JOURNAL OFVIROLOGY, Apr. 1980,p.187-199 Vol. 34, No. 1
0022-538X/80/04-0187/13$02.00/0
Structure of
Replicating DNA Molecules of Bacillus subtilis
Bacteriophage
429
MARTAR. INCIARTE, MARGARITA SALAS,ANDJOSE M. SOGO*
CentrodeBiologia Molecular, Consejo SuperiordeInvestigaciones Cientificas, UniversidadAutonomade Madrid, Madrid 34,Spain
We isolated )29 DNAreplicativeintermediates fromextractsofphage-infected Bacillus subtilis, pulsed-labeled with[3H]thymidine, by velocity sedimentation in neutralsucrose followedby CsClequilibrium density gradient centrifugation. During a chase, the DNA with a higher sedimentation coefficient in neutral sucrose and a lower sedimentation rate inalkalinesucrose than that of viral
029
DNA wasconvertedinto mature DNA. The material withadensity higherthan thatofmature429 DNAconsistedofreplicative intermediates,asanalyzedwith anelectronmicroscope.We foundtwomajortypes of molecules. One consisted of unit-length duplexDNA with onesingle-stranded branch at arandomposition. The length of the single-stranded branches was similar to that of one of the double-strandedregions.The othertype ofmoleculeswasunit-lengthDNAwith onedouble-stranded regionandonesingle-stranded region extendinga variable distance fromoneend. Partial denaturation of the latter molecules showed that replication wasinitiatedwith a similar frequency from eitherDNA end. These findings suggest that 429 DNA replication occurs by a mechanism of strand displacement and that
replication
startsnon-simultaneously
from either DNA end, asin thecase of adenovirus.Bacillus subtilis phage 429 has a double-strandedDNAof molarmass11.8x
106
gmol-' (24) with a proteincovalently linkedtoboth 5' termini (12, 14, 23,34). Theprotein
attachedto theDNA is theproduct
ofcistron3, p3 (23), a virus-codedprotein inducedearly
after infection (6, 13), which is essential for DNAreplication
(5, 11, 30,33).
AdenovirusDNAalso hasaprotein covalently linked to both 5' ends (7, 20). Replication of adenovirus starts at either DNA end and pro-ceeds by astrand displacement mechanism (1, 17, 28, 29). Rekosh et al. (20) have proposed a modelforthe initiation of adenovirusDNA rep-licationinwhichthe terminalprotein would act as aprimer.
In the case of phage
4)29,
nothing is known about theorigin or mechanism of DNA replica-tion. In thisreport we show the isolation and an analysisofthe structure of429
DNA replicative intermediates with an electron microscope. As in adenovirus (17), the two major types of429
DNAreplicating molecules are unit-length du-plex DNA molecules with one single-stranded branch(typeI)and unit-length DNA molecules with onedouble-stranded region and one single-stranded region extending a variable distance from oneend (type II). Partial denaturation of type II molecules shows that initiation of repli-cationtakes place with a similar frequency from
either DNA end. The results presented in this paper suggest thatphage 429 DNAreplication occurs by amechanism of strand displacement initiatednon-simultaneouslyateitherend of the DNA.
MATERIALS AND METHODS
Bacteria andphage.Host bacteria wereB. sub-tilis11ONA
tryf
spoA- su- and B.subtilis168MO-99 [met- thrj]+ spoA- su+3 (18). 029 was the mutant susl4(1242)thatproducesanormal burst anddelayed lysis of the bacteria infected under restrictive condi-tions(5).Reagents and enzymes. [methyl-3H]thymidine and['4C]uracilwereobtained from the Radiochemical Centre.Sarkosyl NL97 was a gift from Geigy Chemical Co., and p-(hydroxyphenylazo)-uracilwas agift from Imperial Chemical Industries. Nuclease-free pronase,
Bgrade,wasfromCalbiochem, and fungal proteinase K,chromatographically purified, was fromMerck & Co., Inc. Glyoxalwasfrom AldrichChemicalCo.
Pro-teinase K-treated429DNA,labeled with['4C]uracil, was agift fromR. P.Mellado.
Isolationof replicating
4)29
DNAmolecules. B. subtilis 11ONA su- was grown at 30°C in definedmedium (6).When thecellconcentration was108/ml, thebacteriafrom15ml of culture were concentrated fivefold in defined medium containing 1 mM amino
acids, uridine(200
jig/ml),
andp-(hydroxyphenylazo)-uracil(100,ug/ml)toinhibit host DNAreplication(4) andinfectedwith mutantsusl4(1242)at amultiplicity of 10. At 50 min postinfection, [3H]thymidine (100 187
on November 10, 2019 by guest
http://jvi.asm.org/
188 INCIARTE, SALAS, AND SOGO juCi/ml)wasadded, and1.5minlatertheincorporation
wasterminatedby adding25mMsodiumazide and by placing the cell culture inaDry Ice bath. After cen-trifugation, the cell pelletwassuspendedin 0.5mlof
a buffer containing 10 mM HEPES (N-2-hydroxy-ethylpiperazine-N'-2-ethanesulfonic acid; pH 7), 0.2
mMEDTA, andlysozyme (2.5mg/ml) and incubated for3 to 4mintoallow thelysisof theinfected bacteria. Abuffer(0.5ml) containing1mMHEPES(pH 7),0.2
mMEDTA, and 2% sodiumdodecylsulfatewasthen added, and the mixturewasincubatedfor1hat37°C
torelease the DNA from the bacterial membrane (R. P. Mellado and M. Salas, unpublished data). The sample wasplacedontopofa39-mllinear5 to20%
sucrose gradient in 1 mM HEPES (pH 7)-0.2 mM EDTA-0.1% sodium dodecylsulfate andcentrifuged for 17 hat 18,000 rpm and 20°C inan SW27rotor. Fractions of about1mlweretaken from the top ofthe gradient, and the acid-insoluble radioactive material wasdetermined inasample.The fractionscontaining bothunit-lengthandreplicatingDNAmolecules,the latter with a sedimentation coefficient higher than that of native DNA, were pooled and treated with glyoxal to prevent base pairing and pronase as de-scribed previously (17). Afterphenol extraction, the sample was concentrated with 2-butanol (26) to a volume of about3ml; Sarkosyl wasaddedto afinal concentration of0.1%, and thedensity wasadjusted with solidCsCltoabout 1.71g/cm3.After centrifuga-tion for60hat40,000rpminaBeckman type65rotor, fractionswerecollected from the bottom of the tube and the totalradioactivitywasdetermined inasample ofeach fraction. Densitywasdetermined in selected fractions of thegradient.The fractions indicated below
werepooledanddialyzed againstabuffercontaining 10mMTris-hydrochloride (pH 7.5)and1mMEDTA. Electron microscopy. The formamide-cyto-chrome cspreading technique (8, 31) wasused, with
80mM Tris-hydrochloride (pH 8)-7 mM EDTA-30
mM NaCl-50% formamide-0.007% cytochrome c in
the spreading solution and redistilled water in the hypophase.
For thepartial denaturation experiments, the DNA solutioncontaining35mMTris-hydrochloride (pH 8),
15mM EDTA,70 mM NaCl, and 2.5% glyoxal was heatedfor 1min at66°C and immediately quenched
in ice water. Formamide and cytochrome c to final concentrations of 50and 0.007%, respectively, were
thenadded, and the DNA was spread over a hypo-phase ofredistilledwater.
The filmwaspicked uponstandard carbon-coated grids, stained with uranyl acetate, and rotary shadowed withplatinum-carbon.
Micrographs were taken inaJEOL 100B electron microscope at 80kV with a magnificationof10,000, determined with a carbon grating replica of 2,160 lines/mm from Balzers Union.
The contour lengths of the DNA molecules were measured with an SAC Digitizer, and the data were processedin a PDP11/45minicomputerwithaDOS/ BATCHoperating system.
RESULTS
Sedimentation
analysis
of 429 DNArep-licative intermediates. Figure 1 shows the sedimentationanalysis of the DNA synthesized during a 1.5-min pulse with [3H]thymidine by phage
4)29-infected
B. subtilis at 50min postin-fection. At neutral pH,amain peak of radioac-tivity cosedimented with mature 429"'C-labeled DNAusedas amarker (Fig. 1A), butthere was also someradioactive material with a sedimnen-tationcoefficient greater than that of unit-length 4)29 DNA. This material disappeared after a chase withan excessof cold thymidine(Fig.1B). When the pulse-labeled material was centri-fuged through an alkaline sucrose gradient, most ofthe radioactivity sedimented at a rate lower than that of mature 429 DNA (Fig. 1C). Since no radioactive material sedimented faster than unit-length4)29DNA, the existence of covalent circles or structures resulting from a hairpin mechanism in the replication of 429 DNA is unlikely. The radioactive material sedimenting slower than mature 429DNAdisappeared after achaseand was converted into unit-length DNA (Fig. 1D). Uninfected bacteria, pulse-labeled and chased under the same conditions as the 429-infectedcells, produced no radioactive material under thepeak of 429 DNA (Fig. 1A to D). The aboveresultssuggested that the material with a sedimentation rate greater thanthat of mature 4)29 DNA inaneutral sucrose gradient and lower than that ofphage DNA in an alkaline gradient consisted of 429 DNAreplicative intermediates. To determine the density of the [3H]DNA which sedimented in a neutral sucrose gradient intheregion correspondingtobothmature429 DNA and replicative intermediates, the frac-tionscollected from apreparativegradient were pooled, treated with glyoxal and pronase as de-scribed above(17), and subjected to equilibrium centrifugation in CsCl. Figure 2A shows that the DNAappearedin apeak withan averagedensity higher than thatof mature 429 DNA. The in-creasedbuoyant densityofreplicating429 DNA molecules could have been due to the formation ofsingle-stranded DNA tails in the replication process (seebelow).Electron microscopy of 429 replicative intermediates. The fractionscorrespondingto mature429DNA andreplicative intermediates, labeledin a 1.5-minpulseat50minpostinfection andseparated ina preparative neutral sucrose gradient as described above, were pooled, treated with glyoxal and pronase, and centri-fugedtoequilibriumin CsCl(Fig. 2B).Fractions 9 to 15, 19 to 24, and 25 to 29 were pooled, dialyzed,and examined with the electron micro-scopeasdescribed above. The DNA in fractions 25 to 29 consisted mainly of mature double-stranded molecules. Fractions9 to15contained J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
REPLICATING
029
DNA MOLECULES 1894
c 0
-E
0
n-0
2
c
0
u 5
E
30
a
-20
,ji:
I
-2 I
E
ob U
~0 la 4._
_V
20 40 20 40
Fraction number
FIG. 1. Neutral and alkalinesucrosegradients of
o29-infected
B. subtilis,pulse-labeledand chased. B. subtilis11ONAsu- (1 ml)wasinfectedwithmutantsusl4(1242) and, after50min,labeled with[3H]thymidine (100t,Ci/ml,46Ci/mmol)asdescribed in thetext.At51.5min,a samplewasremoved, a200-foldexcess of nonradioactive thymidinewas addedto the remainingmaterial, and the incubationwascontinuedfor20min.Afterlysis, all of the sampleswereincubatedfor3hat37°Cin the presenceof1% sodiumdodecylsulfate andproteinase K (200,ug/ml).
