May 0022-538X/83/050405-08$02.00/0
CopyrightC1983,AmericanSocietyforMicrobiology
"Endless" Viral
DNA
in Cells Infected with Channel Catfish
Virus
JOStCEBRIAN,DANIELLE BUCCHINI,tAND PETERSHELDRICK*
Institutde RecherchesScientifiquessurle Cancer,94802Villejuif Cedex, France
Received 7September 1982/Accepted 21 January 1983
Thestate of intracellular viral DNA in cells infected with channel catfish virus
has been studied by the Hirt selective extraction procedure and by restriction
endonucleasedigestion. The sedimentation properties and restriction patternsof
viral DNAin the Hirtsupernatantfraction indicate that themajority, ifnotall,of
the DNAis in the form of linear unit-length (Mr 85 x 106) molecules. However,
restriction digests of viral DNA in the pellet fraction lacked two fragments
corresponding to the molecular ends of unit-length DNA. In addition, there
appeared in HpaI digests of pellet DNAa newrestrictionfragment interpretable
astheproduct offusionbetween theendsofunit-lengthmolecules. The size of the
new fragment requires that fusion occur in such a way that one copy of the
terminallyrepeatedsequences(Mr 12.3 x 106) of the unit-length DNA is lost in
the process. In pulse-chase experiments, radioactivity flowed from the pellet
fraction tothe supernatantfraction, suggestinga precursor-product relationship
for these DNA species. The results are easily understood if unit-length virion
DNAisgenerated by excision from concatemericstructures.
Asextracted from virions, the genome of the
herpesvirus channel catfish virus (CCV) is a
linear molecule of double-strandedDNA with a
molecular weight of approximately 85 x 106(5,
7). CCV DNA is terminally repetitive; that is,
thesamenucleotidesequence (Mr 12.3 x 106)
ispresent atboth ends of the molecule (5). The
repeated sequences are mutually oriented with
direct polarity, a configuration which permits
theformation of circlesor concatemers by
intra-molecular orintermolecular recombination,
re-spectively(4, 17-19). As circlesorconcatemers
are formed in this way from linear DNA, the
molecular endsareeither lost intheformercase,
orin thelattercase,theirfrequency relativeto a
giveninternalsequenceis reduced inproportion
tothe number of unit-length molecules making
upthe concatemer.
When linear DNA isdigested with restriction
endonucleases, the molecular ends generate a
subset offragments in which one end of the
fragment is definedby aninternalrestriction site
and the other end by the molecular end itself.
For CCV virion DNA, in which the terminal
sequences are notaltered by permutation (17) or
inversion (11), this subset contains two
frag-ments. The size and relative position of these
have been determined for the restriction
endo-nucleasesEcoRI,HindIII, HpaI, andXbaI (5).
t Present address: Institut de RecherchesenBiologie Mol6-culaire,75221Paris Cedex05, France.
In this communication, we describe
experi-ments which use the Hirt selective extraction
technique (12)tofractionate intracellular DNA
late in the replicative cycle of CCV. HpaI and
XbaI digests of CCV DNA in the supernatant
fraction contained normalamounts ofend
frag-ments, but these were notdetectable indigests
of viral DNAfrom the pellet fraction. The data
indicate the existence oftwoforms of
intracellu-lar CCV DNA, one of which corresponds to
unit-length genomes and the second to
"end-less" structures such as circles, long
conca-temers, or possibly some combination of the
two. Moreover, pulse-chase experiments
sug-gest that the endless form of CCV DNA is a
precursorof theunit-length genomes.
Results similar tothose presented here have
been previously published by Ben-Porat and
Rixon (2)forpseudorabies virus and by Jacobet
al. (13)for herpes simplex virus.
