of the effect of gliotoxin in cell-free synthesis
clearly show that it is a potent and unique inhibitor of
poliovirus RNA synthesis in these systems. Moreover,
gliotoxin inhibited the activity of purified
3DP°o.
Therefore, two agents that block two different poliovirus proteins involved inRNAreplicationare nowavailable: acompound that interferes with the activity of poliovirus protein 2C,guanidine, and another one that blocks protein 3DPoI, gliotoxin. Futurestudies of the action and the characteriza-tionofthe target for theother inhibitorsofpoliovirus RNA synthesis will complete ourunderstanding of the effectsof these agents.
ACKNOWLEDGMENTS
The expert technical assistance of M. Chorro and M. A. Sanz is acknowledged.We thankS. J. Plotch(Lederle Laboratories, Pearl River, N.Y.)forkindly providingpurified3D°".
P.L.R. isthe holder of an F.P.I. and of a Residencia de Estudi-antes,Ayuntamientode Madridfellowship. Plan Nacional, project numbersB1088-0233andPB90-0177,andFundaci6n Ram6nAreces areacknowledgedforfinancialsupport.
B
E 302
20 B
1625
a 20
15
I
5
0
VOL. 66,1992
on November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.315.560.100.251.2] [image:5.612.60.298.571.672.2]1976 RODRIGUEZ AND CARRASCO REFERENCES
1. Bienz, K., D. Egger, M. Troxier, and L. Pasamontes. 1990. Structuralorganization ofpoliovirus RNAreplication is medi-ated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163.
2. Came, P. E., and L. A. A. Caliguiri. 1982. Chemotherapy of viral infections.Springer-Verlag, Berlin.
3. Carrasco, L., M. J. Otero,andJ.L. Castrillo. 1989. Modifica-tions ofmembranepermeabilitybyvirus infections. Pharmacol. Ther.40:171-212.
4. Castrillo, J. L., and L.Carrasco. 1987.Action of 3-methylquer-cetinonpoliovirus RNA replication. J. Virol. 61:3319-3321. 5. Castrillo, J. L., D. Vanden Berghe, and L. Carrasco. 1986.
3-methyl-quercetin isapotent and selective inhibitor of polio-virusRNA synthesis.Virology152:219-227.
6. Eggers, H. J.1982.Benzimidazoles,p.377-418.In P. E.Came, and L. A. Caliguiri (ed.), Chemotherapy of viral infections. Springer-Verlag, Berlin.
7. Gonzfilez, M. E., F. Martinez-Abarca, and L. Carrasco. 1990. Flavonoids: potent inhibitors of poliovirus RNA synthesis. Antibiot. Chem. Chemother. 1:203-209.
8. Guinea, R., and L. Carrasco. 1990. Phospholipid biosynthesis and poliovirus genome replication: two coupled phenomena. EMBO J.9:2011-2016.
9. Ishitsuka, H., C. Ohsawa, T. Ohiwa,I.Umeda, andY. Suhara. 1982.Antipicornavirus flavoneRo 09-0179.Antimicrob.Agents Chemother. 22:611-616.
10. Koch, F., andG. Koch.1985. The molecularbiology of poliovi-rus. Springer-Verlag, Vienna.
11. Krausslich,H., and E.Wimmer.1988.Viralproteinases.Annu. Rev.Biochem. 57:701-754.
12. Lain, S.,M. T.Martin,J. L.Riechmann,andJ.L.Garcia.1991. Novel catalytic activity associated with positive-strand RNA virus infection: nucleic acid-stimulated ATPaseactivity of the plumpoxpotyvirushelicaselikeactivity. J.Virol.65:1-6. 13. Li, J.-P., and D. Baltimore. 1988. Isolation ofpoliovirus 2C
mutants defective in viral RNA synthesis. J. Virol. 62:4016-4021.
14. Merryman, P., I. A. Jaffe, and E. Ehrenfeld. 1974. Effect of
D-penicillamineonpoliovirus replication inHeLacells.J. Virol. 13:881-887.
15. Miller, P. A., K. P. Milstrey, and P. W. Trown.1969. Specific inhibition of viral ribonucleic acid replication by gliotoxin. Science 159:431-432.
16. Palmenberg,A.C.1990. Proteolytic processing of picornaviral polyprotein.Annu.Rev. Microbiol.44:603-623.
17. Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and-dependentpoliovirusmutantsfromcloned cDNA. Mutations inpolypeptide 2Caredirectly responsible foraltered guanidinesensitivity.J.Virol. 60:793-796.
18. Plotch, S.J.,0.Palant, and Y.Gluzman. 1989. Purificationand properties of poliovirus RNApolymerase expressed in Esche-richiacoli. J. Virol. 63:216-225.
19. Semler, B. L., R. J. Kuhn, and E.Wimmer.1988.Replication of thepoliovirusgenome, p.23-48. In E.Domingo, J. J. Holland, andP.Ahlquist(ed.),RNAgenetics.CRC Press,BocaRaton, Fla.
20. Takegami, T., R. J. Kunh, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451.
21. Tershak, D. R.,F. H.Yin, and B. D. Korant. 1982.Guanidine, p. 343-376.In P. E. Came and L. A. Caliguiri (ed.), Chemo-therapyof viralinfections. Springer-Verlag, Berlin.
22. Tobin, G. J., D. C. Young, and J. B. Flanegan. 1989. Self-catalyzedlinkage of poliovirus terminal protein VPgto poliovi-rusRNA.Cell 59:511-519.
23. Trown, P. W., andJ. A. Bilello. 1972.Mechanism of action of gliotoxin: elimination of activity by sulphydryl compounds. Antimicrob. AgentsChemother. 2:261-266.
24. Van Dyke, T. A., andJ. B. Flanegan. 1980. Identification of poliovirus polypeptide P63as asoluble RNA-dependentRNA polymerase.J. Virol.35:732-740.
25. Wimmer, E., R. Kuhn, S. Pincus, C. F. Yong, H. Toyoda, M.J. H. Nicklin, and N. Takeda.1987. Moleculareventsleading
topicornavirusgenomereplication. J. Cell. Sci. 7(Suppl.):251-276.
J.VIROL.
![FIG. 2.gliotoxin.ofbetweenMaterialsinhibitorconcentrationsthe(0)in[3H]uridine the infection Inhibition of poliovirus and cellular RNA synthesis by (A) Time course of poliovirus RNA synthesis](https://thumb-us.123doks.com/thumbv2/123dok_us/1309183.84195/2.612.115.516.82.318/gliotoxin-ofbetweenmaterialsinhibitorconcentrationsthe-infection-inhibition-poliovirus-synthesis-poliovirus-synthesis.webp)