["4C]labeled
429DNA(6,000 cpm)wasaddedtoeach3H-labeledsample, and theyweresedimented in eitherneutraloralkaline5 to20%1osucrosegradients inanAH650rotorofaSorvall ultracentrifuge eitherfor2h(neutral gradients) orfor2.5h(alkalinegradients) at42,000 rpm and20°C. Fractionswerecollectedfrom the bottomofthe gradient,andthetrichloroaceticacid-precipitable radioactivity wasdetermined. A controlofuninfected cellswaspulse-labeled and chased under thesameconditionsasthe infected B. subtilis. (A) Neutral gradient, pulse; (B) neutral gradient, chase; (C) alkaline gradient, pulse; (D) alkalinegradient, chase. Symbols: * -, 3H radioactivity, infected cells; *--- -A, 3H radioactivity, unin-fected cells; 0---0,"Cradioactivity.Sedimentation is from right to left.single-stranded DNA with or without a small double-stranded regionatoneend;mostof these molecules were shorter than unit-length
429
DNAandwere notanalyzed further. Fractions 19 to 24contained replicative intermediates. Fig-ures 3and4showsometypical electron micro-graphs. Asacontrol, more than 99% of B. subtilis DNA,labeled with
[3H]thymidine
before infec-tion, sedimented to the bottom of a neutral sucrose gradient and did not overlap with the main peak of mature429
DNA and replicative intermediates.As in the case of adenovirus (17), two major typesof structures were found in the peak of
429
DNA replicative intermediates. We will
desig-natethem type I and typeII,in accordance with the nomenclature used for adenovirus. Type I molecules consisted of linear double-stranded DNA,mostlywithonesingle-stranded tail(Fig. 3A),althoughmorethanonesingle-strandedtail wasalsoseen at alowerfrequency (Fig.4A and B and Table 1). Type II molecules consisted of linear DNApartially doublestranded and par-tially single stranded (Fig. 3B). Acombination of type Iand type IImolecules, designated type I/II molecules (17), was also observed. They consisted of linear molecules partially double stranded and partially single stranded, with a single-strandedtailcomingoutfrom the double-stranded region (Fig. 3C). Type I/II molecules VOL. 34,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.508.104.409.68.379.2]190 INCIARTE, SALAS, AND SOGO
O-0
2
E
06_It
S._ 0
a
2
I
E
U
CP
c0
a1
10 20 30 40
Fraction number
FIG. 2. Cesium chloride gradient centrifugation ofpulse-labeled +29-infected B. subtilis. B. subtilis11ONA su (3ml)wasinfectedwithmutantsusl4(1242)and, after50min, pulsed for 1.5min with[3H]thymidine(46 Ci/mmol, at aconcentration of40iLCi/mlin[A] or 100gCi/mlin[B]). At 51.5min, the cells were lysed, treated with 1% sodiumdodecyl sulfate for1hat37°C, and centrifuged in a 5 to 20% neutral sucrose gradient (39 mlof total volume)asdescribed in the text. A marker of429 '4C-labeledDNA(50,000 cpm) was added in (A)beforecentrifugation. Thefractions containing unit-length 429DNA andreplicative intermediates were pooled,treated withglyoxalandpronase, andcentrifuged inaCsCl gradient to equilibrium as described in the text. Fractions were collected from the bottom of the tube, and the trichloroacetic acid-precipitable radioactivitywasdetermined. Thedensityof selected fractions of the gradient was previously determined. (A) Analyticalgradient; (B)preparativegradient.Symbols: *-*,3H radioactivity; --- 0, 14C radioactivity; A--- -A, density.
with two single-stranded tails were also seen, althoughveryrarely (Fig.4C).
Table 1 shows that typeI, typeII,and type
I/
II molecules accounted for about 84% of the totalnumber ofmoleculesscored. TypeII mol-eculeswerethemostabundant(55%), and type Imolecules were present inasmallerproportion (20%). If the total number of single-stranded tailsintypeImolecules is
considered,
the num-ber(63molecules)
increasesto24%of thetotal,
althoughit doesnotreach the
expected
value of 55%.This isprobablybecause type IImolecules are betterunfolded than type I molecules and morenon-analyzablemolecules,which werenotscored, were found for type I than for type II
molecules. Type I/II molecules accounted for 9% of the total. Another classofmolecules ob-served, which we designate type III molecules (14%of thetotal), consistedoflinearDNA
par-tially
double stranded, with single-stranded re-gions at both ends (Fig. 3D), and linear DNA molecules with double-stranded regions at the twoends andaninternalsingle-stranded portion (type IV molecules; 2% of the total). Although in a small proportion, molecules with branch migrationwerealso found (Fig. 4A).Length determination of
#o29
replicative
intermediates.
Length
measurements weremadefortypesI,
II,
I/II, andIIImolecules,since they were present in greateramounts.Figure5A J. VIROL.on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.508.112.407.64.403.2]I.'t,,"..I----,-...':;.-..r,,,"I'm...'....'-..,,.-'"''.It';..---.---
.)...
,.-.;:...--'!....".'.
.'_
"...'
-.--.-..'...'.;
':-.:,.:-..--.%.,! .--..""..S.!._'..-.i..
V.-"'.,,..-.
...,-,;.. '.....A
---:..';j'...! .M.,!.,...:..:..-.!-..'...?...'-'..'..,-'.I...-"...'..-..:-...I.-:,, -''-.,...--"'..'.--..
...il,I..N-.,.I111.--_-.'I....-!...-....
-.:..'...1...,.."I'...,...-''.:?.;....
...-I........
-....".
...1..'.-
... j.. I'%:.....
.'''.;.-.1-:._..:-
...,-...---.%_:..,
-'.
:..:: ,.-,....'A"'-%..,.,.,
.j,,,,&,..:".__'-;';:... .-'_...t--'.t.-...,-'. -!.."...:'..,..'...-'.. '.'...
'...-.....,.,.,-,,.,,V... ...'_,,...-..".,..'''-I...'-!...-.!..----, ,-'...,.-.'\'-..,...:-'...h.'.7-.'...,!...:...-.-.:...';.: -..:."_._-,-...."I.,.,..'..'. t:-...,,,;.&",..%.-....:-..','.',
.:....'..-i;.'-,,...." ."."'.,7..'...,'4..-'....',..:..,..:...-...:..:.?..II..1.'.,I.':'..."...I,-,..'j..1--....-'' '''_...-..'.''....I-.-...I'I1-.'..,-..t.I.-..",'...:'....I...:.,1..'...",-,,,,,.:.I..-..-...-';-:";..,..-
...,.'..."...'..:.;.':1:',."",.. .'. ..k..:.'..-.:.-..;....-...--.."_'I"..1t..--..-."":
'--...,7...':.'..-"...'. ,:-...'.
'.'-'i..
.1-.. ..-..'-.''..-,i....-'fI...,.,....",.IP.: .I"X.".-,.. I:.!..:.;,,....',::...--'-....-!.,....'n:"..;...'i:-..."-".:":,.:,-:;7..;...-'.-....,...-.' '..'..'.':..,-."....,;-.-.."'-,...-..i.1._?:"..I.':Ir..."".:.1_--..:.-.,.:':.-.7.'...'.. i,:-....-,.":... ".'.'-.*''-...1..,-.-.:....-....-.-.--,.'..";.,.',.'--"-.'-.-"2.-_R..-.,,-.'.!...'.I.-.'"'.'...'.I"'.1'...,:..'.'',.;-I
...-
...,-; ...-..--".'-.-.....,."-'...:.--,::' 1%-.-,.'....'..,':'-'-..r: ..-..:.,--.. .:--,\....:...-.-'.---.;::-..-"-';;C....-:;.'!'.1;-';'..,...-..."'.:...-...`....,...""'.... ...t. .:-,..., .'.'....--/...'--t--....-.'..''-A...."...!....-.--'.'...:--..'....,',-..:...'...!,..-9-1.4 .:.. --.,:'....1;-.-,:-,.:...,,..'1_;,:,--.-'..IP.:...: ...1-1--'-,..-,.,..,.,.."..-.1,.,.-..-.-'..'`..-,-'....'.-_:-.-...';...-'..'.'"....:...'.-:.,...-...-.'.-.'....-.-. .?:...:::!.,.:.'.%.,, -_-I.'..-...;...,..;;,-.'.;:-_-;'-.......----....".:..:,..,I..,,i '.----...-..:,..."..;....Ii...";..,.:...---."i':...-'-,...I;"-.-'--...-'.',t..i...'I--.i.,:'.1-."'.,?,-I.."'-.''-.-J-':.'-'.r-_1--'. ;.. ..:.. ... .-.'.... ..,,,,...,._.'II':...:...1.-.-','.. -.'-'..-.'.";;.--...rL.',, ,1 .. .'.'%...,. !...'I.'...71'...,':'!1..:.! I-... .V."... ...!-..:.'tr.I....-''',....-Ill.-.1-.11.4"..-...I-'..-,.'...j'...:',.;..". .:-:..'.:.-- -.. ...1...!!...:...'. L'.'.'...'..--.---,.':..,",.i4.'-;.'-."_.';./.' i." !'...---...z-.,...';.".. -:
i-II...--!"'.'.---',.. ""..:.. --,;,.-:.,..:." ...'..."..-1.'"'''_i '71'I',-'.' ...--7..;. -.,.-'.','.;-.'.,Z.,..."...-...-,:"i...".... ..:".''-
'"--.--'-."