(A preliminary report ofthis work was pre-sented at the Herpes Virus Workshop,
Cam-bridge,
England, in August1978.)MATERIALSANDMETHODS
Cells and virus.Thecontinuous catfish cell line BB
(20)wascultivatedat30°CinEagle minimum essential medium (MEM) supplemented with 5% fetal calf se-rum.Confluentmonolayers in plastic petri dishes
(60-or100-mmdiametercontaining, respectively,3 x 106 to 4 x 106cells and 10x 106to12 x 106cells)were
infected with CCVat amultiplicity of10to20PFUper cell. Virusadsorption was carried out in0.2-ml
(60-405
on November 10, 2019 by guest
http://jvi.asm.org/
406 CEBRIAN, BUCCHINI, AND SHELDRICK
mm dish) or 0.5-ml (100-mm dish) volumes of MEM containing10% (vol/vol) dimethyl sulfoxide for 20 min at22°C. Two milliliters (60-mm dish) or 5 ml (100-mm dish) of MEM without serum was then added, and the infected cells were incubated for the desired time at 22°C under an atmosphere of95%air-5%CO2.
Radiolabeling ofintracellular DNA. Cultures were labeled with [methyl-3H]thymidine (-50 Ci/mmol;
Commissariat al'Energie Atomique, Saclay, France, and NewEngland NuclearCorp., Boston, Mass.) by adding the isotope to the culture fluid toobtainafinal concentration of5 to50 ,uCi/ml, depending upon the experiment. Chaseswereperformedby removal of the radioactive medium, followed by awash with MEM
containing 2 mMthymidine and0.1mMdeoxycytidine (1),with additional incubation in the same medium.
Nonselective extraction of intracellular DNA. The extractionprocedure was derived from the method of Gross-Bellard et al. (10). Infected cell monolayers werelysed with 0.5% sodium dodecyl sulfate (SDS) in 10 mMTris-hydrochloride (pH7.5)-10 mM EDTA (1 ml per106 cells), and the lysate was incubated in the presence of 100 ,ug of proteinase K(E. MerckAG, Darmstadt,Germany) per ml for 12 h at 37°C. After a second addition ofproteinase (100,ug/ml,6hat37°C), thelysate was adjusted to 1% SDS and extracted twice with an equal volume of phenol previously equilibrat-ed with 0.5 M Tris-hydrochloride (pH 8.3)-0 mM EDTA. The aqueous phasewasdialyzed againsttwo
changes of 10 mMTris-hydrochloride(pH7.5)-l mM EDTAandstoredat4°C.
Selective extraction of intracellular DNA. Selective DNAextractionbythe methodof Hirtwasperformed aspreviouslydescribed(12), except thatproteinaseK (100 ,ug/ml) was included in the lysis buffer. The presence ofproteinasewasfound tofacilitate subse-quent resuspensionofethanol-precipitated DNA
(be-low); otherwise, no essential differences were ob-served in the velocity sedimentation properties or
buoyant densities of supernatant ofpelletDNAsif the proteinase was omitted, in agreement with Hirt's
originalobservations (12). Thelysate(1 ml of
extrac-tionbuffer per1 x 106to2 x 106cells)wasincubated
at37°Cfor1h before theaddition, withgentle mixing,
of 0.25 volumesof 5 M NaCI. After being held forat
least 2 h at 4°C, the precipitate was pelleted by centrifugationat27,000 x gfor 30 min. ForCsClor sucrose gradient analyses, the supernatant wasused
directly. For restriction endonuclease analysis, the supematant was first dialyzed at 4°C against two
changes of100volumes of10mMTris-hydrochloride
(pH7.5)- mMEDTA,adjustedto0.3 Mammonium acetate, andprecipitatedwith 2 volumesof95% etha-nol. Thecentrifugedprecipitatewasthensuspended in
asmall volume of theappropriate restriction endonu-cleasebuffer(seebelow).Thehigh-salt pelletfraction wasgentlyresuspended(usually24 h at room
tempera-turewithoutagitation) in theoriginal volume of
extrac-tion buffer(10mMTris-hydrochloride [pH 7.5]-10mM
EDTA)lacking SDS andproteinase Kand
reprecipi-tated withNaClasdescribed above. Pellet fractionsto be subjected to CsCl density gradient centrifugation
were first extracted withphenolasindicated for total
DNA.
CsCI gradient centrifugation. Solutions ofthe
mate-rialtobeanalyzedwereadjustedto adensityof1.7g/
cm3by the addition ofcrystalline CsCl.Centrifugation
wascarried out at 20°C in polyallomer tubes for 72 h at 35,000 rpm in a 50Ti (Beckman Instruments, Inc., Fullerton,Calif.) rotor, or for 20 h at 50,000 rpm in a 65 VTi (Beckman) vertical rotor. For radioactivity mea-surements,samples ofgradientfractions were deposit-ed on GF/C filters (Whatman, Inc., Clifton, N.J.) whichwere sequentially soaked in 10%trichloroacetic acid (10 min) and70%ethanol (10 min) and then dried.
