Vol. 37, No. 2 JOURNAL OFVIROLOGY, Feb.1981,p.683-697
0022-538X/81/020683-15$02.00/0
Properties of Simian Virus
40
Small
t
Antigen
Overproduced
in
Bacteria
CARL S.THUMMEL,TERESAL. BURGESS, ANDROBERTTJIAN*
DepartmentofBiochemistry, University ofCalifornia, Berkeley, California 94720
We constructedaseries ofbacterialplasmidswhich containedtheEscherichia coli lac promoterfusedto a simian virus 40 restriction fragmentcoding forsmall t antigen. These plasmids expressed different levels of intactviral protein de-pendingonthelength of the constructed ribosome bindingsite.Smallt antigen synthesized by themostefficientproducer, HP1,constituted 0.5 to1% of the total cellularprotein. On the basis of extensive characterizationby immunoprecipita-tion, gel electrophoresis, isoelectric focusing, tryptic fingerprint analysis, and chromatographic properties, this plasmid-encoded proteinwasvirtuallyidentical to authentic
simian
virus 40 small t antigen. Partialpurification
of theHP1-encoded and authenticsmalltantigensrevealedthe presence ofboth monomeric andmultimeric forms.
Twoproteinsareexpressedby simian virus40 (SV40) early after infection of either permissive monkey cellsornonpermissive rodentcells.One of these polypeptides, largetumor (T) antigen, is a 90,000- to 100,000-dalton nuclear protein encoded
by
the viral A gene(56).
The SV40 largeTantigen
has beenextensively
character-ized and showntobe involved in the initiation of viralDNAreplication
(52),regulation
of viraltranscription (10,
39), and induction ofneoplas-tic transformation (49, 53). In contrast, small tumor (t) antigen, a 15,000- to 20,000-dalton
cytoplasmic protein
encodedby
the SV40 Fgene,was
only recently
detectedandplays
anasyetundefined roleinthe viral life
cycle
(11, 37, 45).Si
mapping (2) andsequenceanalysis
(38) oftheSV40
early
mRNA's revealedtwotranscriptswhich could be
distinguished
by
the size and location of theirintervening
sequences.Al-though
thesemRNA's share their initial81co-dons,
thecoding
sequencefor thecarboxy
ter-minus ofsmallt
antigen
isspliced
outfromtheprimary transcript
forlarge
Tantigen (38).
Inagreement with these
observations,
bothtumorantigens sharesometryptic
peptides (28, 46)
aswell
as their N-terminal amino acid sequence (36).Furthermore,
deletionmutantslacking
se-quences between 0.54 and 0.59 map unitsonthe conventional SV40
genomic
mapproduce
atrun-catedsmallt
polypeptide,
but retain theability
tosynthesizeawild-type
large
Tantigen (11,
15, 45).These F gene deletion mutants have been used to study the role of small t
antigen
in productive andnonproductive
infections,
but the results were variableand,
in certain cases,difficulttointerpret. Someof the F gene deletion mutants appear to be
partially
defective for lytic growth on monkeycells(57). In addition,small tantigenseems toberequired for the induction ofafully transformed phenotype (5, 15, 45, 48). This function may,however, be dispensable in actively growing cells(30, 43).Finally,thesmall tproteinmayprovide somefunctions
which are required forfullviral oncogenicity, since F gene mutantvirusesinjected into neonatal hamsters inducetumorswhich grow more slowly and have alonger latency than those inducedby wild-type SV40 (27; W. Topp, personal communication). On the basis of these observations, it is likely thatsmalltantigenplayssome role, albeit un-defined, in the viralinductionof both transfor-mationandoncogenicity.In support of the idea thatthe
small
tpoly-peptide
mayberesponsible for certainmorpho-logical changes associatedwiththe transforned phenotype, itwas shown thatmicroinjection of SV40 DNAcoding for smalltantigen (0.735to 0.375mapunits) induced dissolutionofthe cy-toplasmic actin cable networkin ratfibroblasts (19).Injectionof DNAfromanFgenedeletion mutant, however, had no effect on the cyto-skeletal structure. Although these results sug-gestthat SV40 smallt
antigen
isresponsible
for cytoskeletal changes,nofirmconclusions can be made until similar experiments areperformed
withpurifiedsmalltprotein.
Ourunderstandingof the
biological
function of SV40 small t antigen has been impeded by thedifficultiesinvolved inpurifying
theprotein from conventional sources. To overcome this problem, we chose to construct arecombinant plasmid which would produce this eucaryotic683
on November 10, 2019 by guest
http://jvi.asm.org/
protein in bacterial cells.Three features ofSV40 smalltantigen facilitate its synthesis in Esche-richia coli. First,unlike mosteucaryotic genes, the smallt structuralgeneis notinterruptedby intervening sequences (2, 38).Second,theentire coding sequenceispresenton aHindIII
restric-tion fragment which conveniently containsthe smalltinitiation codon 12 basepairs (bp) from
one of the ends. Finally, the only known post-translational modification of theproteinis
ace-tylation of the N-terminalmethionine(36). Con-struction ofaplasmidable topromote transcrip-tion and translation of the SV40 small t gene should therefore allowsynthesisin bacterialcells of a protein which isvirtually identical to
au-thentic smalltantigen.
Several bacterialplasmidshavebeendesigned todirect efficientexpressionofeucaryoticgenes as fusion proteins (16, 29, 32). Backman and
Ptashne (1) havedeveloped a plasmid
contain-ing the lac regulatory sequences which allows
theproduction of intactgene products by
con-structingahybridribosome
binding
siteconsist-ing of the Shine and Dalgarno sequence from
,8-galactosidase
(44)
fusedtoaforeign
initiationcodon. This system has been used to produce phage lambda repressorand croproteintolevels
ashigh as 2.5% of thetotal bacterialprotein(1, 42) and also tooverproducehuman
growth
hor-mone (18). Roberts and his co-workers have
used this system to achieve
synthesis
ofSV40 small t antigeninbacteria(41).
However,
they detected only alow level ofexpression
of this eucaryoticgeneproduct.In thispaper,wereport thequantitation,isolation,
and characterization ofSV40smalltantigenoverproduced
by
E. coli carryingarecombinantplasmid.MATERIALS AND METHODS
Celisand viruses.E.coli strain294 (endoI, rk-, Mk+,B1-)was agiftfrom H. Echols.E.coli294with
the lac il genewasconstructedby conjugation with a
strain ofE.coli whichcarries this geneonanFfactor,
agenerousgift from F. Heffron. Bacteria with drug
resistance were grown in the presence of 30 ,ug of
ampicillin (Sigma Chemical Co.) per mlor20ugof
kanamycin(Sigma)perml.
Plaque-purifiedSV40strain776waspropagatedon
CV-1 cellsasdescribedpreviously (55). CV-1 monkey
celisweremaintained onplastic dishes with
Dulbecco-modified Eagle mediumsupplemented with an
anti-biotic-antimycoticmixture (GIBCO Laboratories)and 5% fetalcalfserum(GIBCO).
Purification ofplasmid DNA. PlasmidDNAwas
purified byamodificationofthetechnique described
by Katzetal.(25). Bacteriaweregrowntoanoptical
densityat 550nmof0.5 inM9medium (33)
supple-mented withCasamino Acids, vitamin B1,and0.05%
glucose and were treated overnight with 20,ug of
chloramphenicol (Sigma) perml toamplifythe
plas-midDNA.The cellsweresuspendedinlysis buffer (50
mMTris[pH 8.0],25% sucrose,1mMEDTA)
supple-mentedwith4mg ofsolidlysozyme per ml. After 20
minat0°C, EDTAwasaddedto0.15 Mfinal
concen-tration, and solid pronase (Calbiochem)wasaddedto
approximately 1 mg/ml. Incubation of the
sphero-plastswascontinued at0°C for 10min, followedby
the addition of 0.1 volume of TET buffer(50 mM Tris
[pH8.0], 60 mM EDTA, 0.3% Triton X-100) to lyse
the cells. After 15minat0°C, the lysate was
centri-fugedat20,000 rpm for 30 mintopellet the
chromo-somalDNA. Thelysate supernatant was then phenol
extractedthreetimes, ether extracted three times, and
ethanolprecipitated. The plasmid supercoilswere
pur-ifiedby bandingtoequilibrium in a cesium
chloride-ethidium bromide gradient (8) followed by isoamyl
alcohol extraction and extensive dialysis against TE
buffer(10 mM Tris [pH 7.8], 1 mM EDTA).
This procedure was scaled down for small plasmid
preparations ("minilysates") from 5- to10-ml cultures.
An RNasedigestion(Worthington Biochemical Corp.)
wasincluded at the pronase digestion step. In placeof
the cesium chloride-ethidium bromide gradient, the
DNAwaspurified by exclusion from a 2-ml Sepharose
CL-2B column equilibrated in TE with 0.1 M NaCl.
This DNA wasconcentrated by ethanol precipitation
and used for restriction enzymeanalysis.