,... ..':%'.-t I-..
:..,.'..---.''-... !'. ,---I-,;...,...'..;'-'....';.'-...,--;...1 ,.-.... .:-..!.:'.".,--.'..--...-...,,-...-.1::; ,---'.'..:-.---...-.. 4.e!, .. --:'.--.-."'.- ."-- -,
..:---.::..'..t..
...%. ..7I.-,....'.-.1.,.I".,:'.''-,. ."'.;...:'..1!.,.',1...,.-..--...,.,.;',.,--_-..-.-..-I,!...--,.1-1I-__..,....%.'--... -'t'..---...:...-,.'. --1: ,'.-,',...:...-....-.-..--t-I...---,II..;.1...-I:-.-'_.'...--':.:1..::i.:...-.?.---...::.r.'.:..''..%:.'. ..'....-..'.'-,-,...-.-''...---....-:. .--.-:....:.-'-':.. ,...----,,,"'.,; -:z....--.-.:. 7'.. -':----.'..
...-...
-.-I...,...'F,--.:.-.,...-.
-'...;1.1':'.....,....-..-c.... Z".. -'.-'-.i--.'--.-....;,..'..'..'C.".--.--...",;,.,:--I..--,-....---,.. ...--.--:-,-... ..-..- !, .--...-... ....
....-...'..,-.":-,..,-...-,.'.....:...-.'....-'.'....r...'...:.,-,-N,....:...7...t..-...-4-...:-,..-..;...'I.!...:...I.1.".-...-...:;...'!-.,.: ....:..!..-''..."....,..;,,...'-%... .---..'....".'.:. --- .t.- -,-...-...1%,-'.."...-'..''...t,1, -. .-.-,,
: .'....---.----...'t ...-...'.-"...
.:'I'''--I.,.--..-..':,..-..--.:-.-...1'..,:::....-,,.',.'...- I...!..;...'-._--,-,'.'-,..'.:'.,-...-..-':..'....--..'.'.-.'..-'.!-'-.'...711'...,.I..;..:-'''-V:...,...-'I'*...J.,....-...-....i,--.-,,-...,-"....'.... --....-....---'..'..-;. '?. Z.!..:i--..-...'t,,.--.4,-."-....:-.Z,-...',...-,,.." .., "..!:-...;'-'.;I!.. ...''..:I:1. 1...-,...'...I-...---z .:--,..';-..'_....--..!'. -.-''....''"''----,'....'...-'..i,."'.:.,...",....,...'-.-:'--'i-.'--.'...,...--Z.-...,",,...'...-.:!..:.,..i..--.'-'--.'::,..,!, ;.'....":...,.;....%.-,...t
%...;....,.,. '.r...:..:'.."-:%:...;...:...."--''7",,-...
...---... ..-...:.I.. ,, -,.,-,
.. .'-,..-'...'-... .... ...--,4... .:
:.".'
...;...:...-:---:'
....,:,.,.....'.t'....-...'..'.I--...---....'...-.....--...'...-. ....;-.;-..-...:,.",!... ..:.4."::.':....1.:-'.',-..-..'...:...--,..'..':'i..:.'...
1'. ..-''':'...-;,..--..1.Ii'...:.:-.:...,,..-.1.'..'i.:. !..-.,...:'... k,-...".''...'I...,-..:...:.... -.:.,.".,:4-";n,:!.:....,.'z.-...,.,,...-....--...-...:._-,-..--.:-,'.."...`"--...---': -.t-,,-..:
..'
.'..-'
''
.'...-.
.:...'
--'-!'.L.7..I -'...' :'....-I-.1.;..,I..'.:..,-.,--. .. :j.'.._-.' .-,...:..';...", ....-,-.'...-,,.-";'-'r!--:...-_.'.--.-.,,'-.,.'-'-'::.'A,,,.-.-.---".."!.-'...-.7.1...'...;-
.--.-"-
";".:'-.'...,-.':
"''
"'
,"
,.---..-.,
,.--.-"..'-....I.:..:';-.
...
--
I.I.-.
.-:,
."
.I..-.. .;;..;..-'. '.--'-_._.;.--'.-...-...-,-::,.-,-...I--.: -...-""'-,..,--,-.:...I,..-...-../...-..'.-.-.."...-.--!,..-,..'.. --...I_---.t."...,. ...-, ...'...:I...:..-...:....,.".--.'.::.. '.-.1...:;,..--:-...-..-I..'-.-".,.,-.::-'..:'".'.':.. .'.' ;;"- -...z'-":..-:.'..'-..-.-I. .-.'. ...";...-... "r...."....,::,.--,-"..:-.,.t.,.."':..-"
:,--.. .--""."...C;'...
1.-:.,:;--9..'-.--,...
..'1.:,.:...:..!,-, ....,..''.. .,r...'_:
.i..,-,i.: ...
..
....;.,.. ,...'.!'.- -,t.;...,". -,:'.6.-...,Z ,.':,._t'.-::x'. .,---.'z'
...:...-'.. :,-..'....-...,.;. ---, .19..I.-.:",-.-,..!.--,--....,....-.--
-.---..I.-
....;.'..'...'...:.-..':---....,.,:,.!..',-...;...'.I...-.-.r"...--:'.)..-:..--,.%--,...--,"...;...I...-..."..-,:'..-....:":..."...-'.---'.-:,,-,,,-:.,,".-...'.,'...-... _.:..-..:.,...-..-.-"..:..:...----"-::I..'. .--..,..I'...%..,-.1-,.,....-.-....!...Z....,,---" :_'-44-"-Y.---.-';..--.".....
,-.-.--.
",..-'..-..,.'Z..-';.- -'A....'...1,.'-..-:-.'-..,k'i".I".......-I-- .-!--..
...'..,:;.;...: .... .... ...:....N ""---;.'...-.i..,,:;-.- -:--..",.:, '...-.''
.."'...-.''-
..'.'.--...
-'....--..,'."":.--...'i...,..- '-.1:-t-::i..-,.:.."..''':'---'.--t-....-'7I---."'....----..ti...\'...-...I.-,-..-. -k."... -:::..I'...'--.'--',-,...-,-..i'.....I...--
.."-!...-'-...:..:...'.'.':r"..----..--'..-II I..".,::.-..-...-,..-:-.:...N.. --,,..'....;..''...'"--.1...'; --.--,,... .,-.'....".:..
1,....,.':.-'-..I...-...I..-...:...-.---!t,.:1.:..1...",.-,...,",-".'-:..;.-'...L,i-I- "-.-....:...'..t...!..:' ..'.."-".-..'..". '.'"'.-,,,.-..-;.:". "...,-...".." ..I::.1:I.-'...-...-..-..'....:;..'.-,-,-. :..!'...I...N..'...;.'... ..1...'--.-.--,.-.-.!.'.-...:....,-,... ....--..:., ..,Y"..I--...:...-...I...1.. t.1..-.-,...".'.,. ..'-.:
'. ,-..-..-.-.' -::?. ...-::... :'.-'.'-? -%-....;-..-;...-';-..'.-'..-..-'-.,:...r..."%'-,..
..-'..".':...,II...t.!-...-.'..%...,.......-...-....I....-..:..:..I.,:....-...I....'.:...'..., .-- ..'-".-.!'...-".:... ""...--.-'."..%..."i.. ....'...-.!...'....4,"...'-... ...-,...-...-,....:.,... ..."',:,,...---i.,-'.--:.--.:-!i?.-...--.7...:....:.,I...'.'....-4'-,."-"-..1..:..:,-,...-,,'....-_-.1.". -;-'j' .' -!,. ..'.'"C4 '...
...!,.'."....-.7...-...iI-..;....
-:..-11-:...-'"1. "I-'"".:-.."
....:-:-
--...;....7....--.--4'.'...'.... :..,.1-1;... .' .-Z.: "'.';-.."...:4 ....1..-:...-.7"...:..., .:;...-'--':....
%...'II.;...t..I...:...
--..-
..I-...-
"I..I...."...I...I...I...-...'I-...I..--
'6...-..
,.1.-,I,...'.
:...
... .....-...-
...%...-,-.,..;...,..1.:.,- ..."..-...".:'-'! .' .'..'..,.... ..i-..':... 1.-..:...--,,,...'I..-:...,.II.... ..' .-.-:! .-. v... ...
...-.,.
.:..::...I...,t'-,.--.:i.- .1- :_1
...:
.--:.. ..,'.,.. .%-'.'::.'...4.:.'.-1.,.'-.---,.-.. .... ...i....I...'-.'... .. .--'. '_'. ...-I.
..%-..-:. ...--,.-!...,..-...:
...I\.,....
.-..'-'-
_:.,.-...-..
'- ..,,'.'...'--.I-.,...,'...,....'--.I..
...I.:-...-I.:'.-'!'.',..'..t-..-;--.-'!.-1..I...:...--'...'.."'-.'--',,-.-"-..,,-,,-...-..."..
...'..-:..:--
.-...:1..---'.II..I---._...."I-.-'--;.. ;1....-;;. .- -'....'. ..,... .. ..I. .-.-1,.:...1-1:-,...-..-'.-:;--.j. '..%-.1..I..:.''.-",.
.., -!...I--....':%-,.-,1..-...'
..."..:;'.:."..'..'...-...:.L...."...!...---.'.'...1 %. .!"...,..'...'---':-.:'.. '.-.",.,I.1;1...: ...'...1..:---."",,_...:!,....--,--, ,,,,I..."...",'..t!)';?,..-Z
1...1'..-,?.1. ...."....4.'1... -7 i-:.;....1:.,...1....i. ." %,.-.,.'...
..
.-.---
...-,--!.--.---.-...--
-....,...:...,.,...:...,...-.;--'v -..,.'...:-.. '...---,.,.'.:i... ...-'..-,'.'-,..'.'.'-,..%'.-,..-...:....;.'" ---I"I...;'.
,.II...I....:...;... --iI..t...'.." ..-:..!t:... ...-.-...-L.II..'.t-'.:'.--. -...; ..-:'
....-"-"----...I.:..-....I..-:...0...-1..'--...:%1,..'-'...1:--
.--:-:"...-...,....