Sucrose gradient centrifugation. Samples (usually 1 ml) werelayered onto 10-ml, 5 to 20% (wt/wt) linear sucrosegradients (in1 MNaCI-1 mM EDTA-10 mM sodium phosphate [pH 7.5]) and centrifuged in an SW41 (Beckman) rotor at 36,000 rpm for 150 min at 20°C. Measurements of radioactivity were performed
asdescribed above.
Restrictionendonuclease analyses. All enzymes were purchased from New England Biolabs (Beverly, Mass.).Incubationbuffers forHpaIand XbaI were as previously described (5).
DNA was digested with an excess (2 to 5 times basedontheactivity specified by the manufacturer) of enzymefor 2 hat37°C. Incubation mixtures included 100 ,ug of bovine serum albumin per ml. Reactions were terminated by adding 1/10 volume of 0.1 M EDTA-2% SDS-70o glycerol-0.2% bromophenol blue, and thedigestion products were electrophoresed
on0.5% agarose horizontal slab gels (27 by 10 by 0.4 cm)at1.5V/cmfor 15 to 20 h at ambient temperature (-22°C). The electrophoresis buffer was 40 mM Tris-hydrochloride-30 mM acetic acid-20 mM sodium ace-tate(pH7.8)-2mM EDTA. Radioactivity was detect-edbyfluorographyasdescribedpreviously by Bonner andLaskey (3),usingpresensitized Kodak RPX-omat radiographic film (14).
RESULTS
Kinetics of thymidine incorporation during CCV infection.Wolf andDarlington (20)
estimat-edthelength of the CCV infectious cycleat22°C
to beapproximately 12h. Inthepresentstudy,
wewishedto place ourexperiments at a time in
the infectious cyclenear the maximum level of
viralDNA synthesis, so wefirstdetermined the
kinetics of thymidine incorporation into CCV
DNA. Confluent BB monolayers at 22°C were
infected with 10to20 PFUof CCVpercell,and,
aftera20-minadsorptionperiod,weresubjected
hourly to 30-minpulsesof
[3H]thymidine.
Imme-diately afterthelabeling period,total DNAwas
extracted andanalyzedbyCsClgradientdensity
centrifugationas described above. The relative
proportion of label incorporated into cellular
DNA (p = 1.700
g/cm3)
and viral DNA (p =1.717
g/cm3
[9])atvarioustimes afterinfection is shown in Fig. 1. Label associated with CCV DNA wasfirstdetectable inthepulsebeginningat 2h postinfection (p.i.) and became maximal
withintheinterval defined bythe 3 h-p.i. and 5
h-p.i. pulses. In the experiments described
be-low, we thereforeused 30-min labeling periods
beginning at 3.5 h p.i. Dixon and Farber (7)
obtained kinetics for CCV DNA synthesis at
30°C whicharesimilar tothosereported here.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
407
!
0
0 1 2 3 4 5 6 7
HOURS POSI-INFECTION
FIG. 1. Kinetics of [3H]thymidine incorporation into CCV-infected cellular DNA. Infected cellular
DNA was extracted and analyzed by CsCI density gradient centrifugation as described inthe text.
Re-sults areexpressed as the viral (A) and cellular(0)
componentsof the total(A) incorporation. The
maxi-mumtotal incorporationwasassignedavalue of100o. The proportion ofcountsinviral and cellular DNAs
wasestimated fromdensity gradient profilessimilarto
those of Fig. 3.
Fractionation of 3H-labeled DNA by Hirt
ex-traction. Hirt(12) developedaselective
precipi-tation procedure permitting the separation of
low-molecular-weight polyoma virus DNA (Mr
3 x 106) from high-molecular-weight cellular
DNA(Mr > 100 x 106). Since itsintroduction,
the technique has been extended to studies of
viruses with genomes of considerably higher
molecular weight than that of polyoma virus,
notably pseudorabies virus(Mr 90 x 106[1])
andherpes simplexvirus(Mr 100x 106[15]).
