JOURNAL OFVIROLOGY, JUlY 1976, p.279-285 Copyright © 1976 AmericanSociety forMicrobiology
Vol. 19,No. 1 Printed in U.S.A.
Excision of Viral DNA
from
Host
Cell
DNA
After Induction
of
Simian Virus
40-Transformed
Hamster
Cells
TAMARA RAKUSANOVA, JOAN C. KAPLAN, WILLIAM P. SMALES, AND PAUL H. BLACK* Department ofMedicine, Massachusetts General Hospital ,* and DepartmentsofMedicine,andMicrobiology
andMolecular
Genetics,
Harvard MedicalSchool, Boston,
Massachusetts 02114 Received forpublication 12December 1975Simian
virus40-transformed hamster cells
wereinduced
toproduce
infectious
virus
by
treatmentwith
mitomycinC
ory-irradiation.
Aportion of the simian
virus-40
DNA,
whichisintegrated
intothe
host cell genome
inuninduced cells,
was
recovered
in apool
of
relatively
low-molecular-weight
DNA
early
after
induction
treatment inthe
absence of DNA
replication. The data indicate that
excision
of the viral
genome occurssubsequent
tothe
induction stimulus.
Simian
virus 40(SV40)-transformed cells
usually
containonly
afew
copiesof the virus
genome per
diploid quantity of cell DNA (2, 9).
Inthe
transformed
celllines
inwhich the
asso-ciation
of
viral and cellular DNA has
been
in-vestigated, viral DNA has
been shown
tobe
covalently linked
tocellular DNA (10). In
gen-eral, infectious
virus is notreleased
sponta-neously from
SV40-transformed rodent cells
and
mustbe induced
by
varioustechniques
(1, 4).We have been
studying clones of virogenic
SV40-transformed hamster kidney cells that
can
be induced
toproduce infectious
virusafter
treatment with
such
agents as UV ory-irradia-tion, mitomycin
C,
orbromodeoxyuridine and
visible light. We have utilized these lines
tostudy
the
mechanism of
induction,
inparticu-lar, the early
events inthe
induction
process (5, 6).One of the earliest
events maybe
excisionof
the viral
genomefrom
itsintegrated
state inhost cell
DNA,
asisthe
case inthe
induction of
lambda DNA from the Escherichia coli genome
during induction
(11). Inthis
paper, we presentdata which indicate that the SV40
genomeis
excised from
itsintegrated
statesubsequent
tothe
induction stimulus.
The cell line studied, THK 22E,
Cl 1-1, AP1(clone
E), has been described
indetail (6).
It is aclone isolated from
aline of SV40-transformed
inbred
hamster kidney cells.
Allthecells of this
clone
containthe
SV40 T
antigen,but
no Vantigen
has been
detected by
immunofluores-cence
studies. Clone
Ecells
contain 0.5 to 1.5SV40genomes per
diploid
quantityof cell
DNAas
determined
by
Cjt
analysis. Since
the DNAcontent
of clone
E cells isapproximately
twicethat of
diploid
hamstercells,actually
1.0to 3.0SV40 genomes per
cell
are present.Clone
Ecells
occasionally
produce trace amounts ofvi-rus
spontaneously.
However, virus yields ofap-proximately
104 to106
PFU/106 cells
areob-tained upon
induction withmitomycin
C ory-irradiation.
Experiments illustrated
inFig.
1 and 2 wereperformed
to examine the association of viralDNA withhost cell DNA in clone E cells.
Cellu-lar DNA was
subjected
tovelocity
sedimenta-tion in alkaline sucrose gradients;
32P-labeled
SV40 DNA was added to a parallel gradient as
amarker
(Fig. 1). SV40
DNA contentwasmea-sured in the fractions of thegradient
containing
high-molecular-weight cellular DNA and
com-pared
with theSV40
DNA content oftotal,
unfractionated cellular DNA that was
ex-tracted from cells cultured in the same
experi-ment (Fig. 2). SV40 sequences were
determined
by
following
thekinetics ofreannealing
of32P_
labeled
SV40
DNA inthe presence ofunlabeled
cellular
DNA;
thetechnique was performedby
the method ofGelb et al. (2), and the
results
were
calculated and
expressed according
toOzanne etal. (9). From the degree of
accelera-tion of the annealing reaction achieved by a
given amount of cellular DNA, values of 1.6
SV40
genomes/cell
wereobtained
inthis
exper-iment forboth clone E total cellular DNA and
fractionated
high-molecular-weight
cellular DNA.These
results indicate that in clone Ecells the
SV40
DNA islinked to cellularDNA
by alkali-stable bonds.