...-..-:'-.'1".-..::..---...-..I_..--.1...:..'-'. .I--.,:.-...I--';."'.;-...--'...-I...,...---: "..'....::...--:.:.'..'...I!:...-.---....-."-:,...,-.--.---...,..":'-!--.'..-..,....".-....-'.-.-,..-,,-,'I....';'... .'.'"....
I.,,;---...,:.
.',.. ...-.---..-...;...,Z....,....-.-.
-".-..."...-..;-
-'...-.-.'
...-,-..:'..---'
..";' ,,, t. '-..!'..'...%.I...'...1'...'-'---:,-..-...!...:-,I..1:...". -:...:...I'I...,....-.I:..:'I- "-.,:-..
...'..!.'; .'..'
....
....:..;-.-.-'''!'_ ..-I-.; -...-.,
.-....I...-!....I...::";'..,.,:,-.i-...i...t..I...,::....,I....:..
'.t...:-...:.
:'.'*...7.I...'...:',..:....,..!...:'..I..-...:,.;,I'.:.1
..'..:.
:. :'. '<- "-r'I.I...,i.... ..:..':- ... ...I.-'....3,.1- ,<1.
..
:. ..'. .:.. ..
I-:...".
.I...'.-.'t7.-.,,'-:-.- ...,---.-.-- ...'T;1.; '..!_..
... ..'--'-'. -' ." '-'. .:,,,
..--...--..-' -'..
...-...I...'I....I.:...,..,-.",-...:..,-....1... ..:'--,J.;4.'..'...
--,...--'..:1...-:!,",.',...-...-...'.:. ..I..--;A.,,.,.'- ..-.!-.. :.',:- j...,.:,.;..
...-:,.-,----..:,...'... .- .:.-'.-...,.";...-..:.. ...,-,.-- :."'.,--,-.. ....--... ..".--I-..'....-,.f.,
.---:-.-..-:-..!.:.:.,,-.j...,
II...:....,.-:...'--.-.,--.I...--...:...-.'.-..-.IZ!..- '....,.: ...,,,
....:!....'%.,'.-,-....;::...: ..:..I.Z...-i': :.,
.-...-,-,
''.
-.'-
-,
"'..-1...-'...,.,:..,-.::_...-.'--- -,,"
..,...--- -..,:.-,Y...,:.-I... ....
... .--.;..: ,!
...,--.:.:-. -.I..:..:....''.-.:-11M---':.'---..:---I-'AM...;I,--"--'--,,--...:.-...%-...'..
..i..1-.-
--.:...."'..-:;..-...'..",-,,-."'..,-...-
...-...,..-...
:...i...-
.,...:.--,--'-..-,...'-..-,.;'..I...%.-....;.- .:.-: "...,...-.,---..--,I,-,.-,.---,-'-!.,.-,,----."..:-,-",,--..'..;...-'..- ,..''.'. I,",-,.,-.-. :.'..:---..,....:..-.1....'.
..
"-.---
'-... -"-f..-- ...-"'.;..'..-,.---...--.'--;- :.--1'1-_-:11.. ...-,,..'
,---.,"'-.'g;..
......'
'..-"-'.,.. ..,.X:..-.1--:..1-.I.....'%'. -...... if..I.-.,...I.t. .:...:...,....--..--...'-V...---A....-, ..."...-:.'.'.-'--..'.'.:-."_':!'-..-..."..-.,.-.-.1:Ii..-....
_...
;..'..--'1-:-..1:'...:...,.;-..'... -...-.i.-.'-.,-.".-.1....'' I.'...i ..,.."...:..,.. 7-.,:...", ..I; e...
....
... W:I:---i..:...,...-_."I.,'-..-; -1.-.',-...-i...-,!:-.-'.."';.::;L.?.:.' .'.'. ..'.
..1 ..--"'.-'----,' .;.'.-' -'.'-,
.'-,I.1."....",.:!.!....'...-..:...-I' ':
..:..'.--
..''.-.'-:--,."..._'.-:-.'-.'.,,...'_'.-:- -j'-;.-.-...,......:-.,...- i'..%."..,.,.L:... ... ..--1 '.'....'-.Z'.';-.:;..-- .,-.-.,. :!.'.
--:.,:.1:...'-...--.. .:...'....-'..'. ."....:..:...-,
--..--191
on November 10, 2019 by guest
http://jvi.asm.org/
.. ....";,.,-,621.j-,.' ;j-_,.,t.;i,,,.-,'-.,!._t-.---"!..",--",..,.-..-.Ikl3x1.1....;6.,_... - -.. ...!,",..jT.,. .,t-,?,;, .`.---'--.43,-,.1.,.,.,.,;,-'-.A,.-.,.;..-'..,-'t... ,.--..;-...-..Y...ni .,l.":'..: .:-'-`-..-.. ...!..-..:-.-.
'-....i..--,-...
.,-,..;. % -t....-". %., A-(.:-"i"!.,F..'...4'.1'j,.-.%... ..,-- ",..-...-.41"
."--'.,-I,--.q...-'..
,-,.,;.r.0. _.:O.-tj-,.,:..t., """'-..,,
-..
-r;_,;,.-..._
..?,.,.:.,., ,;,-,-:'-!,;.-,-.; c'),Q.i.... .i t...;...-".- ...;j;",..
,'.Yf',- V...., -..'.X'&, .. ,...-% .1-..,.., -. .f:?...:...-;.,. .I...1, .: .- ...-..-...-..
R-.-'i;l,ii,-It. z,,;.,.:I'.'...e..Ii,..,..f,,...Z,-,.il,.-,ri.--..r 1i--,v:1.1-I..".nx4q'v'`!,'A.- --'.4" -fy,.'-:.,-" 2-1..-:-".-I!.,., --,-%...,-V.W.,.:3'.7,:.,i. ,...--,--,.!,..-...'..,..,,,..:!... .,': .:..., .. ... ... .l.i'-t`!.-:jf,... -, -?.-.--..-:.-..I.. :;...
.,;,.,Lt..1,.."...";-.l.,,'..-.-.-.t"!Z.,;j.---2.k';.i.,.,..,1, .Jl-.,-.,.,J,z..i.0;2-.',,!.l,:4.k;-,'-,-;.Y&n---.-,;i.----.. 41,Po.,.."..:k..'..-...,-..,;.,v-&,ky.,;e."',...1147-1 j1-,I...,t,1..".I.-.'
..vilj-
.#rl-l--.,.----Z,!t, ,---.N-,-'.'.Y.'A.j,,?.-t-...;1.---,-t-,-_.--...-_.Ii--,,.__,tn;.-z"oZ, --,!II'.1..,-','.,,-.:.. Y.i-.&;,".,-,.;-.;4,m,-...-,f-s:'j,;,-,(.-j...,.._'.1.:--"---,-'-,-.,.?;..%,.l .-.,: ..-....;-,,,--.tll,L-;...-",J...!-"..'.--,q.t%-1.ft...;,.._.'i.,..--,..-....I.,0..2.":..-:..."".t ..,!.-,-_.,.-.'-..,...- ...,..;-..-!. ..--.i,..;.--.:-::..:,....".:...::..., .;;,X,.-.1.,;4...,..!,q.-...k ".--...",----..v,.,.A.---t..;..-I,_,.,4.,..iZ..:.Pw,,.--,.-.fj:l-,---.l-.-I.7-'.,." _. -...,j.-...-.1. -,'? ','.,'.'--"4,-:...:J ...,..---It:.".;' -...,:: -. --....
" ." .1A ,,
-41.,--i.;;.., -, -m , .: ....
...tff..-- -p't., "I.. ...;..-..i,,S.,_ .--: ....:-
-..-.i4wfN ..!.c..j-
,,-1-- _....,6..:,x..n _,.. 1, " ...,:..,t"...'.-;i.-..;....-...,... "'.-...".Y,-.'._.. .."....,'. ....--;.;...:.;,'...'.. --,...
...v.-...Y
...-..."..,.-...,...
7.",--4-t,'...;I, _,::.,r,,.,.---.!:.,;,.-.,!.:h,,.-,..v.A..-.,,-.!,.V...,..!..., ...;,'_::,-.'.-.,,,,,.--' ..,-... '...ia'.! "....,.: -.'..1. I.. :-.,'ft.t;..j...,.p.,,.1,.1, .f.,%*Z.4,".1-.1,--.3,_,.'..,?,..,_41i,,:,. ....'t'..,:.,.a-.-.ll-.l.-;.l',..,L.-,,-.;-..."j.",...-t..-..:!-:':,-.;-..-:-.;,.-..;r,...-tl ...I..
IV..,.,Z_.SU!. ,,'-lz.*il:'-,W:--t.-_A-,.,j...1 ,., -'!., .,.,.,...-.. ":...,..' i. ,-... -....,1...'-.,..". :.,.:.',-..
..-!'-'. 1,&.-,.. -*.--.F.-*;..,-i; .-,
?i
...- --...": ";,1, :.,...-`. ....-,P-)'%,%-,f'f,-., .%'-" I.'
%',,.-V...
1; k..-.fi.,'S I-, -I'....C...
-1.; ,,.7"i -.y...,...-.
-'.-'-t-.";.,.-
.!-N".t-.,'
.-.I--o'I,.&'.;.-;l.-Y.J. ..1 .N.,nr,:.'-`:.g-.--',!-:-.-,-:-l.%,,:...'..Ir,.7"';-:...I-V..1...I.l.i,7.,.... ., ...K_-...I.---"-'....z.,-.-.. ...:.-;....-..".-"..; ..,..:..,...,.-I;-?.:1 --:.`.-.I'.'.,-.!'..'-,-'---.- ...-.',-'!,!'.7,.lr-_-, 'j,....4 .A.!..-I. izil-P. _ ! .:f.--l-;;--,.-!f;..-:!-....k,..,"-,,,.,:.
,-
""
','-...
%.-,.:..:....I."...T._I- ...;,...1.--..,..,.-,.--,z",Z";-,0,---,,i., ..,.,-;-'-.-'----.:P-.,1,.!(..,,....-..l.f-., .,....".!._..,,. ...4A.".,'. --'--'--A;`..-c'6...,.-... C.,,,
...I..
... _-,--"-,.,tr. ._.E.-,..,--,.-N';lf'-p:.L,.-..I ".1'11-i "..."t.-.,'!--.,.,
"...!.. .."...-..-x!-;-M.:.-9-;`.r.,..."'....1 1.