Inapplyingthe HirtproceduretoCCV-infected
cells, we observed that successive extractions
continued to liberate labeled material into the
supernatantfractionindecreasingamounts(Fig.
2). Of the total radioactivity recovered in the
supernatants of four serial extractions, -50%
wasreleased in thefirst extraction, -25%in the
second, -10%o in the third, and -5% in the
fourth; furtherextractions failed torelease
sig-nificantamountsofradioactivity fromthepellet.
In a representative experiment, -30%o of the
total label incorporated in a 30-min pulse was
found in the combined supematants,and -70%o
remainedpelletassociated. When pulse-labeled
uninfected BB cells were extracted in thesame
way(datanotshown), >95% oftheincorporated
radioactivity remained associated with the pel-let.
Toinvestigate the partitioning of radioactivity
between viral and cellular DNAs, successive
supematants andthe exhaustively extracted
pel-let were centrifuged to equilibrium inCsCl
gradi-ents. Roughly90% of the radioactivity in each
supernatant banded at the density of viral DNA (Fig. 3A), whereas about 60% of the pellet radioactivity had the density of viral DNA (Fig.
3B). Thesupernatants and pellet were also
char-acterized by velocity sedimentation in 5 to 20%
sucrose gradients (Fig. 4). Sedimentation
pro-files ofradioactivity in successive supernatant
fractions were typically heterodisperse (Fig. 4A
and B) and covered a range of sedimentation
coefficients from -50S to -20S. Unit-length
CCVDNAextractedfrom virions sediments at
-53S(P.Sheldrick, N. Berthelot, and S.
Chous-terman,unpublished data). Similar experiments
with the final pellet fraction (not shown)
re-vealed that 80o of the radioactivity sedimentd
to the bottom of the gradient.
We next examined the behavior of
pulse-labeledsupernatantand pellet DNA during a 4-h
chase with excess nonradioactive thymidine.
The results (Fig. 5) show that as the chase
progressed, radioactivity was lost from the
pel-let andaccumulated with similarkinetics in the
supematant. In controlexperiments not
present-ed,the presenceof excess thymidine from 3.5 h
p.i. on did not measurably affect the yield of
infectious virus from afull growth cycle
(mea-suredat 24hp.i.). Velocitysedimentation
analy-At5
0
1 2 3 4
EXTRACTION
FIG. 2. Successive Hirt extractions of CCV-infect-ed cells. Infected cellswere labeled with
[3H]thymi-dine for 30 minat3.5hp.i.andextractedasdescribed in thetext.Resultsareexpressedasthe percentageof totalradioactivity extracted.
VOL.46, 1983
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.490.44.230.58.290.2] [image:3.490.269.414.435.626.2]408 CEBRIAN, BUCCHINI, AND SHELDRICK
I?
I'½
0t 20 40 60 SO 0 40 60 S0
FRACTION FRACTION
FIG. 3. Buoyantdensity gradientsofsupernatant andpelletDNAs.(A) Combined supernatant fractions from theexperiment shown inFig.2.(B) Pellet fraction after four successive extractions (Fig. 2). As a buoyant density reference, [14C]thymidine-labeledCCV virion DNA (arrows)wasadded to the gradients. The gradient bottoms are totheleft.
04
V
M.
I
FRACTION FRACTION
FIG. 4. Velocity sedimentation ofsupernatantDNA.Sucrosegradients (5to20%)of the first (A) and second (B) successivesupernatantfractions shown inFig.2. Arrows indicate thepositionof'4C-labeledCCV virion DNAaddedtothegradients.Sedimentation wasfromrighttoleft.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.490.60.444.54.333.2] [image:4.490.60.445.371.640.2]INTRACELLULAR
t:
0
50~~~~~~~
0 ,
0 t 2 3 4
CHASE PERIOD (HOURS)
FIG. 5. Partitioning ofradioactive CCVDNA be-tweensupernatantandpelletfractionsduringa pulse-chase. Infected cells were pulse-labeled as in the
legendtoFig. 2 andincubated fortheindicated times in the presence of excess thymidine (see text). Radio-activity in
CCV
DNA was determined by densitygradientcentrifugation(as inFig. 1). Values for each
timepoint were obtainedfrom four (combined)
super-natantfractions(0)andfrom the final pelletfraction
(0).