Construction ofplasmids. Restriction enzymes
werepurchased from Bethesda ResearchLaboratories
(BRL) orNewEngland Biolabs. All digestions were
performed in the buffers recommended by BRL at
37°C for0.5to 10 h. Ligations wereperformed with
T4 DNA ligase (BRL) for12 hat20°C in ligasebuffer
(66 mM Tris [pH 7.6], 10mM MgCl2, 1mM
spermi-dine, 15 mM dithiothreitol [DTT],200
jg
ofbovineserum albumin [BSA, Miles-Pentex] per ml, 1 mM
ATP).Horizontal gel electrophoresis was performed
byusing0.7 to 1.4%agarose (Sigma type II) in TBE
buffer(45 mMTris base, 45 mM boric acid, 1.25 mM
EDTA) or Loen buffer (36 mM Tris base, 30 mM
NaH2PO4,1mMEDTA). The DNA wasvisualized by
staining with ethidium bromide and UV transillumi-nation.
Plasmid pGL101 DNA was linearized by digestion
withPvuII; theenzyme was inactivated by heating at
65°C for 5 min, and the digest was stored at -200C.
SV40DNA, prepared as described previously (23), was
digestedwith HindIII and HpaII, andfractionated by
1% agarose gel electrophoresis in TBE buffer. The
1,169-bp B fragment was excised from the gel, and the
DNA waseluted byelectrophoresis into TBE buffer
andpurifiedandconcentrated byDE52-cellulose
chro-matography and ethanol precipitation. The DNA was
resuspended inTEbuffer and stored at 5°C for
sub-sequentinsertion intopGL101.
(i)pSV250.Gel-purified SV40 HindIII B fragment
wastreated with large fragment DNApolymerase I
(BRL) at370C inthepresence of 10 ,uCi of
[a-32P]-dTTP(Amersham), all four dNTP, and nick
transla-tion buffer (50mM Tris[pH7.8], 10mM MgCl2, 1 mM
DTT, 100,tgofBSA per ml). The reaction kinetics
werefollowed by trichloroacetic acid precipitation. By
30minDNAsynthesis had almostplateaued, and the
reaction wasstopped at 60 min by phenol extraction,
three ether extractions, and ethanol precipitation. A
10-foldmolarexcess of thisblunt-endedSV40 DNA
on November 10, 2019 by guest
http://jvi.asm.org/
OVERPRODUCTION OF SV40 SMALL t ANTIGEN 685
wasligated with T4 DNA ligase to 0.2 ug of
Pvull-digestedpGL101 DNA ina
15-pl
reaction volume. Theligation products were then digested with an excess of
PvuII tolinearize recircularized pGL101 DNA, and
this mixwas usedto transform E. coli 294cells by
standard procedures (9). The bacteria were spread on sterile nitrocellulose filters (Schleicher and Schuell)
on L agar plates containing ampicillin to select for
transformants. Theresultant colonies were transferred
to a fresh nitrocellulose filter, and these duplicate
colonies were grown, treated with chloramphenicol,
lysed, and probed with nick-translated SV40 DNA
(40)asdescribed by D. Hanahan (personal
communi-cation). Clones which contained SV40 sequences were identified by autoradiography and picked, and small
culturesweregrown forpurification of plasmid DNA
as described above. Plasmids containing the SV40
HindIII B fragment inserted into pGL101 in the proper
orientationwereidentified by digestion withHaeIIH,
Southern blotting (47), and hybridization with nick-translated SV40 DNA. One clone containing a plasmid
with theSV40 smalltgenefusedtothe lac promoter
wasselected, and the plasmid was designated pSV250.
(ii)pSV240.SyntheticHindHII linkers
(Collabora-tiveResearch) werelabeled with
[-y-'P]ATP
(Amer-sham) and T4 polynucleotide kinase (New England
Biolabs). Phosphorylation was then completed by the
addition ofanexcessofcold ATP (1 mM final
concen-tration), and the reactionwasterminatedby phenol
extraction, three ether extractions, and ethanol
precip-itation. A 30-foldexcessof thesephosphorylated
link-erswasthenligatedtoPvuII-linearized pGL101 DNA
withT4 DNAligase.Proteinwasremovedby phenol
extraction,and thepGL101DNAwasseparated from unincorporated linkers by Sepharose CL-2B
chroma-tography. The excluded fractionswerepooled, ethanol
precipitated, and digested with HindII to remove
excesslinker sequences. Afterphenolextraction, ether
extraction, and ethanolprecipitation, 0.3 yg of this
modifiedpGL101DNAwasligatedtothegel-purified
SV40HindIHBfragment,and E. coli294cellswere
transformed with the recombinant molecules, as
above. Aclone containingplasmid DNA with the SV40
insert in the proper orientation, as determined by
restrictionmappingwithTaqIandSouthern blot
hy-bridization, wasselected, and theplasmidwas
desig-natedpSV240.
(iii)HP1.pSV240DNAwasdigestedpartiallywith
HindIIItoformpredominantlylinear molecules. BAL
31nuclease (BRL) was then titratedtoremoveonly a
fewbp from the ends ofaDNAduplex by digesting
thepSV240partial digests in the presence oftracer
end-labeled DNA for30minat30°Cin BAL31buffer
(12 mMCaCl2,12mMMgCl2,0.6MNaCl,20mMTris
[pH 8.1]). Theamountof BAL31nuclease whichwas
required to remove 95% of the end label, but not
reduce thesize ofthe pSV240DNA as detectedby
agarosegel electrophoresis,wasselected foruse on a
preparative scale. BAL31nucleasetreatmentof the
pSV240partial digestwasterminatedby the addition
of EDTA to 40 mMfinal concentration, and linear
moleculeswerepurified by 0.7% agarose gel
electro-phoresisinLoenbuffer, excision of the linear DNA,
electroelution, andDE52chromatography. These
mol-ecules were recircularized with T4 DNA ligase and
used to transform E. coli 294. AmpiciUlin-resistant
colonies were picked randomly, and the plasmid DNA
wasdigested with HindIII and PstI to identify
plas-mids missing the HindIII site between the lac
pro-moter and SV40DNA.Those clones were thentested
forsmalltantigen expression as described below.
All manipulations were in accordance with the guidelines of the National Institutes of Health and
usedP2-EK1 containment.
Labelingofproteins. Bacterial clonesweregrown
in 5ml of M9 medium supplemented with amino acids
(minusmethionine), thiamine, 0.5% glycerol, and
am-piciUin. Atanoptical densityat 550nmof 0.4,50 to
200jiCi of[3S]methionine (Amersham) was added,
and the culturewasleftat37°C for2minfor
pulse-labelingor 1hfor steady-state labeling. The tube was
then chilled to stop the pulse, and the cells were
immediatelypelleted by centrifugation. The bacteria
weresuspended in 0.5ml of TE, 0.1 M NaCl, 0.1 mM
DTT, and50pg ofphenylmethylsulfonyl fluoride
(Sigma) per ml and sonicated, and the lysate was
centrifugedtopeletthecellulardebris. Inductionwith
isopropyl-,8-D-thiogalactopyranoside was
accom-plished by the addition of this inducer to1 mMfinal
concentrationinthe growth medium one generation
beforeharvesting thecells.
CV-1 monkey celswere infected with 10PFU of
SV40 per cell and leftat37°C untilapproximately 10%
of thecells showedcytopathiceffects. Thecellswere
thenwashed twice withphosphate-bufferedsaline and
incubated with DME medium minus methionine for 1.5 to 2 h at370C.From0.1 to 1mCi of[3S]methionine
wasthenadded, and theplateswere left for2hfor
pulse-labeling or 12h for steady-state labeling. The
cellswerethen washed twice withphosphate-buffered
saline andlysedby the addition of1ml of buffer B(50
mMTris [pH8.0],120mMNaCl,0.5%NonidetP-40),
and thecellulardebriswaspelletedbycentrifugation.
Extractswerestoredat-800C ifnotusedimmediately.
Immunoprecipitationandgelelectrophoresis.
Cellextract wasdiluted into bufferB,andan
appro-priateamountofantibodywasadded,usually5to 10
pl.
After2 to 4hat0°C,a10%suspensionofFormalin-fixedStaphylococcusaureusCowan I (ATCC 12598)
(26) in buffer Bwasadded,usingtwovolumes of 10%
S. aureuspervolume ofantibody. After1 to 2hat
0°C,theS. aureuscellswerepelleted by
centrifuga-tion, washed three times with bufferB,andsuspended
insamplebuffer (0.5 M Tris[pH6.8],5%
,B-mercap-toethanol,3% sodiumdodecylsulfate[SDS],10%
glyc-erol, 0.05% bromophenol blue).The resuspended S.
aureuscells were boiled for 2 minand pelleted by centrifugation,and thesupernatantsamplebufferwas
loaded onto an SDS-polyacrylamide gelwitha 7 to
18%gradient ofacrylamide (51).
Purification ofradioactive smalltantigen.A
10-ml culture ofHP1cellswaspulsedfor5min with
500pCiof[3S]methionineasdescribed above. This
crude extractwasthen subjectedto
immunoprecipi-tationwith 200plof hamster anti-Timmunoglobulin
G, and theprecipitatedproteinswerefractionatedby
preparative SDS-polyacrylamide gel electrophoresis
as described above. The band of small t antigen,
identified byautoradiography, was excised, andthe
proteinwaselutedbyelectrophoresisdirectlyintoa
VOL. 37,1981
on November 10, 2019 by guest
http://jvi.asm.org/
dialysis bag containing0.2mg ofBSA. The SDSwas
removed by dialysis againstincreasing concentrations
of urea, upto 8M, followedbydecreasing
concentra-tions ofurea inthepresence of 1% BSA. After final
dialysis against 1% BSA, precipitated material was
pelleted by centrifugation, andthe purified small t
antigen (ca. 107cpm)wasusedfortrypticfingerprint
analysis orradioimmunoassay.