I.
I,;.l:_,.:.,
--.,...'... T...I .-i..,.j:,"e.,. e." -- ...'!.? !, .,'. .,..?.V.'-.;.,C.,..,..j:..,.. ;...
i1,..,,,--t.. fd. ---"o. .;! .v 11,-,''--,.'.-..".,-.,,. ...-..
,-,.,-... zt ....'. ..._.;e -I-.11'..,-.'
,--,....',-:-,--....--..-,,..-1. .'...-..- --"..-;)-..,I..,;,.l;....'-
,....-.
I.....-",,-.-..---!,Z.I,."e.,..-Z,'."...":.I.cl...:..'L.-.'.',41.:k4.1 1:.i:".I....!..4.-'..;-le";---.-,.,.-- I .",%."..1;. ..'...,j.,,-...: ...-I., -: ;.-..7--.,..:!.-.--.
, ...; -1,...:-..: -.
.* ,.'.'-At ":-'- "' ..'-'--t
"./...-,.."--., 21;.7-ir.'!t7,::-,.t.; Z..." 1:....Z.,..,"Si-.. ....,.-3.Ls!'.'.:.., .41-, -,-.:$i-.',._',!.-.k... -f;,.:.,4."...1%,,-z"',-r..-I"i;!Z...,... ;.`-,..,...-, -.0-.,-t-.-?t.;,--; -,,,...-:..'..;..-.... .:.;..., ...,
-.-.-.- ,'-p;..ir,---'.'ir.;...:j.,.,-,--s-.-A-r--,
....''-
I.,J_'.t-,.,.I--;.l,-.,,:.-j-_ik,f.... .r,r...,,'..'..."'.:rt.---.,,l',,,-;-,-,....!..."....ll..'.,.,.. ...I...,,,-.--.- ..-.:-.W..-.::-'A.t,., -4.i;1--.i-:jx"! !-..:.,..;.!,:-..,...-.1. '.. ,-'.,.-.-.
..O.-.'.._,.,,...-.,...-t...,...!;-...:.Z.:..,.,;,-..,.,-..:,,...:.:.". ., -..",-; -,,I!
.-,
1-1-..., "..1.. ..i( ..,.''.;.'P.--," . " 17...': .:...11 ..."-..-..,M...._.,;-..,."...',!.t..-1."-...
,,.
...I:,..,...-.,....,...:... A.t...I.,.e,.,;,k,-l"..,.,.,,..,,.,::.". -, ... ", .;...",...,f--.,-..'..-.:..I.:,,','--t'",.-.-,.-,",-!"-.-"I....:,'....."'..;it! 'i-- -V-,4;"; -, .. -1%, ,.,. .., ...,
-lj,."j.&..., 1;.'.', ... .,-- I" ,!...-41.:il..;. - ,- -,-J.- -
.--..,. ....i..."...1-?,.-....'-- "-.... .,..,-.j, -'..--_--,...-I.., ".%,-;-;:-. L.. .-!i'...-:.:..,..,-".",... .--,,
ft:.,.-.!-I,... ".)._:!..iwl-.';.,:f,j,-.... "t... ".
, ...-.
._..'. -.7.-,....
.-,-I:C7--.
)-;,i.:...- ;.,...,..--:-..--.l.'ll ",,, "'..
..,.. -4.-,..-;.-.1 .."...!..-...;,:. _..."..":...t.-....""-..."!-.':,:,-.. ..,... ..I.. ...
...> "-...I --,-, r...;.. -. I.. i1....!...
12. -,;.--T.-!..-t,-.,-.,.,..:;... '7",.,-.,..,,-I--..-.".1,-,-",j;...
-'."
-,-I--,
I ".:..,-,4 '.." ;-,. -%.0.,r,.7.I-_ -...:...
..,'J",.'.7..,...;g.. -..,,Z.4; ".?-jl -.., .. .-"-.,-., ,I...,...;,,.,l-,;'.'-::-.,-." i1.-;':,-'-..Z." -:'-.-.'-'..-;.---... .:, ; .1.'...-. ;. .1. ,--.,pr.-,,,
"". .-N...,..'.-!. -.t ...:C-.:.f;27_". .,,:.,.,-,,.;-,.,..'.. ::.-t.l...,;: -..-.1 :..., ....-,'t-.-.--' -.e.ag,, ...,..,,;,".,..', N.,;-2:t:-.-," .;
:_--.,.----'..--
.,..:...,
-!...
.'..,-.
..--.
I..",-... n.: ,-.-' -,;, .;.--.!.-...,
...-.
..
'-.;! -..--!-...-...i--1. '..._.-._.,.A... ..,....it-..;.,-..,....'...:..., ..,,...L.;..:;;,
.'...,...-..
..;...
....,
''!:...
...,
.,.-.
......
..1-..... .,.Y.-...,.."... !.--.-,-,..l..-.,%'-.--. .... v't-'. -.!,
-...
...,.:,.%. .I..
_, :_.:... ".
... -.'
.-,,.-
".,...'.,.'....Z.A."..1I ...;. ::!.... ..'. ,-.-.1 .,, . .,1,,,-,". ;-.1..'L .11 ,- ...::
....
..-...
...'...-:--
..1. -J---... ....,,,,-..."..".'...-I'l,',',,.-! ,,,.-..".-.-T...,..7,":.-,.-..-!', --.-!---,--..:i-l-,...::.,:.-"-.;-J-;-:,i.4..,.I,. _."..,t...
Z: i.-'.".','---:-'..i.':,.--. ,:-... ....;
-:4....-.,(Y--&-.."%,
,.,-;-,,-,-:-o-.,,,...
,:.--,.-
..-:-....,.-..:
...
-,--,,.,.,.-,-I ,". -,.-,?!..-,-.-
-..---...
-. .'-!.:.,.:.. ..'.,...-Q- -- -.."-.;.. 1:-.,..,.-."Z'.::,:.,:... .--,-.,::--.,.4:.-;....-...';"....,. I".. ..--.",...-_-,,I:....,-'-...:-t?:.-. ..
..-,.%- 1,.-I...Y....":...-:-.`,--- ..".,,
'...I....
... '...-....,
.:.-,
-,,5I-,-..
O.,.-l-l,--.-Z--.C!.-,,.t.,..," '.'-:-L! fZ...-,,4." ... ",.,.
... :...
,.,....
.; ...I...,,...'....., -..,
.-...,
,1.-`--,-...'..-,I.-
.-'.-.',....
-_--,-_;..--,...,;,
I-...
..-.. ..- ....4-;N.j..,;-,.._,. ..."._;..."...':. -,..--;'-I.- ....2,
....".I..
....: .14".";.-,,._.- .. ..;..,-.-,.-..'. "..i.;,'.. _;,...-,;.,..-,,';...,...:;...,".,Z ,-.'..,.i... -. .. -..!'...'.. ,."-..,1.".: -t" :. V,I-". ...,-,'.f,.".-.'... :, ;-t,.!.Z.-.--,.%,,-.21. '.). .!... 4.. "..ir.-.--'...
-F:,tl._,.._,:. ...,:-.., .i;,....::-...
..I.... ...-.,.__.;)_.; ,:.. ,...#. ;..:,.. ..."...: '...,.
.%..;"..-."t'.. ..1"....l;."I' i '."...I..-_:,.-,-;F?:-"-,.,.-,?-...!...t.i.:".I.- -,.'--Iv..-...1, -....-t..-r,` %.,-"...!..!.. .;.. ...-'.,;:
_.. ..-..'....;".,.-:..f:-,-...,..-...-'.-.I...;....-il.-...',...,._q---l.-_-l. -...!-,.,.f.'j.-.-..-.-!-,--"...I!...4.1.--Q--...'--'-.I., ..'...'..._!7..,I, '--.ff'-j,_-l.-'l.,. ....A...-.!-.'.1, ,-`_-_-...-..-,-..,-:,-.,:,.,"--'. ...,,-.-i..,...,,,--..,:.-2, "il.:...::.,-.;,..,..L.,..Ii"...'...,...--.-,--,-:,.,.:..-.,.-.P,..,...'.,,..:I-.,:"..,...,--,::.!...,,"..l.:..l.-'.%-..,-:...-'.s.:,;...'!;:,..."...-,.,-., :...'.-'t---..!.-.,,_--...-,;.4, .4t -.i:...,..,.'..".-_.-,.,.-".-:.-.-j"'..,.:.:..."'...r:i-!;---,-.--,;'-?."!---.-...,'!---.;,-- ...,...",..,--!---,"-.,-:.-.-_:.:: .'.!--:..'. .;----:---.---.-,.!,,'..,.;,'...ti...-:',."....,....:.,:-.:...-;.::..,... ..-,':.-'.'-,
.- ,-,.:,.-.... .,.' -i-.:.,.",...-..;1.- .4, ;..,I -, .. ..'.- ..:.-::.. .,".- "....('..-.,'.7..;;:..:.,-.4-.,",';.:.-1-;.:.. ..
-.i."..,.: P.11'.:,.-...,-.,L., 1,-_..,-,,-,-.:..;t;'-....%"...'..,...".-..;..,-....t.-....- .. .,-L'-...,..-':,.':.. '.--:.,'i;k,"..;:...:..-":-...:-/ -'..;'..,,':',-"...-I...-.---.,....---'.-':,,, '...-,..;...,
'...;...,....--....,.,:,.:,.,.,_-!.1,,.::..:,...;.., ,-,-.,-,!----..l.,... ,."... .1.I1..,.1... ...---....".-.-...-"---'.;'.'-...:-.----:... ..;""-r-r-.'._-..,,.---".;:. .:,-....-,-.-,:,--;.-,,.-..,-..-,1:...;"-..-'...;;.,.'...'.!.%.,.!..,-:-...--,-.:. ...,..:..:-11-..._...'.-' ".-". .:?...%...t...:....,..t.-,-;,.,- "..:.."f, "--_j..,..%...--.'.,Cv:.11-I--,';-..i.1..,..,,:.,._.-.-..:.:...,'.-'...,.,-:..-...,..---.--,--...-I..."::-.'.----.. t.,.--
---....,... ...:f.I-,;.:..,.,..'-....-...!.i.,;,--,-'--I.-I.--;-i.. ;--.;--...'---'.-.-;..,'.-.,.-.-.! .". .- -C:.,(,eI,....-,..-..,.,: .... !...-._...--.-' ..7-,.:...." -..-: t... .,_..,....", ,:,..-:4-,l.',.T.$'.-;;:...- ,'-:-:_, ,.!-,.-i-.--...l-.-:,_:,-....,%;:.-.:...-.. ,-.,,.. .. ._.r,.-, -::...'.' ..--'.