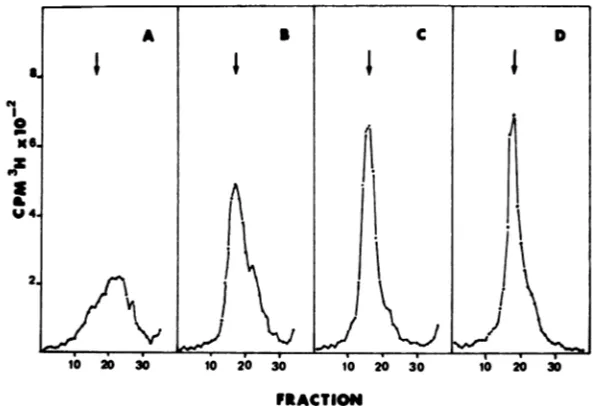
sisof the supernatantfraction preparedat
vari-oustimes duringthe chase(Fig. 6) showed that
the major part, if not all, ofthe accumulating radioactivity sedimented at -53S, the position
ofunit-length CCV DNA.
Restriction endonuclease analysis of
superna-tant, peUlet, and nonselectively extracted DNAs.
In restriction endonuclease digests of linear
DNA, the ensemble of fragments contains a
subset produced on the one hand by internal
restriction sites and on the other hand by the
molecular ends of the DNA. For unit-length
CCV DNA, two fragments appeared in this
subset. The present experiments were carried
outwithHpaI, whose recognition sites lie
out-side the terminally repeated sequences, and with XbaI, one of whose recognition sites lie within
these sequences (see Fig. 9 and reference5).
CsCl gradient-purified CCV DNA from the
supernatant and pellet fractions was digested
with each of these enzymes, and the resulting
fragmentswereresolved by agarose gel
electro-phoresis (see above). The fragment proffies of
restricted supernatant DNA are
indistinguish-able from those of CCV virion DNA digested
with the respective enzymes (Fig. 7A through
D), supporting our conclusion that this fraction
contains principally unit-length viral genomes.
Withpellet-fractionDNA,however, quite
differ-ent results were obtained. The profile of XbaI
digests lacked band Faltogether, and, as
esti-mated from densitometer tracings of
photo-graphic negatives, theintensityof the 2x molar
band (D, E) relative to bands, C, G, and H
diminishedtotheequivalentof -1.1 molar(Fig.
7 G and H). Similarly, in gelsofHpaI digests,
bands B andC wereno longer detectable (Fig.
7Eand F).
Visual inspection of the gels permitted us to
FRACTION
FIG. 6. Velocity sedimentation ofsupernatantDNAduringapulse-chase. Thefirstsupernatantfractions of a
30-minpulseat3.5 hp.i. (A) and after chases of 1, 3, and 4 h (B, C, and D, respectively) were sedimented in S to
20osucrosegradients. Arrows and direction of sedimentation are as in Fig. 4.
46, 409
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.490.60.213.53.253.2] [image:5.490.93.390.439.642.2]410 CEBRIAN, BUCCHINI, AND SHELDRICK
A B C D
Bw -:
B"
D"....Cwe&..
E _w
E
FF
F*%M
E :F
A_ L
E E
F'
H
G G
J
10-fold lower than in virion
(or supernatant)
DNA. A similar estimate can be obtained by
comparing
HpaI
fragments
BandC(Mr
= 17 xB 106 and 13.2 x 106, respectively) with HpaI
fragment
G(Mr = 1.5 x106).
Inadditiontotheabove changes, a new band (B') appeared in
profiles of HpaI digests for pellet DNA (Fig.
7F). The molecular weight
(Mr
18 x 106) ofthis new
fragment
isprecisely
thatexpected
E from a fusion (via overlap of the terminal re-F- peats) of theHpaIfragments B and C of virion
DNA (seeFig. 9). Thiscanbecalculated by the
followingequation: fragment B (17 x 106)plus
fragment C (13.2 x 106) minus the terminal
G repeatsequence
(12.3
x106) equals
17.9 x106.