Radioimmunoassay. The sample to be assayed
(upto 20
p1)
wasaddedto100pl
ofradioimmunoassaybuffer(phosphate-buffered saline,0.1%NonidetP-40,
0.1%BSA)containing0.03to0.05tdofhamster anti-T
antibody. After 20 minat00C, 1,000 cpm (2to10ng)
ofpurified radioactive smalltantigenwasadded,and
the immune complex formation was continued for
another20min.Thecomplexwascollected with2
pl
ofa 10% suspension of S. aureus Cowan I cells by
incubationat00Cfor10min.The cellswerepelleted
bycentrifugation andresuspended in 10
pl
of water,and radioactivity was quantitated by scintillation
counting. Theamountofantibodyusedin theassay
wasdeterminedby titrationastheamountrequiredto
precipitate50 to70%of the pure radioactive smallt
antigen.
To assayextractfromSV40-infected monkeycells,
large T antigen was first removed by precipitation
with5
p1
of anti-D2serum.Thesupernatant from thatimmunoprecipitationwasthensubjectedtothe
stand-ardradioimmunoassaydescribed above.
Column chromatography of small t antigen.
(i) HP1. For preparative purposes, HP1 cells were
growntolatelog phase inarichmedium(0.7%
Casa-minoAcids, 0.7% tryptone[Difco Laboratories],0.7%
yeast extract, 0.7% brain heart infusion, 20 mM
K2HPO4[pH7.2],0.5%glycerol,20,ug ofampicillin per
ml). Thecells were lysed by sonication in10mM Tris
(pH7.8)-imMEDTA-0.2 MNaCl-0.1mMDTT-50 Mgofphenylmethylsulfonyl fluoride per ml, and the
lysatewascentrifugedat100,000xgfor1h.Nucleic
acidswereremoved fromthe S-100by the addition of
fresh 1% protamine sulfate(Sigma)to0.2%final
con-centration andremoval of theprecipitate by
centrifu-gation. The supernatant was then brought to 60%
saturation with ammonium sulfate (Schwarz-Mann),
and the precipitate wascollected by centrifugation,
resuspended in the appropriate buffer, and loaded
ontoagel filtration column. The columns were
equil-ibrated in either20 mMTris(pH 7.8)-i MLiCl-0.1
mM DTTor 20 mM Tris(pH 7.4)-50 mMNaCl-0.1
mMEDTA-0.1mMDTT.For alarge-scale
prepara-tion (15 g ofHP1 cells), 20 ml of the resuspended
ammonium sulfate pellet was loaded onto a Biogel
A1.5mcolumn (80 by 3.5 cm), and 5-ml fractions were
collected,togenerateaprofile similar to that seen in
Fig.7.
(ii) SV40. One hundred 100-cm plates of CV-1
monkeycellswereinfected with 5 PFU ofSV40 per
cell,andthecellswere harvested afterapproximately
3daysat370C.AfterswellinginDounce buffer (0.01
M sodiumphosphate [pH 7.0], 0.01 M NaCl, 4 mM
MgCl2, 0.1mMDTT,50ug ofphenylmethylsulfonyl
fluoride perml), the cells were disrupted by 20 strokes
in atype Bground glass homogenizer and centrifuged
at 5,000 rpm for 5 min to pellet the nuclei. The
supernatantcytoplasmic fraction was brought to 60%
saturation with ammoniumsulfate, and the precipitate
was pelletedbycentrifugation and resuspended in2
ml of20 mM Tris (pH 7.4)-50 mM NaCl-0.1 mM
DTT. This was fractionated onaBiogel A1.5m column
(60 by 1.5cm) equilibrated with thesamebuffer, and
1.2-mlfractionswerecollected. In all cases, the nucleic
acids andprotein were monitored by absorbance at
260 nm and280nm,respectively. The smalltantigen
wasdetected byradioimmunoassay.
RESULTS
Construction ofplasmidsdesignedto ex-press SV40 small t antigen. We have used theplasmidpGL101toachieveexpression of the SV40 small t gene in bacteria. This plasmid consists primarily of pBR322 DNA sequences between the EcoRI site and the PvuII sitewhich contain thegenecoding forampicillin resistance. Spanning theEcoRI-PvuII
junction
is a95-bp
restrictionfragmentcontaining theE.coli UV-5 lac promoter, operator, and ,B-galactosidase ribosome
binding
site(24).Thisfragment
is suf-ficient to promote efficient transcription and translation ofany genewhichhasits initiation codon fusedtotheribosomebinding site in the properorientation.Digestion ofSV40 DNAwith therestriction enzymeHindIIIgeneratesa1,169-bp
fragment,
designated theB
fragment,
whichcontains the entire small t gene with the initiation codon located 12 bpfrom one ofthe ends. This frag-ment was purified by preparative agarose gel electrophoresis and then insertedintothe PvuII site ofpGL101 by usingtwodifferentprocedures (Fig.1).The first recombinant plasmid was con-structed by
modifying
the ends of the SV40 HindIIIBfragment.
E.colilarge fragment
DNA polymerase I was used to synthesize a DNA complementto the single-stranded endsoftheHindIII
fragment
togenerate blunt ends.Plas-midpGL101DNA wasthendigested withPvuII, andthe modifiedSV40restrictionfragmentwas inserted with T4 DNA
ligase.
Because hybrid molecules do notcontainanyPvuIIrestriction sites, the ligation products were digested exten-sively withPvuII before transformation to re-duce the background ofrecircularized pGL101 DNA.The circular recombinants were then usedtotransform E. coli 294, andtransformantswere selectedby growthinthepresence of
ampicilhin.
Bacterial colonies containing SV40 sequences were identified by a modification of the Grun-stein andHogness (21)screeningprocedure. Sev-eral suchcloneswereselected, and theplasmid DNAs were analyzed by restriction enzyme mapping and Southem blot hybridization to confirm the presence and determine the orienta-tion ofthe inserted SV40HindIIIBfragment.A
on November 10, 2019 by guest
http://jvi.asm.org/
OVERPRODUCTION OF SV40 SMALL t ANTIGEN 687
EcoRI
Pldc
PvullAMR I3
CutwithPvufl
Ligate onHindmlinker Cut
t g HindM
SV40HindNB Fragment
Mixond ligote
P/vc
9 ~~~~~Ligate
t_
I
with PvuIl
P/Ic HlndHM
t~~~~~
IPartial cut withHindM
and treated with BAL 31
t
SV40HindmBFragment dNTP+lorge t Ifrogmentpo/I
Mixondligate
FIG. 1. Schematic representation ofplasmid construction designed to express SV40 small t antigen.
Preparation of the DNA and construction of the hybrid plasmids is described in the text.
clonecarrying the smalltgenefusedtothe lac promoter in the proper orientation was selected, and the plasmidwasdesignatedpSV250.
Roberts et al. (42) have demonstrated that overproduction of proteins utilizingtheplasmid lac promoter system can be achieved by ran-domly varying the distance between the bacte-rial ribosome binding site, postulated by Shine andDalgarno (44), and the initiation codon of the inserted gene. A change of asfewas 3 bp canaffect thelevel of expression byasmuchas 20-fold (42). Withthisin mind,we constructed
a
plasmid
whichcontainedarestriction sitebe-tweenthe Shine andDalgarnosequenceand the smalltAUG codon. This recombinantallowed directmanipulationof the basesequence within thiscrucialregion.
PvuIIwasusedtolinearizepGL101 DNA,and HindIII linkerswere
ligated
tothe plasmid by using T4 DNA ligase. The SV40HinduI
B fragment was then inserted into this HindIII site, and the hybrid molecules were used totransformE. coli294. The transformantswere
screened for colonies containing SV40 se-quences, as before. One clone with the SV40
insert in theproper orientation wasselected,and
the
plasmid
wasdesignated pSV240.This plasmid has two HindIII sites, one of whichlies betweenthe bacterialShineand
Dal-garno
sequenceand the smalltinitiation codon.PlasmidpSV240DNA wassubjectedtopartial digestion with HindIIItogenerate apopulation ofmolecules,someof whichwere cutonlyatthe hybrid ribosome
binding
site. These molecules were then treated with nuclease BAL 31 to decreaserandomly the distance betweenthetwosequences. Linearmoleculeswerethen
purified
by agarose
gel
electrophoresis, recircularized withT4 DNAligase,
and usedtotransformE.coli 294.
Forty
transformantscontaining
themodified
pSV240
molecules were selected and screenedbyrestrictionenzymemappingof plas-mid DNA for loss of theHindIII sitenear the lac promoter. Fourteen such cloneswere iden-tified and tested byimmunoprecipitation
for theirabilitytoproduce small tantigen. Asex-pected,avarietyof different levels of
expression
was detected. Three clones, carrying
plasmids
designated HP1, HP2, and HP3,
appeared
toproduce
substantially
moresmalltprotein
thanVOL. 37,1981
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.485.45.442.69.378.2]didpSV240. Sincetheseplasmids exhibited ap-proximately thesame high level ofproduction, HP1 was arbitrarily selected for quantitation andcharacterizationofthe viralprotein.