.--,-,.-. -- -I ...:?.:.-.,":,I.:.;-.---'-'.':,'..,,f'-'","
...,
.,----.,.-"..-...,...-..i.,...-...1..,...r %;.. .,-,-i..,..-i'f.,'.11: .'---`_...,..--.;.,.,: ;;.."':..-.--.-.;.1." t; .:.,'-..';.-'."'..'...' .'.:.14.---..-`J.--,-,...-...,! -;.'...,. .,.
.A.. ,;.; t,I ...'... .-;; -..--.%'..!-'...-.'; ...Z.%,.,.... j...'.. ..1,_.."..'....: V...'... ..."...t, ....-'.-'.-,,.,; ;...,-..!-....-.;fzpi_-.--._-t#-11.--.;.r.,--`,...-,C-, "__.,
'....
-;,!,:,
.'..'....;
...;.-,,. .j:,.;.-;-....--...II...ij,--,,---_,-:--.--:-
_- .'-....,_. ...,--..;f-,.-,--,:
:,-,.-.-.'.- ...-.., ,,.. 11,...,.... ...".""",?....-'.-.;."---';..-,-"-,I"..-, "'!,'.,...:.-,-.-.,...!....,!,.-..;.;-,:...::.,:..,1...-!-.'.,...1..-.:,-,,,...,.--,eit."...T.; ....,..',-,--,.,..-.'..'.. `...-..-.;d....,-.,---:,-.,...%.,----:"',.v...;.""..:...,-...,,.k-lz`...'...?...:"....,,,.1..4..'.-:.-.Z..7.,...-...,:.,.:-.,1.:.-: ;..,IF,.:,...--:-..!%....,..,.:.:.":.:..,-`....-,-..;,.."::.,II-.,::..,.:!.',,x.;:.--..;..I. -'j."."'.-"t.,... ..:...-,.C.,,..7."..;.-J.::..1,,.----!.-,;,.!.,:.:,,,%r.-- -.:,:,'..:.)..'....;...,.--..."..,.,W-,-,.-,--.-:'.:.,,-..._..;._--,-.-.-,---....;.".;,"..,;..-..?.'-1.Z..,....- ,--.','... -,.
-.---1:A ...-.'..:..- ", i7%.... .:,,-...-...-!... .L'..'.,t'`,,--.!.'-_!-...;,..:,;....,_-.:...7...",,, -:,: ,--..-,;,_:_-.,- -, ... --.'.'i'... .' .1,., ;,:.." ..'..- -:--,t,:"-,
.! ".I,0... ..l.,
_:.. ...
.., ..-.:.. "'W".-"..-,.'...I-1 1. '_ ...,.;--..j.:11.-.:. .; ....-,
.---I...Ii.!__".
-..-j..,...,;'l..."..-....'...".:. ;...l...-,..---%--_..---_,II.-.... .1`'.......-.1. .",.:,.I-.1.7I.:-....:,....'....,,.- ...A..,.."`-..--'.,...-,.. 1:....,...,.-...,Z..."'....,...---!;.,-.;"..,.1.;..."....-...,..;....--.-..:...,..e..-!:t...."r,.j-...".. ..L.1.-..-;."-;:!:.,...--.',t..,.-'.":,. '.'.-:....--...:---4'.)",...;-),-,,::., ....,:..,;... ,..',;-,
..-,,.l.: `.' -, :1. ....:,,I1, I,..,,,.",::..--....;,-t.:;,:.-!.'.,'...., .-.,I ;..-.'?....,"...!.!:'.-...-"...,. .,...;.-.,..:. ... ,C....-a-.1. .:,,. ,,,-"--,,
--I._,-.-I..,-....,...,-, --el.- -11'..-:,.':!,..L,. ....- ,. "-...,.'.,....,.-.-11....,.:,--..-:".I.L.,....'.--.. ....-:--...l",-,-.1-s,.,..,-:-,I ..,i..,-'--.---..-..:,...""'... ,.,...."--.. .- .-...F..-...,.":...-...-.I-_.._.,-.!--.-;.-,...:(.-...,;.'...'-.,..,...,.."-'..-.,.1.:.'-,-`.;...,.:.,.'t..,"'-,-,,;...,.",...'...'.":-'j.".;-..,,,-..,.:...,'...,!.,.-...-...,. ....,-.-,.'.. ,.-.:.:...-."..-11.,- .-...;.,---i,:._,,-...-....---..!,.",--...I.:?..:.'...ll.:_..,,,i...1I,...1 ...::"...-"..'.--.,.,.-:...-.,...-...,..11....,-...j-..'-.--'..!-''A";
., .----.---1.-,,..
,-...:..,., -I..-1..,-f-...
.:.. ...,.--1. : -,,,..:...-..,. ...:-...
:...:.:..:..-,I..'.'-..'...:...'. ...,;:..-, .'r -,.,-,.-.-.-% ." .; ...;-.",,..;,z-...
,.. .-.
It, ;.-,..--
,,--..,.1.1.,:.,:-....,....:-,..,-1, ..j-:-,---"---'.-'. ,.j:-,-..,..-.:.i,.,...!...-....,". ..-.-,.. .. -.,. ,:-...
--.--...:.!....,;..;.;..-L. .:...- %,I'. ..--:,.f,. ..-,.,.-...--"-.'.---., .-
---....,.....---.1-;7, ....,....
...---,"-....-,,,,--.I-iI.,.--.-' ..,.. .. .,.,...
-...
.-..,,..---.!....--".-.'.-...-"...-I ,....""'..1...-,'. --,-.;t..f ;.,--,.;'.'!.',,...,..;:....,---:.V.---,-,.::.,:;-,,----.",...A..!., ..:
...,
..?.-...,- .. ...-....!!..7_-,.!:,"...'. " ...,..,-,-., .,.:.';;--..."..:. ..,.!..,:..-..._-.,.--....-_. - 1. I--'I.:.-I"., ,..-..,:..
,:...-7. ..,;.-... '!...,.;... ..:. ..'...-:.V...:...-....'...I.'.-'.-,:.--.:4-".-.-,.,'!....,..:--t..,;. :.. ...., ...,.,; .... .:
..-...-:..,....1... ..
.., ---"--.'.-., :.,...
FIG. 4. Electronmicrographsoftype IortypeI/II replicating 429DNA moleculeswith tuwosingle-stranded tails. (A) Type Imoleculewith two single-strandedtailsmovingin opposite directions; (B) type Imolecule
uwithtwosingle-strandedtailsmoving in thesamedirection; (C) typeI/IHmolecule withtwvo single-stranded tail.smovinginthesamedirection. Bar=0.5tim.
192
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.508.75.458.54.619.2]REPLICATING 429 DNA MOLECULES 193
shows ahistogram ofthe
length
of the double-stranded region in type Imolecules,
with an average value of 7.02±0.47/im.
Thislength
wasTABLE 1. Approximatefrequenciesof the different
typesofreplicating 129DNAmolecules %of
ana-DNA molecule No.
lyzable
mole-cules' Type IOnesingle-strandedbranch 43 20
Twosingle-strandedbranches 10
TypeII 145 55
TypeI/II
Onesingle-strandedbranch 22 9 Twosingle-strandedbranches 1
TypeIII 36 14
Type IV 6 2
"About28% ofthe totalnumberofmoleculeswere non-analyzable.
30
20
0 E
0
E
z
10_
2 4 6
(a+ b), pm 8
greater than that
(6.26
± 0.18 um)reported
previously for429 DNA (24),which was deter-mined using ethidium bromide as a spreading agent
(16).
Thisdifference wasprobablydue to thedifferent techniqueused heretospread the DNA. Figure 5B shows the ratio between the length of the double-stranded region with the valuemoresimilartothat of thesingle-stranded tail and the totaldouble-stranded DNAlength. Asexpected,there wasarandom distributionof theintersectionsof thesingle-strandedtails with theduplex
regionin type Imolecules.Thelengthof the single-stranded tails in type I and typeI/II moleculesand thecorresponding double-stranded regions was also determined. Figure5C shows that when the ratio of single-to double-stranded DNA length was plotted, there was a maximum at a value of 1. These data suggest that initiation of replication occurs at or near the end(s) of 429 DNA. The distri-bution obtained, mainly toward the positions with a ratio smaller than 1, wasprobablydue to the difficulty in visualizing clearly the ends of single-stranded DNA regions and to a
suscepti-0.2 04 0.6 0.8 0.5 1.0
a/(a b) c/a 1.5
FIG. 5. Length distribution, positionof the growing points, and length of single-stranded branchesintype
Ireplicating 4)29DNA molecules. Replicating 429DNA molecules were isolatedand prepared for electron
microscopyasdescribedin thetext.Inthedrawingsofthe differenttypesofmoleculesin this and thefollowing figures, the heavy linerepresentsdouble-strandedDNA and the light linerepresentssingle-strandedregions.
(A) Length of thedouble-stranded DNA in 56type Imolecules. (B) Position of thegrowing points intypeI
molecules. Thehistogramrepresentsthe ratio between the length of the double-stranded regionwithalength
similartothatof thesingle-strandedtail (a) and that of the total length of duplexDNA (a+b).Fifty-sixtype
Imolecules withatotalof 67 growing pointsweremeasured. Thedeterminationof the growing points of18
typeI/II moleculeswasalso includedin the histogram. In thiscase,theratiobetweenthelengthof the
double-strandedregion with a length similartothat of the single-stranded tail andthatofthe total length of the
DNA(double and single stranded)wasdetermined.(C) Length ofsingle-stranded branchesintypeImolecules.
Thehistogramrepresentstheratio between the lengthof thesingle-strandedtail (c) and that of the
double-strandedregion withasimilarlength (a). Fifty-sixtypeI molecules withatotalof 67 tailsweremeasured. The
length of thesingle-strandedbranches of18typeI/II moleculeswasalsoincluded inthehistogram.