For another project,we have recentlycloned
fragmentB' in the HpaI site of cosmid pHC 79.
Restriction endonuclease site maps(M. Lacasa
H and J. Cdbrian, unpublished data) agree
com-pletely with the presentassignment.
Thefusion fragment HpaI B' is more
conve-niently observed in digests of intracellularDNA
A B c D E F
[image:6.490.56.240.62.377.2]J
FIG. 7. Restriction endonuclease digests of viral DNAfrom a Hirt extraction of CCV-infected cells.
Infectedcellswerepulse-labeled (30 min)at3.5 hp.i.,
and viral DNAwasselectivelyextracted andpurified byCsCldensitygradientcentrifugation. Supematant
(lanes B andD) andpellet (lanesF andH)DNAswere digestedwithHpaI(lanesBandF)andXbaI(lanesD andH). Digestsof14C-labeledCCV virion DNA with therespectiveenzymes(HpaI, lanes AandE;XbaI,
lanes C and G) were included for reference. Condi-tions forelectrophoresisandautoradiographyare giv-enin thetext.
roughly estimate an upperlimit for the relative
concentration of the missing fragments. For
example, inthe XbaIprofile,fragment F, witha
molecularweight of5.6 x
106,
was notdetect-able under theconditions of filmexposureused, whereas fragmentJ, at 0.65 x 106, was clearly
visible (Fig. 7H). Since the molecular weights
(and supposedly the amount of[3H]thymidine)
ofthese fragments differby a factor of10, the
concentration ofXbaIfragment F must be less
than1/10thatofXbaIfragment J; otherwise,the
bandwould bevisible.Moreover,invirionDNA one copy of XbaI fragment J is present per genome, which meansthattheconcentrationof
ends (strictly speaking, one end for XbaI) per
genome equivalent in pellet DNA is morethan
mow,...
C _ .. ..
[image:6.490.279.423.315.595.2]D t IIiw;
FIG. 8. Restriction endonuclease digests of total DNA from nonselective extraction ofCCV-infected cells. HpaIdigests of DNA from a nonselective
ex-traction (see text) of infected cells pulse-labeled (30
min) at 3.5 h p.i. (lane B) and chased with excess
thymidine for 1 (lane C),2(lane D),3(lane E), and4h
(lane F). HpaI-digested, "4C-labeledvirion DNAwas
includedas amarker(lane A).
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
INTRACELLULAR CHANNEL CATFISH VIRUS DNA
VIRION DNA
TR TR
C B
.~~~, . ^
D E A F
a a apA I.3
Hpa I
XbaI
INTRACELLULAR DNA
B B
A E A E
A
p . a a Ip ..a _
HpaI
Xba I
FIG. 9. Restriction endonuclease site map of virion and intracellular CCV DNAs. Thick lines represent the M =12.3 x 10 terminal repeat sequences referred to in the text. Only the restriction fragments relevant tothe
presentstudy arelettered.
from a nonselective extraction (see above),
since it andfragmentHpaI B form adoublet in
gel profiles (Fig.8B). Achase with
nonradioac-tive thymidine was also incorporated into the
experiment shown in Fig. 8, and, asthe figure
shows, thegelprofilesevolvedduringthecourse
of the chase. Ina30-minpulse,only band B' (as
in pellet DNA) was apparent, but asthe chase
proceeded its intensity decreased whereas the
intensities of bands B and C increased. These
results areinagreement with theprevious
sug-gestion that pellet DNA isaprecursor to
super-natantDNA.
Certain bands other than thosejustdiscussed
werealso found to have modifiedintensities in
gel profilesofpelletandtotal DNA.Thesewere
XbaI band A(Mr= 29 x
106)
andHpaIband A(Mr = 31 x 106), both of which are clearly
underrepresentedrelativetovirion and
superna-tantDNAs in the gels of Fig. 7and 8. We have
investigatedthismoreclosely withBglII, which
gives smaller fragments (Mr < 15 x 106;
S. Chousterman andM. Lacasa, personal
com-munication) in thegenomic regions covered by
XbaIband A andHpaIbandA. Inthiscase(not
shown), no reduction in the intensities of the
corresponding bands was observed. We
there-fore think it likely that this effect is due to
nonspecific breakage of intracellularDNA
dur-ing the isolation procedures used (see above).