Synthesis of SV40 small tantigenin bac-teria.We have two antibodies available to de-tect the early gene products of SV40. One of theseantibodieswasisolated fromthe serum of hamsters bearing SV40-induced tumors and is specific for thevirallycodedlargeTand small tantigens (3, 13).The otherantibodywasmade by injecting rabbits with
purified
D2 protein isolated from HeLa cells infected with the ade-novirus-SV40hybridvirusAd2+D2(54).The D2 protein lacks theaminoterminalregionoflarge T antigen which is shared with the small t polypeptide (22). Antibodies directedagainstD2 will therefore react onlywith large T antigen-specific determinants and notcross-react with small tantigen (54). SincetheSV40HindIIIB fragmentcontains one-half of thelargeT struc-tural gene as well as the entire smallt coding sequence,itcouldpotentiallypromotethe syn-thesis oftruncatedlarge T-related proteins. The useof the anti-T and anti-D2seraallowsusto distinguish these proteins from SV40 small t antigen.TodetectSV40-relatedproteins expressed in bacteriaand comparethem with authentic SV40 smalltantigen,bothSV40-infectedmonkey cells andHP1bacterial cellswerepulse-labeled with [35S]methionine,andtheextractsweresubjected to immunoprecipitation with anti-D2 and anti-T sera. A control preimmune serum was also used toidentify proteinswhich were nonspecif-ically precipitated (Fig. 2A,lanes1 and 2). The immnunecomplexes werecollected on protein
A-bearing
S.aureuscellsandthenfractionatedby
SDS-polyacrylamide
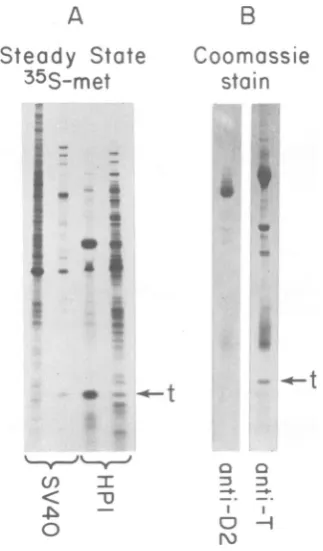
gelelectrophoresis. Asex-pected, the rabbit anti-D2 serum precipitated only the SV40 largeTantigen from the infected monkey cell extract,whereasthe hamsterserum precipitated both the large T and small t anti-gens (Fig. 2A,lanes3and5).No proteins in the HP1 extract werespecifically precipitatedwith the rabbit anti-D2serum, but the hamster anti-Tantibodyselectively precipitateda 12-kilodal-ton(kd) protein and a 15-kdproteinwhich co-migratedwiththeSV40smallt antigen (Fig. 2A, lanes4 and6). These resultsdemonstrate that theHP1-encoded smalltprotein is the same size and has the same antigenic determinants as authentic SV40 small t antigen.
Furthermore,
with the exception ofthe 12-kd protein, there appear tobenoadditional SV40-relatedproteins synthesizedinHP1.
Sinceour abilityto detect small t antigen is dependent on the use of anti-T antibody, we wereinterestedin
determining
theefficiency ofourimmunoprecipitations.SDS-polyacrylamide gel electrophoresisofacrude extract prepared from pulse-labeledHP1 cells allowed us to vis-ualizedirectlya15-kd protein whichcomigrated withSV40 smalltantigen (Fig.2C,lane4). After anti-Timmunoprecipitationandgel electropho-resis of the supematant fraction, however, none of this protein could be detected (Fig. 2C, lane 3). Furthermore, the pattem displayed by the supematant was almostidentical to that of an extract prepared from E. coli cells carryinga control nonproducer plasmid (Fig. 2C, lane 5). These results indicate that our immunoprecipi-tation technique is capable of selectively and quantitatively removing small t antigen from a crude cell extract.
Quantitation of small t antigen produced byrecombinantplasmids.PlasmidspSV240, pSV250, andHP1 were constructed toprovide different levels ofexpressionofsmalltantigen. To compare the relative amounts of small t proteinbeingproducedby these clones, bacteria carryingpSV240, pSV250, and HP1 were pulse-labeled with[3S]methionine,and the crude ex-tracts weresubjected to anti-T immunoprecipi-tation with an excess ofantibody, followed by SDS-polyacrylamide gel electrophoresis (Fig. 2B). Plasmid pSV250 was found toproduce two-fold more small tantigenthan didpSV240,and HP1 produced approximately 25-fold more of thisprotein than didpSV240. Asexpected,the relative levels of small texpressionvaried con-siderably with changes in the structure ofthe hybrid ribosomebindingsite.
Havingdemonstrated that HP1 is ahigh pro-ducer of SV40 small tantigen,wewereinterested in quantitating the amount ofsmall t protein present in bacteriacarryingthisplasmid. Since
pulse-labeling
reflects the rate of synthesisrather than absolute amount, HP1 cells were labeled understeady-state conditions. A crude extract containing 106 cpm of labeled protein wasthensubjectedto
immunoprecipitation
with an excess of anti-Tantibody (Fig. 3A, lane 3). By excising the band of antigen from the gel, 12,000 to 25,000 cpm could be recovered. Since thesmall tpolypeptidecontainsapproximately threefold more methionine residues than does anaverage protein (14), we calculated that small t antigen represents 0.4 to 0.8% of the total labeledproteinin the extract.Toconfirm thisquantitation,we immunopre-cipitatedsmall tantigenfromacrude extract of HP1cellsanddirectlyvisualizedthe proteins by Coomassie blue staining (Fig. 3B). Approxi-mately 2 to 3 ,tg of small t antigen can be
inmmunoprecipitated
from 300,ug ofprotein. Thisis equivalent to 0.6 to 1% of the total cellular protein,or40,000 to80,000moleculesofsmall t
on November 10, 2019 by guest
http://jvi.asm.org/
OVERPRODUCTION OF SV40 SMALL t ANTIGEN
A
B
;0i;0fXFv-1;
0;,-0AAt.ira
X -W tE;*;y 0
;- titSt ff?.^-7. 00
*:0i.i.i
aX v 0;; +-.SS:T..
00 a.ff SX ffff00.:i:(Vit:}7.tit-.;d03Si,
tU0fff W- .-'d il 0 _
ff *-', iV;0 r ;
f<-si.X*''it0<,.?W;.';
t00S0i;0000 f-t t: 0&X.> 0!S00 00000 |w-WEE-E
o O a
O D :
-:
_-_ I I
O O -|
N
n1)D 'D
Ir
<
(J)VJCJ).
<<
.4
O
00
FIG. 2. Characterization ofsmalltantigens byimmunoprecipitation andgelelectrophoresis.SV40-infected
monkey cells and clonespSV240,pSV250,andHPIwerepulse-labeled with[35S]methionine,and the crude extractsweresubjectedtoimmunoprecipitationwith theantibodies described below. Theimmunecomplexes
werecollectedonprotein-A bearingS.aureuscells(26), fractionatedon a7to18%gradient ofpolyacrylamide
containingSDS(51),and visualizedbyautoradiographyasdescribed inthetext.(A) Labeledextractsfrom
SV40-infected monkey ceUs (lanes 1, 3,and5)and HP) cells(lanes 2, 4,and6)weresubjectedto
immunopre-cipitation with control rabbit anti-hamster serum, rabbit anti-D2 serum, and hamster anti-T antibody,
respectively. (B) Samples containing2x 106cpmofextractfrom SV40-infected monkey cells and 1.5x106
cpmofextractsfrom pSV240,pSV250,andHP) cellsweresubjectedtoimmunoprecipitationwithan excessof
anti-Tantibody.(C)Lane1depictsananti-Timmunoprecipitate from SV40-infected monkeycells. Asample
containing4x 105 cpmofextractfromHPI cells wassubjected toanti-Timmunoprecipitation, and the
precipitated proteinsaredepictedinlane2;1 x 105cpmof the supernatant from thisimmunoprecipitation
was runin lane3. Asample containing105cpmofHPI cellextractwasruninlane4,andanequalamount ofradioactiveprotein from apSV240-like clone, carrying aplasmid withthe SV40 insert in the wrong
orientation,was runinlane5.(D)HP) DNAwasused totransformaderivativeofE.coli 294whichcarries the laci'geneandthusoverproduceslacrepressor(34).Anti-Timmunoprecipitationandgelelectrophoresis
wereperformedonceUseither withorwithout induction withisopropyl-,B-D-thiogalactopyranoside.
antigenperHP1bacterial cell.
Wethencompared thelevelofexpression of
HP1 with that ofcelLsproductively infected with
SV40. Infectedmonkeyceliswerelabeled with [3S]methionine under steady-state conditions.
Acrude extract was prepared, and 106 cpm of
labeled protein wastreated with an excess of
anti-T antibody (Fig. 3A, lane 2). By excising theband ofsmalltantigen from the gel,
approx-imately 0.03to0.06% of the totallabeled protein could berecovered.