TypeI molecules A B C
a b , 20
C
-10
10
5
VOL. 34,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.508.53.245.137.299.2] [image:7.508.104.395.338.536.2]194 INCIARTE, SALAS, AND SOGO
bility to degradation greater forsingle-stranded than fordouble-stranded DNA.
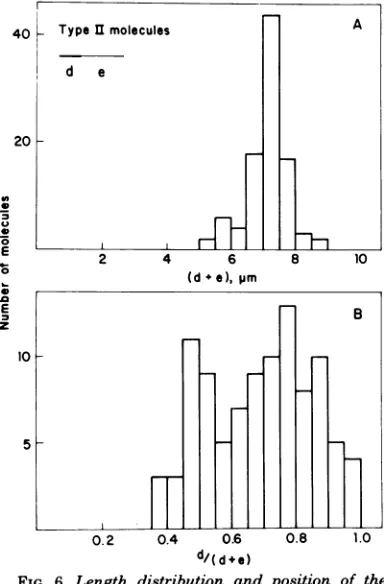
Figure 6A shows the length distribution of type II andtype I/II molecules. The lengths of double- plussingle-stranded regions (excluding the single-strandedtails in typeI/IImolecules) gave an averagevalue of 7.15±0.70jim,similar tothatobtainedfortype Imolecules.The point of transition between double- and single-stranded regions in type II and typeI/II mole-cules wasrandomlylocated (Fig. 6B), although molecules with a longer duplex DNA region were moreabundant than thosewith a longer single-stranded region.Thisresultwasprobablydue to thefactthatmolecules with longsingle-stranded DNA regions were mainly present in the peak correspondingtofractions9 to 15 ofFig. 2B.
The length ofthe double- andsingle-stranded
40
20
0
.0
E
z
0.2 0.4 0.6 0.8 1.0
d(d+e)
FIG. 6. Length distribution andposition of the growing pointsin type IIreplicating 429DNA mole-cules. Replicating 429DNAmoleculeswereisolated
andprepared forelectronmicroscopyasdescribed in
the text. (A) Length of the double- and single-stranded unbranchedregions in 74 type II and 23
type I/II molecules. (B) Position ofthe transition
betweendouble- andsingle-strandedregionsintype II molecules.Thehistogramshows the ratio between
thedouble-strandedregionandthatofthetotal DNA
lengthin 74typeIIand 23typeI/II molecules.
regionsin type IIImoleculeswas 7.13±0.45 ,um (Fig. 7A), similar to the values obtained for type Iand type II molecules. The length of each of thesingle-stranded regionsintype IIImolecules isshown in Fig. 7B. Since no defined maximum was detected, there seemed to be a random distributioninthetransition points between sin-gle- anddouble-strandedDNA regions.
[image:8.508.68.260.279.571.2]Partial denaturation map oftype II rep-licating DNA molecules. The small number of type I molecules with two single-stranded tails,eachgrowing from a different end (6 out of a total of 56), does not permit the conclusion thatreplication can be initiated ateitherDNA end.Totest thispossibility,partialdenaturation ofreplicating DNA molecules was carried out, andthe mapoftype IImoleculeswasobtained. Figure 8A shows the partial denaturation map of
029
DNAtaken from the work ofSogo et al. (25). It can be seen that the right end was always open, whereas the left end was closed in most cases. Figure 8B and C shows thepartial dena-turation maps of type II molecules in which the partiallydenatureddouble-stranded region,cor-15
10
a 5
E
0
.0
= 15 z
10
5
TypeMmolecules
f g h
r-
,1
2 4 6 8 10
(f+g+h), pm
B
-~~~~~
f andh, pm
FIG. 7. Length ofthe total DNA andofthe single-strandedregions in type IIIreplicating DNA
mole-cules.Replicating 4)29DNA moleculeswereisolated andprepared forelectronmicroscopyasdescribedin
the text. (A) Total DNA length (f+ g + h) in 51
molecules. (B) Thelength ofeach ofthetwo single-strandedregions (fand hseparately)in 51 molecules
wasplottedinthehistogram. (d+e), pm
J. VIROL.
I 2 3 4 5
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.508.273.468.328.586.2]REPLICATING p29 DNA MOLECULES 195
responding to the initiation ofreplication, could beclearly assignedto theleft (Fig. 8B) orright (Fig. 8C) DNA ends. A totalof 32 and 23typeII molecules were foundto initiate at the left and right DNA ends, respectively. Figure 9 shows electronmicrographs of partiallydenaturedtype IImolecules withone end closed (Fig. 9A) and
oneend open(Fig.9B),correspondingtotheleft and right ends of p29 DNA, respectively.
DISCUSSION
Pulse-chase experiments suggest that the pulse-labeled radioactive DNA with a sedimen-tation coefficient greater than that of mature
50%
0 20 40 60 80 100
[image:9.508.114.394.150.583.2]DNA length (units)
FIG. 8. Partial denaturationmaps of type II replicating DNA molecules. (A) Partial denaturation map of
029
DNAtakenfromthe work of Sogo et al. (25). (B) Partial denaturation map of 32 type IIreplicating DNA molecules with a denaturation degree of 25%. The double-stranded region, corresponding to the initiation of the replication, can be assigned to the left end of the DNA. (C) Partialdenaturation map of 23 type II replicating DNA molecules with a denaturation degree of22%o.
Inthis case, it can be seen that the double-stranded region corresponds to the right end of the whole genome. Five maturec29
DNA molecules, partiallydenatured,wereincluded in (B) and (C) to determine clearly the ends of thehistograms. VOL. 34,1980
on November 10, 2019 by guest
http://jvi.asm.org/
196 INCIARTE, SALAS, AND SOGO
FIG. 9. Electronmicrographsofpartially denaturedtypeIImolecules.(A)Partiallydenatured molecule in
which thedouble-strandedend isclosed,correspondingtoinitiationattheleftendofthegenome.(B) Partially denatured molecule in which the double-stranded end isopen,correspondingtoinitiation attherightendof
thegenome. Bar=0.5 Lm.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:10.508.71.455.71.614.2]REPLICATING 429 DNA MOLECULES 197 DNAconsists of replicative intermediates.
When the DNA with a buoyant
density
in CsCl greater than that ofphage429 DNA was analyzed with the electron microscope, type I, type II, and type I/II molecules were found. Length measurements of these molecules sug-gest for 429 replication adisplacement mecha-nismsimilartothe onepostulatedfor adenovirus type 2 withinitiationator neartheendsof the DNA (17).Evidencethat initiation of4)29DNA replication maytake place ateitherend of the DNA hasbeen obtained by partialdenaturation of type IIreplicatingDNAmolecules.Sincetype I DNA molecules with two single-stranded branches correspondingtothedisplaced paren-talstrandsinitiating simultaneously atthe two DNA ends are found very rarely, it may be suggestedthatinitiationof429replicationtakes place at or near either DNA end, but non-si-multaneously.For thereplication of the displaced parental strand, two possible nonexclusive mechanisms were postulated for adenovirus type 2 (17): (i) before the parental strand has been completely displaced, initiation would takeplace at the op-posite end oftheDNAmolecule; (ii) theparental strandwould be completelydisplaced,giving rise to a double-stranded DNA molecule and a sin-gle-stranded DNA molecule (Fig. 10A). Since the 3' ends of the two DNA strands ofadenovirus
A
OR
types2and5have the same sequence of 102 and 103 nucleotides, respectively (2, 27), the initia-tion of the
replication
atthe 3' end of the dis-placed strand could take place through circle formation by hybridization ofthe complemen-tary sequence at the 5' and 3' termini of the DNA (17). In any case, a newly synthesized molecule of the terminal protein of adenovirus DNA could recognize the 3' ends and prime replication by the covalent attachment of the initiatingnucleotide (20).In phage 429 DNA, there is no evidence for the existence of a large region ofinverted ter-minal repetition, as in adenovirus DNA. Dena-turation ofprotein-free 429 DNA and renatur-ationat a low DNA concentration didnotgive rise to the formation of single-stranded circles (A. Talavera, personal communication) under conditions in which circles were produced with adenovirus DNA (9, 32). The partial denatura-tion map of429DNAindicates that under con-ditions in which the right end is open, the left endremainsclosed (25). Nevertheless, the exis-tence of a small region of inverted terminal repetition cannot be ruled out. A nucleotide sequencedetermination of both 429 DNA ends isbeingcarriedout toanswerthisquestion.
An alternative mechanism to initiate 4)29 DNA replication would be that anewly synthe-sized molecule of the terminal protein of 429
FIG. 10. Possiblemodels for 429DNA replicationandforthe role of the terminalproteinin initiation of
replication. The model isanadaptationof those proposed by Rekoshetal. (20) andLechnerandKelly(17), consideringreplicationonlinearDNA(A)orcircularDNA (B).The dotsatthe 5' ends of the DNArepresent
the terminalprotein. Thedotsfollowedby pNrepresenttheprotein linked tothe initiatingnucleotide (or deoxynucleotide) thatisproviding the free 3'-OHgroup toelongatethe chain. Continuous lines represent
parentalDNA, anddiscontinuouslinesrepresentnewlysynthesizedDNA. (1) and (2)arealternativemodels toinitiate thereplication of thedisplacedstrand.Onlytheinitiation of replication ofoneoftheDNA strands
hasbeendrawnforsimplicity.
B
I
/ OR
NpS
..I /,
'0
O~~~~~~~~
©
P+*pN
I
4,/
pOF
I
VOL. 34,1980
*,O/V
0 -0. 11
lq.le 09
on November 10, 2019 by guest
http://jvi.asm.org/
[image:11.508.92.399.384.581.2]198 INCIARTE, SALAS, AND SOGO
DNA, p3, which we have shown is needed for the initiation of replication (R. P. Mellado, M. A. Penalva, M. R. Inciarte, and M. Salas, sub-mitted for publication), primes replication not by interaction with the 3' end of the DNA as proposed in the case of adenovirus (20), but rather by interaction with the parental protein (Fig.10). This is consistent with previous results showing that a functional parental protein is needed to carry out429 DNAreplication (23). In this case, it mustbeassumedthat before the parental DNAstrand has been completely dis-placed, initiation would take placeat the oppo-site endof the DNA molecules (Fig. 10, possi-bility 1A or B). However, if the replication is initiated on the completely displaced parental strand, some kind ofprotein-DNA interaction mustbepostulated. Ifthere were asmallregion ofidenticalsequence at the ends of429DNA, a circle could beformedby an interaction of the protein bound at the 5' end of the displaced strandwiththe 3' end(Fig. 10, possibility2A or B). Having a circle, a newly synthesized mole-cule ofproteinp3couldinteract withthe paren-talprotein subunit and initiate replication from the3' end. Ifthis werethecase,single-stranded circles should be found asreplicative interme-diatesif no protease treatment were includedin theisolationprocedure.