Largerfragments would be affectedtoa greater
extentby such breakage.
DISCUSSION
By applying the selective extraction
proce-dureof Hirt (12)toCCV-infectedcells, we have
obtained evidence fortheexistence oftwo
class-es of intracellular viral DNA which differ in
several important respects. DNA released into
thesupernatantfraction sedimentedatthesame
rate (-53S) as did DNA extracted from CCV
virions (Fig. 4). This result and the fact that
restriction endonuclease digest profiles (Fig. 7)
were indistinguishable from profiles of virion
DNAindicate that supernatant DNA consists of
unit-lengthCCVgenomes.
Pellet DNA, on the other hand, besides the
fact that it doesnot enterthesupernatant under
theextractionconditions,differed from
supema-tantDNAin that itsedimentedtothebottom of
gradients in which virion DNA sedimented
ap-proximately halfway. Inaddition, theXbaI and HpaIfragmentsthatarediagnostic of the
molec-ular ends of theunit-length CCV genome were
not detectable (<1/10 the expected
concentra-tion) indigests of pelletDNA (Fig. 7). By this
lattercriterion,therefore, pelletDNAis endless.
Thepresenceofanew"fusion"fragment(B')
which replaced the missing end fragments in
HpaIdigestsofpelletDNA(Fig. 7) isimportant for two reasons. First, it effectively eliminates the possibility that end fragments are missing
from pellet DNA because they areattached to,
forinstance,aproteinwhich couldprevent them
from entering the gel. This phenomenon has
beenfoundtoapplyunder certainconditionsto
the end fragments of the adenovirus genome
(16). Second,thesize ofthefusionfragment(Mr
= 18 x 106) requires that the fusion leading to
loss ofthe molecular ends takeplacethrough a mechanism in which one copy per genome
equivalent ofthe terminal repetition of12.3 x
106 is also lost, rather than by a "blunt end"
fusion in which case a new fragment with a
VOL.46,1983 41
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.490.59.421.52.252.2]412 CEBRIAN, BUCCHINI, AND SHELDRICK
molecular weight of 30 x 106(HpaI fragment B
plus HpaI fragment C) would be formed. In
XbaI digests of pellet DNA (Fig. 7), nofusion
fragment is expected, because arestriction site
lieswithin the terminalrepeat(Fig. 9).
Fusion with loss of a copy of the terminal
repetition couldoccurbygeneticrecombination
betweenrepeats (4,17, 18)orby exonucleolytic
removal of complementary strands from two
repeats with subsequent annealing of the
un-paired strandsto regenerate asinglerepeat(e.g.,
reference 19). Whatever theprecise mechanism,
fusion of intragenomic repeats would lead to
unit-length (less oneterminalrepeat) circle
for-mation whereas concatemeric structures (in-cluding multimeric circles) would result from
intergenomic fusion. Subsequent encapsidation
of unitlengthgenomescould thenproceedviaa
replicative mechanismof the typeproposedfor
bacteriophage T7(19) whereby the terminal re-peats are "restored" tovirion DNA.
Either unit-length circles or concatemers
could accountforourresults. However, unless
heldin placebyaproteinase K-resistantlinkage,
unit-length circles would not be expected to
remain in thepelletfraction. Norhave webeen
ableto detectcircular molecules by direct
elec-tronmicroscopicexamination of CsCl
gradient-purified pellet DNA (N. Berthelot, personal
communication). Admittedly, theseare not
ab-solutely irrefutable argumentsbecause of
possi-bletrappingeffects inhigh-molecular-weight
cel-lularDNA, buttheydo leadustofavor the idea
thatpellet DNA consists principallyof
concate-meric forms (or"rollingcircle" forms[8],which
wouldnotbedistinguishable fromconcatemers
in thepresentexperiments)of theCCVgenome.
Similarconclusions derived from restriction
en-zymeanalysishavebeenreached for the
replica-tiveforms ofDNAinpseudorabiesvirus(2) and
herpes simplexvirus (13) infections and for the
intracellular DNA ofbacteriophage SPOt (6).
ACKNOWLEDGMENTS
We thank Madeleine Laithier forprovidingcell cultures and virus stocks.