Sequencedetermination of thehybrid ri-bosome binding sites. The nucleotide
se-quenceof50bpencompassingthehybrid
ribo-somebinding site ofpSV240,pSV250,HP1, HP2,
and HP3 was determined by the Maxam and
Gilbert(31) sequencing method (Fig. 4).As
pre-dicted, pSV240 contained one HindI linker
between thelacpromoterand theSV40insert,
andpSV250wasmiing3bpof the linker. HP3
is a12-bp deletionmutant ofpSV240,whereas
HP1 and HP2 are missing an identical 11-bp sequence.Thedistancefromthe Shine and Dal-garno sequencetothe smallAUG codon is thus
reduced from 20bpin pSV240to 9 bpin HP1
andHP2and 8bp inHP3. This indicates that
C
12
345
D
0L N
120
K-I
IO0K-1
85K-1
iXi
20K~
i. .-4A^12K-#'
I +
)
c-OG)
VOL. 37,1981
689
on November 10, 2019 by guest
http://jvi.asm.org/
690 BURGESS, AND
B
Coomassie
stain
() In
< X
a I I
N
FIG. 3. Quantitation ofsmalltantigen produced
byHP). (A) Samples containing106cpmofextract
from SV40-infected monkey cells andHPI cells
la-beled understeady-stateconditionsweretreated with
anti-Tantibody,and theprecipitatesweresubjected
togel electrophoresis. The endlanescontain 6.5 x
104cpmofcrudeextractfromthese cells. The
immu-noprecipitatedsmalltantigens,shown in thecenter
lanes, wereexcised andquantitatedasdescribed in
thetext.(B)An unlabeled crudeextractofHP) cells
containing300A gofproteinwassubjectedto
immu-noprecipitationwith10 1l ofrabbit anti-D2serum or
hamster anti-TIgG. Theprecipitated proteinswere
fractionated byslabgelelectrophoresisanddirectly
visualizedby stainingwithCoomassiebrilliant blue
(Bio-Rad Laboratories). These samples were not
heatedbefore gel electrophoresis.
therearesequences inpSV240 thatreduce the efficiencywith which themRNA canbe bound
by the 30Sribosomal subunitor,possibly,which
attenuatetranscription.
Characterization ofthe small t antigen produced by HP1. Our motive for constructing
a bacterial plasmid which overproduces SV40 small tantigenwas tofacilitate purification of the protein for functional analysis. We were
therefore interestedin characterizingthe HP1-encodedprotein andcomparing it with authentic SV40small t antigen. Having determined that
the size and antigenicity of the HP1-encoded
andauthenticsmalltantigenswereidentical,we
nextused themore sensitivetechnique of
isoe-lectric focusing to characterize the physical properties of thesetwoproteins. Crudeextracts
prepared from both
[3S]methionine-labeled
HP1 cells and E. coli cells carrying a control
nonproducer plasmidweresubjectedto
isoelec-tric focusing in the first dimension and then SDS-polyacrylamide gel electrophoresis in the second dimension (Fig.5A andB). Under these conditionsitwaspossibletoseparatebothsmall
tantigen and the 12-kd protein frommostother bacterial proteins. The HP1-encoded antigen
wasfoundtomigrateintheisoelectricfocusing dimension withanisoelectricpoint of 7.2to7.4, identicaltothereported isoelectric point of
au-thentic SV40 small t protein (Fig. 5C) (12). In addition,amixture ofsmalltantigen isolated by anti-Timmunoprecipitation from both SV40-in-fected monkeycells and HP1 cells comigrated
during two-dimensional gel electrophoresis (Fig. 5D). Therefore, the size and the charge of the HPl-encoded small tantigenwere identicalto
that of the authenticvirally coded protein. Upon isoelectric focusing,weobservedcharge
heterogeneity in small t antigen isolated from both SV40-infectedmonkeycellsand HP1cells.
Inmostcasesthree differentspecieswere
iden-tified, although more forms could be resolved
withalongerexposure.
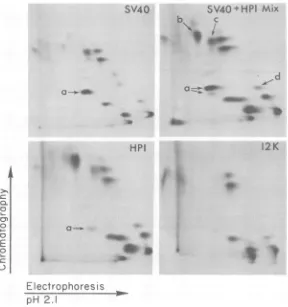
We performed tryptic fingerprint analysis of the smalltantigen expressed in HP1 and
com-pared itspattemwiththat of theauthentic SV40 small t protein. Small t antigen was purified
from pulse-labeled SV40-infected monkey cells and HP1 cells by anti-T immunoprecipitation
and SDS-polyacrylamide gel electrophoresis. These purified radioactive antigens were then
oxidized with performic acid and subjected to
digestion with tolylsulfonyl phenylalanyl chlo-romethyl ketone-treated trypsin. The peptides
were fractionated by electrophoresis atpH2.1
in one dimension and ascending
chromatogra-phy in the second dimension. Allexceptoneof
thetryptic peptides comigrated in both dimen-sions(Fig. 6). The unique peptide corresponded
totheN-terminal fragment of smalltantigen in thefingerprintpatterndescribed by Linkeetal. (28). The altered mobility of the HP1-encoded
small tpeptide could, therefore, be due to the absence of N-terminalacetylation knownto oc-curineucaryoticcells(36), although other
post-translational modificationsof the HPl-encoded protein cannot be ruled out. As observed by
other investigators (46), some of the peptides seem to be over-represented in the digest of
small tantigen isolated fromonesourcerelative
to the other. However, even tryptic digests of
identicalproteins exhibit such quantitative
dif-A
Steady State
35S-met
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.485.70.232.71.347.2]OVERPRODUCTION OF SV40 SMALL t ANTIGEN 691
PLASMIDS smollt
SD HindM linker
pSV240
--AC[AG
G AAACAG TCAGTTTGCAAAGAGGAT----TGITCClTITGTC
GTTCGAJAACGTTTCTACCTA--3bp
SD 7
pSV250
--ACIAGGAAACAGAGCTTTGCAAAG
GAT----TGI
CTTTGTCTCGAAACGTTTCTACCTA--HPI,HP2
llbp
SD 'lZ7
--AC GGAAACAGCCAG G
GAT--
--TGITCCTITTGTCGGTCTACCTA--LEVELOF EXPRESSION (molecules/cell)
<2,000
3-6,000
40-80,000
12 bp
SD
`
HP3
--ACGGAAAACAGCCA
T GAT-- 40-80,000--TGj[CCTTTGTCGGTTACCTA--FIG. 4. Nucleotidesequenceof the hybrid ribosome binding sites. Plasmid DNA was digested withHinfI,
end-labeledbyapolynucleotide kinase exchange reaction with[y-32P]ATP(7),and then cleaved withPstI.
The 890-bpfragment containing the hybrid ribosome binding site for small t antigen was purified by gel
electrophoresis and sequencedaccordingtothetechnique of Maxam and Gilbert (31). The ribosome binding
sequence,hypothesizedby Shine and Dalgarno (44), is abbreviated to SD, and the SV40 sequence is drawn in
bold letters. Thedirectionof transcription isfromlefttoright.
ferences, suggesting that this variabilityis inher-entinthe technique.
The structureof the 12-kdprotein synthesized byHP1 wassimilarlyanalyzed by tryptic finger-printing. Three, andpossiblyfour, smallt pep-tides were absent in the 12-kdprotein pattern (Fig. 6, peptides a, b, c, and d). Peptides a, b, and chave all been mapped tothe amino ter-minus ofsmalltantigen(28). The12-kdprotein may therefore be considered to be an amino-terminal deletion of small t antigen. Further analysis willbenecessary toascertainthe exact structureofthis12-kdprotein.
Chromatographic properties
of theHP1-encoded small t
antigen. Having
achieved ahigh level of production ofintact small tantigen, we began working on the purification of this proteinfrom E. colicarrying theHP1 plasmid. Since no biochemical
activity
for the small tprotein hasyet beenidentified,wedependedon anti-T antibodies as a specific probe for the presenceoftheantigen.
Initially,
acomplement fixationassaywasattempted, butsmall tantigen wasfoundtobe inefficientinfixing complement, evenwhenitwaspresent inlarge
amounts.We thereforedeveloped
a sensitive and rapid ra-dioimmunoassay. One unit ofsmall t antigen (approximately 5 to 10 ng) was defined as the amount ofprotein required to give 50% inhibi-tion ofcomplex formation between pure radio-active small tantigenand alimitingamount of anti-Tantibody.Our initialattemptatthepurificationof HP1-encoded small t antigen involved subjecting a crudepreparation of HP1 extract togelfiltration chromatography. Surprisingly,about 50% of the
antigen was found to elute with an apparent molecularweightof greater than 106 (Fig. 7).
SDS-polyacrylamide gel electrophoresis and direct visualizationoftheantigenby Coomassie blue stainingwere used toconfirm the presence of the small tprotein in the excluded and in-cludedfractions(Fig. 8A).Approximately three-fold more 15-kd protein was visible in the ex-cluded fraction than in the included fraction. This result could be accounted for if two-thirds of the cross-reacting antigen in the included fractionwerethe 12-kdprotein.