Besides type I, II, and I/II replicative inter-mediates, another class of molecules is found both in
029-
and inadenovirus type 2-infected cells, which consists of linear DNA partially doublestranded withsingle-stranded regionsat both ends. Inthe caseofphage429,
the single-stranded regions ofthese molecules (type III) have arandomlength.Althoughthese molecules are14%of thetotalin429and 9% of thetotalin adenovirus (17), their presence must be ex-plained.Apossibilityto accountfor thistype of molecule is that if acircleis formed to initiate thereplicationof thedisplaced strand,the inter-action between theproteinand the 3'endof the DNA could take place occasionally atrandom with internalregionsof themoleculearound the 3' end. Therefore, type III moleculeswould be abortive type II DNA. Anotherpossibility
for explainingtype III molecules is thattheyarise byrecombination between two type II molecules in which each of the displaced parental single strands isbeingreplicatedintheopposite direc-tion. Thisrecombination eventwould also pro-duce abortive DNA molecules.It iswell establishedthatin vitro the
protein
linked to
429
DNAisabletointeract withitself,
givingrise to the formation ofa
variety
of cir-cular structures and concatemers (19, 23). The same result is obtained in the caseof the ade-novirus DNA-proteincomplex
(3, 15, 21, 22).Girardetal. (10)have analyzed with an electron microscope the replicative intermediates iso-lated bytreatment ofadenovirus-infectedHeLa cell nuclei with Sarkosyl and have shown the existence of a majority of unit-length circular DNAmolecules. It is, however,difficultto estab-lish whether the circles are true intermediates in DNAreplication or whether they are formed in vitro during the isolation and processing of thesamples.
Thefollowingargument, however, favors the idea that both
429
DNA and adenovirus repli-cation might take place on circular DNA (Fig. lOB):ifreplicationoccurred onlinear DNA and the chances toinitiate replicationatboth ends are similar, type I DNA molecules with two single-stranded branches being displacedin op-posite directions would probably be more fre-quent thanhas been found both in429
and in adenovirus type 2 (17)replication since in linear DNA both endswould be free to initiate repli-cation (Fig. 10A). However, if replication oc-curred on DNA kept circular through protein-protein interaction, there could be some con-straintforthesimultaneousinitiation at the two 3' ends (Fig. lOB), and, therefore, the above moleculeswould be rare.ACKNOWLEDGMENTS
WeareverygratefultoE.Vifnuelaforhelpfuldiscussions
and for critical reading of the manuscript. The help ofM.
Lozano andM.Caballero intheprocessing of the data from
the electron microscopeisgratefully acknowledged. This investigation was aided by grants from Comisi6n
Asesora para elDesarrollo de laInvestigaci6n Cientificay
Tecnica, Comisi6n Administradora del Descuento Comple-mentario (Instituto Nacional de Prevision), and Direcci6n General de Sanidad.
LITERATURE CITED
1. Ariga,H., and H.Shimojo.1977.Initiation and termi-nation sites of adenovirus 12 DNAreplication.Virology
78:415-424.
2. Arrand, J. R., andR.J. Roberts.1979.The nucleotide sequencesattheterminiof adenovirus-2 DNA. J. Mol. Biol. 128:577-594.
3. Brown, D.T., M. Westphal, B. T. Burlingham, U. Winterhoff, andW. Doerfler. 1975. Structure and
compositionoftheadenovirus type2core.J. Virol. 16: 366-387.
4. Brown,N.C.1970.6-(Hydroxyphenylazo)-uracil:a
selec-tive inhibitor of host DNAreplicationinphage-infected Bacillussubtilis.Proc.Natl. Acad. Sci.U.S.A. 67:1454-1461.
5. Carrascosa,J.L.,A.Camacho,F.Moreno,F. Jime-nez,R. P.Mellado,E.Vinuela,andM.Salas. 1976.
Bacillus subtilis phage 429: characterizationof gene
productsand functions. Eur. J. Biochem. 66:229-241. 6. Carrascosa, J. L., E. Vinuela, and M. Salas. 1973. ProteinsinducedinBacillus subtilisinfected with
bac-teriophage429.Virology56:291-299.
7. Carusi,E. A. 1977. Evidence forblocked 5'-termini in human adenovirus DNA.Virology76:390-394. 8. Davis, R.,M.Simon,andN.Davidson. 1971. Electron
microscopeheteroduplexmethodsformappingregions J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 34, 1980
of base sequencehomology in nucleic acids. Methods Enzymol.21:413-428.
9. Garon, C. F., K. N. Berry, and J. A. Rose. 1972. A
uniqueformof terminalredundancyin adenovirus DNA
molecules. Proc.Natl. Acad. Sci. U.S.A. 69:2391-2395. 10. Girard, M., J. Bouche, L. Marty, B.Revet, and N.
Berthelot. 1977. Circular adenovirus DNA-protein complexes frominfected HeLa cell nuclei.Virology83: 34-55.
11. Hagen, E. W.,B. E. Reilly, M. E. Tosi, and D. L
Anderson.1976.Analysisofgenefunction of
bacterio-phage 4)29ofBacillus subtilis:identification of cistrons
essential forviralassembly.J. Virol. 19:501-517. 12. Harding, N., J. Ito, and G.S. David. 1978. Identification
of theprotein firmly boundtothe ends ofbacteriophage 429DNA.Virology84:279-292.
13. Hawley,L. A.,B. E. Reilly,E.W.Hagen, and D. L. Anderson. 1973. Viral protein synthesisin
bacterio-phage 4)29-infected Bacillussubtilis. J. Virol. 12:1149-1159.
14. Ito, J. 1978.Bacteriophage 4)29 terminal protein: its
as-sociation with the 5' termini of the 429genome.J. Virol. 28:895-904.
15. Keegstra, W.,P.S.VanWielink,and J. S.
Sussen-bach.1977.The visualization ofacircularDNA-protein
complex from adenovirions. Virology76:444-447. 16. Koller, Th.,J. M.Sogo,and H.Bujard.1974.Amethod
forstudying nucleic acid-protein interactions with the electronmicroscope. Biopolymers13:995-1009. 17. Lechner, R. L., andT. J.Kelly, Jr.1977.Thestructure
ofreplicating adenovirus2 DNAmolecules. Cell 12: 1007-1020.
18. Moreno, F.,A.Camacho,E.Vinuela,and M.Salas.
1974.Suppressor-sensitivemutantsandgeneticmapof
Bacillus subtilisbacteriophage4)29. Virology62:1-16. 19. Ortin, J., E. Vinuela, M. Salas, and C.Vasquez. 1971.
DNA-protein complexincircularDNAfromphage429.
Nature (London) New Biol.234:275-277.
20. Rekosh,D. M.K.,W. C.Russell,and A. J.D. Bellet.
1977. Identificationofaprotein linkedtothe ends of adenovirus DNA. Cell 11:283-295.
21. Robinson,A.J., andA.J. D. Bellett.1974.Acircular DNA-protein complexfromadenovirusesand its
pos-sible role in DNA replication. Cold Spring Harbor
Symp. Quant. Biol.39:523-531.
22. Robinson,A.J.,H. B.Younghusband, and A. J. D.
REPLICATING 429 DNA MOLECULES 199 Bellett. 1973. A circular DNA-protein complex from adenoviruses. Virology 56:54-69.
23. Salas, M., R. P. Mellado,E.Vinuela, and J. M. Sogo. 1978. Characterization of a proteincovalently linked to the 5' termini of theDNA of Bacillus subtilis phage
)29. J. Mol. Biol. 119:269-291.
24. Sogo,J.M.,M. R.Inciarte,J.Corral,E.Vinuela,and
M. Salas. 1979. RNA polymerase binding sites and transcription map of theDNAof Bacillus subtilis phage 4)29.J.Mol. Biol. 127:411-436.
25. Sogo, J. M.,P.Rodeno, Th. Koller, E. Vinuela, and M. Salas. 1979. Comparison of theA-Trich regions and the Bacillus subtilis RNA polymerase binding sites inphage 429 DNA. Nucleic Acids Res. 7:107-120.
26. Stafford,D.W.,and D. Bieber. 1975.Concentration of DNA solutions by extraction with 2-butanol. Biochim. Biophys. Acta 378:18-21.
27. Steenbergh,P.H.,J.Maat, H. VanOrmondt, and J. S.Sussenbach. 1977. The nucleotide sequence at the termini of adenovirus type 5 DNA. Nucleic Acid Res. 4:4371-4389.
28. Sussenbach, J. S., and M. G.Kuijk. 1977. Studies on
themechanism ofreplication of adenovirus DNA. V. The location of terminiof replication. Virology
77:140-157.
29. Sussenbach, J. S., and M.G. Kuijk. 1978. The mecha-nism ofreplication of adenovirus DNA. VI. Localization ofthe origin of the displacement synthesis. Virology 84: 509-517.
30.Talavera, A.,M.Salas, and E. Vinuela.1972. Temper-ature-sensitive mutants affected in DNA synthesis in phage029 of Bacillus subtilis. Eur. J. Biochem. 31: 367-371.
31. Westmoreland,B.C.,W.Szybalski, and H. Ris. 1969. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy.Science 163:1343-1348.
32. Wolfson, J., andD.Dressler.1972.Adenovirus-2 DNA contains an inverted terminal repetition. Proc. Natl. Acad. Sci. U.S.A. 69:3054-3057.
33. Yanofsky, S., F. Kawamura, and J. Ito. 1976. Termo-labile transfecting DNA from temperature-sensitive
mutantofphage4029.Nature(London) 259:60-63.
34. Yehle, C.D. 1978.Genome-linked protein associated with
the 5' termini ofbacteriophage429DNA. J. Virol. 27:
776-783.