This workwassupportedby generalgrants fromtheCentre Nationale de la RechercheScientifique and a specialgrant ATPCNRS 655-2108.J.C.was aFellowof theLigue Nation-aleFrancaisecontrele Cancer.
LITERATURE CITED
1. Ben-Porat, T., A. S.Kaplan, B. Stehn, and A. S. Ruben-stein. 1976.Concatemeric forms of intracellular herpesvi-rusDNA.Virology69:547-560.
2. Ben-Porat, T., and F. J. Rlxon. 1979. Replication of herpesvirus DNA. IV. Analysis ofconcatemers. Virology 94:61-70.
3. Bonner,W.M., and R.A.Laskey. 1974. Afilmdetection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur. J. Biochem. 46:83-88. 4. Campbell,A.1962. Theepisomes. Adv. Genet.
11:101-145.
5. Chousterman, S., M. Lacasa, and P. Sheldrick. 1979. Physical map of the channel catfish virus genome: loca-tion of sites forrestriction endonucleases EcoRI, HindIII, HpaI, and XbaI. J. Virol. 31:73-85.
6. Cregg, J. M., and C. R.Stewart. 1978.Terminal redun-dancy of"highfrequency of recombination" markers of Bacillus subtilis phage SPOl. Virology 86:530-541. 7. Dixon, R. A. F., and F. E. Farber. 1980. Channelcatfish
virus:physicochemical properties of the viral genome and identification of viralpolypeptides. Virology 103:267-278. 8.Gilbert, W., and D. Dressler. 1968. DNAreplication: the rolling circle model. Cold Spring Harbor Symp. Quant. Biol.33:473-484.
9.Goodheart, C. R., and G. Plummer. 1975. Thedensities of herpesviral DNAs.Prog. Med. Virol. 19:324-352. 10. Gross-Bellard, M., P. Oudet, and P. Chambon. 1973.
Isolationofhigh molecular weight DNA from mammalian cells. Eur. J. Biochem. 36:32-38.
11. Hayward, G. S., R. J. Jacob, S. C. Wadsworth, and B. Rolzman. 1975. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in therelative orientationsof their long and short segments.Proc.Natl. Acad. Sci. U.S.A. 72:4242-4247. 12. Hirt, B.1967.Selective extraction of polyoma DNA from
infected mouse cell cultures. J. Mol. Biol. 26:365-369. 13. Jacob, R. J., L. S. Morse,andB.Rolzman. 1979. Anatomy
ofherpessimplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in thegeneration of the fourisomeric arrangements of viral DNA. J. Virol. 29:448-457.
14. Laskey, R. A., and A. Mills. 1975. Quantitative film detectionof 3H and14Cinpolyacrylamidegels by fluorog-raphy.Eur. J. Biochem. 56:335-341.
15. Pater, M. M., R. W. Hyman, and F. Rapp. 1976. Isolation ofherpes simplex virus DNA from the "Hirt superna-tant."Virology75:481-483.
16. Sharp, P. A., C. Moore, andJ. L.Haverty. 1976. The infectivityof adenovirus 5DNA-protein complex. Virolo-gy 75:442-456.
17. Streisinger,G., J. Emrich, and M. M.Stahl.1967. Chro-mosomestructureinphageT4. III. Terminalredundancy andlength determination. Proc. Natl. Acad. Sci. U.S.A. 57:292-295.
18. Thomas, C.A. 1966.Recombination ofDNA molecules. Prog. Nucleic Acid Res. Mol. Biol.5:315-349. 19. Watson, J. D. 1972. Origin of concatemeric 17 DNA.
Nature(London)NewBiol. 239:197-201.
20. Wolf, K., andR. W. Darlington. 1971. Channel catfish virus: a newherpesvirus of ictalurid fish. J. Virol. 8:525-533.
J.VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
![FIG. 1.gradientintoDNAcomponentsThemumwasthosesults Kinetics of [3H]thymidine incorporation CCV-infected cellular DNA](https://thumb-us.123doks.com/thumbv2/123dok_us/1446753.97137/3.490.269.414.435.626/fig-gradientintodnacomponentsthemumwasthosesults-kinetics-thymidine-incorporation-ccv-infected-cellular.webp)