We havetried4 MNaCl,0.2 M
,8-mercapto-ethanol,6 Murea, avariety of
detergents,
and extremesin pHin anattemptto dissociate the small tproteininto itsmonomerform. None of theseagents, either aloneorincombination,
wassufficienttodisruptthe smallt
complex.
How-ever, with extremedenaturationconditions such asthose employedduring SDS-polyacrylamide gel electrophoresis, we found the antigen mi-grating as a monomeric species. The small tmoleculesarethereforenot
covalently
boundtooneanother, except
perhaps by
labile disulfide-typebonds.Wearecurrently pursuing
alternate conditions to achieve dissolution of the high-molecular-weightcomplex.
Having established that small t antigen was
present in HP1 cells ina
high-molecular-weight
form, we were interested in
characterizing
the gelfiltrationproperties
of authenticSV40small tantigen.Acrudecytoplasmic
extractof SV40-infected monkeycellswasfractionatedby
aga-rose A1.5m
chromatography,
and the small tprotein was found to elute in a broad
peak
extending from
approximately 500,000
daltonsVOL. 37,1981
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.485.96.392.77.229.2]692 THUMMEL, BURGESS, AND TJIAN
IEF
llJ
CD
uI
, ':
X W i f,&S 0, * '
'#E i t40" .0::
.|i::
*< * =
0:"i:i" i : i.0, f,+,. ziS.iS:-:
hiR:::::D:
:,'-'i ::: ::' :::::*.
t:.ff
,,, ::04:: (:
.:'iE't9 i.iMW =
A:-:,::Xi':...::7
,,.ft
.SaWBs:.tA,C,i'
::'iN-'''Vit;
''i":0.'
.00>4s4'0'StCE;
i@'X,
d7
...7:-,,¢-,,;
.S:-i
,,R,,,,w0E$:
..RE
¢,>v.-: m.'i =%Q, i, ,04
.'' -:
:::-:E:. .
i- iS.i vii 0
.... ... .;,
-7a;-0;v: 000 0- Bx
.. *t:f)000; ff ff .;
.:
::::
:Xt000;C00. f;
:::
:? ..*A:
::i..7 Z E:
1, -*.it E
f ,:
::
, .00 7d, S*;i
W
::, ,l:
::
FIG. 5. Two-dimensionalgelelectrophoresisofsmalltantigenfrom SV40andHPI.Thefollowingsamples
from[35SJmethioninepulse-labeledcellsweresubjectedtoisoelectricfocusingin onedimension and
SDS-polyacrylamidegelelectrophoresis (PAGE)in thesecond dimension(35).Panels:(A)7x105cpmofanextract fromcellscarryingapSV240-like plasmid, with theSV40insert in thewrongorientation; (B) 7 x105cpm of
HP1 extract;(C)2x 103 cpmofimmunoprecipitatedsmalltantigenfrom SV40-infectedmonkeycells;(D)A
mixture of2 x 103 cpm each ofauthentic SV40 and HPl-encoded small t antigen purified by anti-T
immunoprecipitation.
downtomonomeric size(Fig.7). Onceagain,gel electrophoresis of anti-T immunoprecipitates from several column fractions confirmed the presenceofsmalltantigen in a high-molecular-weight form (Fig. 8B). P. Tegtmeyer, T. Spill-man, and F. R. Schuetz (Cold Spring Harbor Symp. Quant. Biol.,inpress)havealsoreported size heterogeneity of authentic SV40 small t
antigen isolated fromlytically infectedmonkey
cells. The behavior of the HP1-encoded smallt
protein as a high-molecular-weight complex is therefore not unique to the bacterial system. The larger small t complex and high ratio of oligomerstomonomers inthe bacterialextract
maybe due to the increased intracellular
con-centrationfound in HP1cells.
Itis alsopossiblethatsomecomponentof the bacterial cell isresponsiblefor the shift ofsmall J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:10.485.89.394.61.490.2]OVERPRODUCTION OF SV40 SMALL t ANTIGEN 693
t antigen to a very high molecular weight. To testfor this,anextract from E. coli 294cells was mixed with a cytoplasmic extract from SV40-infectedmonkeycellsandsubjected to gel filtra-tion chromatography. Thesmall telution pat-ternlooked identicaltothat frominfected mon-keycellsalone, indicating that, under these con-ditions, no bacterialcomponents are contribut-ing to the formation of the high-molecular-weightcomplex.
We haveused phenyl-Sepharose chromatog-raphyaswellas avarietyof ionexchange resins such asDEAE-cellulose, Bio-Rex 70, phospho-cellulose, and hydroxyapatitetofractionate fur-ther the smalltpolypeptide. Inmost cases,the antigen was found to behave heterogeneously. Todate we have beenabletoachieve a 100-fold purificationof thisprotein, andwe arecurrently
X1%
-c
0.
01I...
0
E-00
pursuing alternative means of
purifying
the HP1-encodedsmall t antigen tohomogeneity.DISCUSSION
We have maximized the expression of SV40 smalltantigen by constructing a bacterial plas-mid whichfuses the SV40 F gene to the E. coli lac promoter-operatorsequence.Recently,other investigators havesuccessfullyusedsimilar tech-niques to achieve expression of various procar-yotic andeucaryotic viralgenes (1, 41, 42). We have modifiedtheirprocedureby first inserting theforeigngene into a vectorcarrying the lac promoter and then reconstructing the hybrid ribosomebindingsiteby treatment with BAL 31 nuclease.This enzyme has asingle-stranded en-donucleaseactivityand also acts
exonucleolyti-Electrophores
is
[image:11.485.97.385.265.572.2]pH
2.I
FIG. 6. Trypticfingerprint analysisofsmalltantigen fromSV40 and HP1. Proteins labeled with
[MS]-methioninewerepurified byanti- Timmunoprecipitation, SDS-polyacrylamide gel electrophoresis, and
elec-troelutionasdescribedin the text. Samplesweresubjected toperformic acid oxidation anddigestedwith
tolylsulfonyl phenylalanyl chloromethylketone-treated trypsin (Worthington) asdescribedpreviously (46).
Samplesof1 to5 1l(ca.50,000to100,000 cpm)werespottedonBrinkmanglass-backedcellulosethin-layer
plates, subjected to electrophoresis atpH2.1 (8% acetic acid, 2%formic acid) for 1,000 V-h, and then
chromatographed in the second dimensionfor4to5h inbutanol-pyridine-acetic acid-water(97:75:15:60).
Theplatesweredriedandautoradiographed. Thearrowsindicatepeptidesdiscussed in thetext.
VOL. 37,1981
on November 10, 2019 by guest
http://jvi.asm.org/
694 THUMMEL, BURGESS, AND TJIAN
.0 0
4
-r
I
0
0 N
4
l
0
4.
= E C,)
0
N
-r
0 40
4
I:
, )
.E
= oi
[image:12.485.64.456.56.264.2]40 60 80 100 120 140 160 180 20 30 40 50 60 TO
Fraction Fraction
FIG. 7. Agarose A1.5m columnchromatographyofextractfrom HPI cells andfrom SV40-infectedmonkey
cells. Crudeextractsfrom15gof HPI cells(1,300mgofprotein) andonehundred 100-cmplates of
SV40-infectedmonkeycells(12mgofprotein)wereprepared and subjectedtoBio-GelA1.5mcolumnchromatography
asdescribed in the text. Small tantigen was detectedby radioimmunoassay by using anti-D2serum to
preabsorb large Tantigenin theinfectedmonkeyceUfractions,asdescribed in thetext.Nucleic acids and
proteinweredeterminedspectrophotometrically byabsorbanceat260nmand280nm,respectively. Thepeak
of smalltantigeninfraction 25 of the infected monkey cell profile couldnotbe detectedby gelelectrophoresis
andCoomassiestainingand thus may represent residuallargeTantigen notpreabsorbed bythe anti-D2
serum.
callytodigestprocessivelyfrom thetermini of
a DNA duplex (20), leaving blunt ends which
canberejoined byT4DNAligase.These prop-erties ofBAL31 nuclease make it ideal for the construction of"expression plasmids" and forin vitro deletionmutagenesis in general.
By transforming HP1 intoaderivative ofE. coli 294 whichoverproduces lac repressor, we haveconfirmed thatexpression of smallt anti-gen is under the control of the lacregulatory sequences. Theexpressionof smalltantigen in this host was effectively abolished. However, uponinductionwith
isopropyl-,8-D-thiogalacto-side, maximum levels of expression could be achieved (Fig. 2D). In contrast, when this gra-tuitous inducerwasaddedto aculture of HP1 in its normal E. coli 294 host, only a slight increase inexpression could be detected. This is
to be expected as there are approximately 10 molecules of lac repressor present in a wild-type E. coli cell (17), and these few molecules can have little effecton the30 to 40HP1 plasmids which are present in a transformed bacterium
(6).
Under ourgrowth conditions, HP1 produced 40,000to80,000molecules ofsmallt antigenper' cell, orapproximately 1,000 to 3,000 molecules ofsmall tantigen per plasmid. Thus, HP1 was functioning at20 to60% of its maximum
theo-reticalefficiencyincomparisonto afully induced
,8-galactosidase
gene(1).
Figure 3A demonstrates that the
concentra-tionofsmalltantigeninan extractof HP1 cells was 10-fold higher than that in an equivalent
amount of SV40-infected monkey cell extract.
The quantitation of small tantigen inthis eu-caryotic systemis
imprecise,
because detection of the protein can be achieved only by immu-noprecipitation (13). However, assuming that 0.05% of the totalproteininthe SV40-infected monkey cells is small t antigen,then 1 liter of HP1cellsatlate log phase containsasmuchof the antigenas canbe recovered from 1.2 x 105 plates of SV40-infected monkeycells. This level ofproduction should facilitate the purification of thisproteintohomogeneity.The small t antigens produced by HP1 and SV40were found tobe virtually indistinguish-able on the basis of electrophoretic mobility, antigenic determinants, isoelectric point, and trypticfingerprintanalysis. The only detectable difference between theseproteinswasthe slower migration of theHP1-encoded N-terminal tryp-tic peptide during ascending chromatography. This difference ismostlikelyduetothe absence of amino-terminal acetylation in the bacterial protein. Like manyeucaryoticproteins (4),small
t antigen isolated from SV40-infected monkey
on November 10, 2019 by guest
http://jvi.asm.org/
OVERPRODUCTION OF SV40 SMALL t ANTIGEN 695
A
1 2 3 45 6
B
1234
[image:13.485.44.236.63.297.2]HPI
SV40
FIG. 8. SDS-gel electrophoresis of
high-molecular-weightandmonomericsmalltantigen. Samplesfrom
gel filtration columnfractions containing
high-mo-lecular-weightand monomericsmalltantigenwere
fractionated by SDS-polyacrylamidegel
electropho-resis either beforeorafter immunoprecipitation. The
proteinswerevisualized by staining withCoomassie
brilliantblue. (A) Samples containing150U ofsmall
tantigen fromHPI cellsaseithera
high-molecular-weight complex (lanes Ito3)orinmonomericform
(lanes 4 to 6) are depicted. A sample of the total
protein (lanes1and4),thesupernatantofananti-T
immunoprecipitation (lanes2 and5),and the
immu-noprecipitated proteins (lanes3 and6)were
electro-phoresedinneighboringlanes.(B) Samples
contain-ing 200 U ofsmall t antigen from SV40-infected
monkeycells which elutefrom gel filtrationas a
high-molecular-weight complex (lanesI and2)or as
mon-omeric antigen (lanes 3 and 4) were subjected to
immunoprecipitation with anti-D2 serum to
preab-sorb thelargeTantigen(lanes1 and3).The
super-natant from that immunoprecipitation was then
treated with anti-Tantibody toprecipitate smallt
antigen (lanes2 and4). Large Tantigen has been
proteolyzed toits 89-kd and 84-kdformsduetothe
extractionprocedureused(46).
cells is acetylated at its amino terminus (36). Functionalanalysis of the purified HPl-encoded protein will reveal whether this modification is required for biological activity.
We found thatpSV240, pSV250, and HP1all produced a 12-kd SV40-coded protein in
addi-tiontothe 15-kdsmalltantigen. Tryptic finger-print analysis of the 12-kd protein revealed a
structure identical to that of small t antigen,
with the exception of several amino-terminal
tryptic peptides.Since theexpressionof the
12-kg protein could be regulatedwith lac repressor, itstranscriptmustalso derive fromthelac pro-moter.Theexpressionofthe 12-kd protein may be due to protein synthesis initiation at an in-phase AUG codon located within the small t gene. In fact, there is a potential AAGGAGG Shine andDalgarnosequencelocated upstream from the fourth and fifth in-phase AUG codons of the smalltcoding sequence (38). The stable duplex that thissequence could forn with the 3' end of E. coli 16S rRNA may favor internal initiationoftranslation(44, 50).
The 12-kdproteinappears to represent a for-tuitous amino-terminal deletion of SV40 smallt antigen. It should be possibleto construct other deletion mutations in the small t coding se-quence ofHP1 by using site-directed in vitro mutagenesis techniques. Thesemutant smallt proteins maybe useful for studying the func-tion(s) of this viraltumorantigen.
In spite of the high level of production af-fordedby HP1,thepurificationofsmalltantigen fromthis clone isnotstraightforward, duetothe aggregation properties of the protein. We are currently employingunconventionaltechniques which shouldallow us to
purify
thisprotein to homogeneity. Wehave beguntoraise multiva-lentandmonoclonal antibodiesdirectedagainst small t antigen to determine its intracellular location inSV40-infected and transformed cells. Inaddition,weintendtopurify
the high-molec-ular-weightcomplex
ofsmall tantigen,
mono-mericantigen, and12-kdproteinandtesttheir biological activities by microinjectioninto mam-malian cells. Finally, wehopeto testthese dif-ferentspecies for potential enzymaticactivities.
ACKNOWLEDIGMENTS
WethankTony Pawson and Tsu-Hsun Kung for helpwith thetryptic fingerprintanalysis,Taffy Mullenbach for tissue culturepreparation, Gail Lauer formaking pGL101available, MichaelKligmanfor criticalreadingof thispaper,and Judith Pike forpreparation of themanuscript.
This workwasfundedby Public Health ServicegrantCA 25417 from the National Cancer Institute, by the Cancer ResearchCoordinating Committee, byaBiomedical Research Support Grant, andby partial supportfrom Public Health ServicegrantES01896 from the National Institute of Environ-mental Health Sciences.
0 LITERATURE CITED
1. Backman, K., and M. Ptashne.1978.Maximizinggene
expressionon aplasmid usingrecombination in vitro. Cell13:65-71.
2. Berk, A.,and P.Sharp.1978.Spliced earlymRNAs of simianvirus 40. Proc. Natl. Acad. Sci.U.S.A. 75:1274-1278.
3. Black,P. H.,W. P.Rowe,H.C.Turner,and R. J. Huebner. 1963.Aspecificcomplement-fixing antigen present in SV40tumor andtransformed cells. Proc. Natl.Acad. Sci.U.S.A. 50:1148-1156.
4. Bloemendal,H. 1977. The vertebrate eye lens. Science 197:127-138.
VOL. 37,1981
on November 10, 2019 by guest
http://jvi.asm.org/
5. Bouck, N., N. Beales, T. Shenk, P. Berg, and G. DiMayorca. 1978. Newregionof thesimian virus 40
genomerequiredfor efficient viral transformation. Proc. Natl.Acad. Sci. U.S.A. 75:2473-2477.
6. Boyer,H.W.,M.Betlach,F.Bolivar,R.L.Rodriguez, H. L.Heyneker, J. Shine, and H. M. Goodman. 1977. In R. F. Beers, Jr., and E. G. Bassett (ed.), Recombinant molecules: impactonscience andsociety,
p.9-20.RavenPress,Publishers,New York. 7.Chacanos, G.,J. H.vandeSande,and R. B.Church.
1975.Endgrouplabelingof RNA anddouble-stranded DNAbyphosphate exchange catalyzed by bacterio-phageT4induced polynucleotidekinase. Biochem. Bio-phys.Res. Commun. 66:962-969.
8.Clewell,D.B.,and D. R. Helinski. 1969.Supercoiled circularDNA-protein complexinEscherichia coli:
pu-rification and induced conversion toanopencircular DNAform.Proc. Natl. Acad. Sci. U.S.A. 62:1159-1166. 9. Cohen, S. N.,A.Chang,andL. Hsu. 1972. Nonchro-mosomalantibiotic resistanceinbacteria:genetic trans-formationof Escherichia colibyR-factor DNA. Proc. Natl.Acad. Sci. U.S.A.69:2110-2114.
10.Cowan, K.,P.Tegtmeyer, and D. D.Anthony. 1973.
Relationshipofreplicationandtranscriptionofsimian virus 40DNA.Proc. Natl. Acad.Sci.U.S.A. 70:1927-1930.
11.Crawford,L.V.,C. N.Cole,A.E.Smith,E.Paucha, P.Tegtmeyer,K.Rundell,and P. Berg. 1978. Or-ganizationandexpressionofearlygenesofsimianvirus 40.Proc.Natl.Acad. Sci. U.S.A. 75:117-121.
12. Crawford, L.V.,and P. Z.O'Farrell. 1979. Effect of alkylationonthephysical propertiesof simian virus 40 T-antigen species.J. Virol.29:587-596.
13.Crawford,L.V.,D. C.Pim,andD. P.Lane. 1980. An immunochemical investigation ofSV40T-antigens. 2. Quantitationofantigensandantibodyactivities.
Virol-ogy100:314-325.
14. Dayhoff,M.O.,R. E.Dayhoff,andL. T. Hunt. 1976. Compositionofproteins,p.301-310.In M.0.Dayhoff
(ed.), Atlas ofproteinsequence and structure 5. Na-tional BiomedicalResearch Foundation, Washington, D.C.
15. Feunteun, J.,M.Kress,M.Gardes, and R. Monier. 1978.Viable deletionmutantsinthe simianvirus40 early region. Proc. Natl. Acad. Sci. U.S.A. 75:4455-4459.
16. Fraser,T.H.,and B. J.Bruce. 1978.Chicken ovalbumin issynthesized and secretedby Escherichia coli. Proc. Natl.Acad.Sci. U.S.A.75:5936-5940.
17. Gilbert, W.,and B.Mueller-Hill. 1966.Isolationofthe lacrepressor. Proc.Natl. Acad. Sci. U.S.A. 56:1891-1898.
18. Goeddel,D.V.,H. L.Heyneker, T. Hozumi, R. Ar-entzen,K.Itakura,D.G.Yansura, M. J. Ross, G. Miozzari,R.Crea, andP.H.Seeburg.1979.Direct expression inEscherichia coli ofa DNA sequence
codingforhumangrowth hormone. Nature (London) 281:544-548.
19. Graessmann, A.,M.Graessmann,R.Tjian,andW. C.Topp. 1980.Simian virus 40small-tproteinis
re-quiredforlossof actin cable networks inratcells.J.
Virol. 33:1182-1191.
20. Gray, H.B., D. A. Ostrander,J.L. Hodnett, R. J.
Legerski, and D. L. Robberson.1975.Extracellular nucleasesof Pseudomonas BAL 31. I.Characterization of single strand-specific deoxyriboendonuclease and double-stranddeoxyriboexonucleaseactivities. Nucleic Acids Res.2:1459-1492.
21. Grunstein, M.,and D.S.Hogness.1975.Colony hybrid-ization:amethodfor the isolation of cloned DNAs that
containaspecificgene.Proc. Natl. Acad.Sci. U.S.A. 72:3961-3965.
22. Hassell, J. A.,E. Lukanidin, G. Fey, andJ.
Sam-brook. 1978. Thestructureandexpression oftwo de-fective adenovirus2/simian virus 40 hybrids. J. Mol. Biol.120:209-247.
23. Hirt,B. 1967.Selective extraction of polyoma DNA from infectedmousecellcultures. J.Mol. Biol.26:365-369. 24.Johnsrud, L. 1978.Contactsbetween Escherichia coli
RNA polymerase anda lac operon promoter. Proc. Natl. Acad. Sci. U.S.A.75:5314-5318.
25. Katz,L,D.T.Kingsbury, and D.R. Helinski. 1973.
Stimulation by cyclic adenosine monophosphate of plasmidDNAreplication and catabolite repression of theplasmid DNA-protein relaxation complex. J. Bac-teriol.114:577-591.
26. Kessler, S. W. 1975. Rapid isolation of antigens fromcells withastaphylococcal proteinA-antibody absorbent. J.
Immunol.115:1617-1624.
27. Lewis, A. M., and R. G. Martin. 1979. Oncogenicity of simianvirus40deletionmutantsthat induce altered 17-kilodaltont-proteins. Proc. Natl. Acad. Sci. U.S.A. 76: 4299-4302.
28. Linke, H. K., T. Hunter, and G. Walter. 1979. Struc-turalrelationship between the 100,000- and 17,000-mo-lecular weight T antigens of simian virus40(SV40)as
deducedby comparison with the SV40-specific proteins codedby the nondefective adenovirustype2-SV40 hy-brid viruses. J. Virol. 29:390-394.
29. Martial, J. A., R. A.Hallewell,J. D.Baxter,andH. M. Goodman. 1979. Human growthhormone: comple-mentaryDNAcloning and expressionin bacteria.
Sci-ence205:602-607.
30. Martin, R. G.,V. P. Setlow,C. A.Edwards,andD. Vembu. 1979. The roles of the simian virus40 tumor antigens in transformation ofChinese hamster lung cells.Cell 17:635-643.
31. Maxam, A. M., and W. Gilbert. 1977. Anewmethodfor sequencing DNA. Proc. Natl. Acad. Sci. U.S.A. 74:560-564.
32. Mercereau-Puijalon, O.,A.Royal,B.Cami,A. Gar-apin, A. Krust,F.Gannon,and P.Kourilsky.1978. Synthesisofanovalbumin-likeprotein byEscherichia coliK12 harbouring arecombinant plasmid. Nature (London) 275:505-510.
33. Miller, J.1972.Experimentsinmolecular genetics.Cold Spring HarborLaboratory, Cold Spring Harbor, New York.
34. Mueller-Hill, B.,L.Crapo,and W.Gilbert.1968. Mu-tantsthat makemorelacrepressor.Proc.Natl. Acad. Sci. U.S.A.59:1259-1264.
35. O'Farrell, P. H. 1975.Highresolutiontwo-dimensional electrophoresis ofproteins. J. Biol. Chem. 250:4007-4021.
36. Paucha, E.,A.Mellor,R.Harvey,A.E.Smith, R. W. Hewick, andM. D.Waterfield.1978.Large andsmall
tumor antigens from simian virus 40 have identical aminoterminimappingat0.65mapunits.Proc. Natl.
Acad.Sci.U.S.A.75:2165-2169.
37. Prives, C., E.Gilboa, M. Revel, and E. Winocour. 1977.Cell-freetranslation of simian virus 40 early
mes-senger RNA coding for viral T-antigen. Proc. Natl. Acad.Sci. U.S.A.74:457-461.
38. Reddy,V.B.,B.Thimmappaya,R.Dhar,K.N.
Sub-ramanian,B. S.Zain,J.Pan,P. K.Ghosh,M.L. Celma,andS. M.Weissman. 1978.Thegenomeof simian virus 40.Science 200:494-502.
39. Reed,S.I.,G.Stark,and J. C.Alwine.1976. Autoreg-ulationofsimian virus 40geneAby Tantigen.Proc. Natl.Acad.Sci.U.S.A.73:3083-3087.
40.Rigby, P.,D.Rhodes,M.Dieckmann, and P. Berg. 1977. Labeling deoxyribonucleic acid tohigh specific activityin vitrobynicktranslationwithDNA
polym-eraseI. J.Mol. Biol. 113:237-251.
41. Roberts,T.M.,I.Bikel, R.R.Yocum,D.M.
Living-ston,and M.Ptashne. 1979.Synthesisof simian virus
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
OVERPRODUCTION OF SV40 SMALL t ANTIGEN 697
40 tantigen in Escherichiacoli.Proc.Natl. Acad. Sci. U.S.A. 76:5596-5600.
42.Roberts, T.M.,R.Kacich,and M.Ptashne. 1979. A general method for maximizing the expression of a clonedgene. Proc. Natl. Acad. Sci. U.S.A. 76:760-764. 43.Seif,R., and R.G. Martin. 1979. Simian virus 40 small
t antigenis notrequired for the maintenance of trans-formation but mayactasapromoter (cocarcinogen) during establishment of transformation in resting rat cells. J. Virol. 32:979-988.
44. Shine, J., and LDalgarno. 1974. The 3'-terminal se-quence ofEscherichia coli 16Sribosomal RNA: com-plementarity to nonsense triplets and ribosome binding sites. Proc.Natl. Acad. Sci. U.S.A.71:1342-1346. 45.Sleigh, M. J., W. C. Topp, R. Hanich,and J.F.
Sam-brook.1978. MutantsofSV40with an altered small t protein are reducedin theirabilitytotransformcells. Cell14:79-88.
46. Smith, A.E., R.Smith, and E. Paucha.1978.Extraction andfingerprint analysis of simian virus40 large and smallT-antigens. J. Virol. 28:140-153.
47. Southern, E. M. 1975. Detection ofspecific sequences among DNAfragmentsseparated by gel electrophore-sis.J. Mol. Biol.98:503-517.
48. Steinberg, B. M., and R. Pollack. 1979. Anchorage independence:analysis of factors affecting the growth and colony formationofwild-typeand dl54/59mutant SV40-transformedlines.Virology 99:302-311. 49. Steinberg, B. M., R. Pollack, W.Topp, and M.
Bot-chan.1978.Isolationand characterization ofT
antigen-negative revertants from aline oftransformedrat cells containing onecopy of the SV40 genome.Cell 13:19-32.
50. Steitz, J.A.,and K. Jakes. 1975. How ribosomes select initiatorregionsinmRNA:base pair formation between the3' terminusof 16S rRNA and the mRNAduring initiation of proteinsynthesisinEscherichia coli. Proc. Natl.Acad. Sci. U.S.A. 72:4734-4738.
51. Studier, R. W.1973.Analysis of bacteriophageT7early RNAs andproteinsonslab gels. J. Mol.Biol. 79:237-248.
52. Tegtmeyer, P. 1972.Simian virus 40 deoxyribonucleic acidsynthesis: theviralreplicon. J.Virol. 10:591-598. 53. Tegtmeyer, P. 1975. Function of simian virus 40 gene A
intransforming infection.J.Virol.15:613-618. 54. Tjian,R., A. Robbins, and D.Lane. 1979. Inhibition of
SV40 T antigen enzymatic activity by amonospecific antibody, p. 637-642. In R. Axel, T. Maniatis, and C. F. Fox(ed.), Eukaryotic gene regulation, vol. 14. Academic Press,Inc., New York.
55. Todaro, G.,and K. I. Takemoto.1969."Rescued" SV40: increasedtransformiingefficiency in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 62:1031-1037. 56. Tooze, J.1980.Themolecularbiologyoftumorviruses.
Part 2, DNAtumorviruse.Cold Spring Harbor Labo-ratory,ColdSpringHarbor, New York.
57. Topp, W.C. 1980. Variable defectiveness for lytic growth of the dl 54/59mutantsofsimian virus 40. J.Virol. 33: 1208-1210.
VOL. 37,1981