Excision of the SV40 viral genome from
theintegrated
state wasdetermined
byfollowing
the movement of SV40 DNAfrom its
associa-tion with
high-molecular-weight host cell
DNAto a
pool
oflow-molecular-weight
DNA afterinduction. Clone
E cells were treated witheither mitomycin C or
y-irradiation
and wereharvested
atdifferent
times after induction.High-molecular-weight
cellular DNA wassepa-rated from a pool of
low-molecular-weight
279
on November 10, 2019 by guest
http://jvi.asm.org/
I 7
6
"-.
\N4
K
t3
I..
'1
BOTTOM
SV40
DNA
form
I
I
I . I'
I
I%-
-A~.T
IusT'4-SV40 DNA
form I[
It
II
I I I
I'I
I' I I
I I
0 12 14 16 18 20 22 24 26 28
Fraction
number
TOP
14
12 3s
10
t3
N"
8 '$.
\
N"6 | (-3
4
a_
2
1
C0r)
FIG. 1. Fractionation of cellular DNA on alkaline sucrose gradients. Clone E cells were seeded in Falcon
plastic plates (150 by22 mm) and grown in Eagle minimal essential medium (Gibco) with a fourfold
concentrationof vitamins and aminoacids, supplemented with 10% fetal bovine serum,2mMglutamine, and antibiotics.[3H]thymidine, 0.05 ,Ci/ml (specific activity,20 Ci/mmol; New England Nuclear Corp.) was
included in the mediumfor 24 h. At the end of the labeling period, the cells were harvested with a solution containing 0.01 MTris-hydrochloride,pH 7.4, and 0.4% sodium dodecylsulfate. Theresultinglysate was
layered on top of analkaline sucrose gradient (10 to
30%o)
in0.3 N NaOH-0.01 M EDTA-0.5 MNaCl.After storageovernight at 4 C, the gradients were centrifuged(17,500rpm, 16 h, SW25.1 rotor, Spinco model L centrifuge). To aparallel gradient, 32P-labeledSV40 marker DNA (specific activity,106countslminper Ag,prepared as described in reference 6) was added. At the end ofcentrifugation, the radioactivity ofan aliquot of eachfraction wasassayed, and the bottom three to five fractions were pooled, neutralized, and extensively
dialyzedagainst 0.01xSSC(0.0015MNaClplus0.00015 Msodium citrate) containing 0.01 MEDTA;this
fractionated high-molecular-weight cellular DNA was used for determination of SV40 DNA content by
C,t
analysis. Symbols: 0,[3H]thymidine(cellular DNA); 0, 32P-labeledSV40 DNA marker.
DNA, including free viral DNA, by
the Hirtextraction
procedure
(3),
and
the amountof
SV40
DNA sequences in the Hirtpellet
and
supernatant fractions
wasdetermined
by
meas-uring the rate of reassociation of 32P-labeled
SV40 DNA in the presence of
unlabeled DNAs
extracted
from the Hirt fractions.Experiments
were
carried
out to ensuretheefficiency
of theHirtfractionation
procedure
inseparatinginte-grated
from freeviral
DNAinclone Ecells.
Inuninduced control
cells, virtually
all thehigh-molecular-weight cell
DNA,
containing theco-valently bound SV40 DNA,
wasrecovered
in theHirtpellet.
Whenpurified 32P-labeled
SV40DNA was
added
to cultures of clone E cells atthe time
of
harvestand
the Hirt extractionwasperformed, approximately
86 to93% of freela-beled viral
DNA wasrecovered
inthe
Hirt supernatant fraction.Another factor to
be considered
in excisionexperiments
relates
tothe
amountof cell
DNAfound in the Hirt supernatant fraction.
Some
cellular
DNA isalways
present inthisfraction,
and
induction
treatments increase the amountby
causing DNAbreakage. Therefore,
onemight also expect a proportionate increase in
the amount of viral DNA in the Hirt
superna-tant fraction after
induction.
To monitor theeffect of induction treatment on the size of
cel-lular DNA, we
determined
the partitioning ofcellular DNA between the Hirt pellet
and
su-pernatant fractions in each
experiment
by
in-cluding
cultures that had beenlabeled
with[3H]thymidine
for 24 h prior to induction inI
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.505.131.408.65.347.2]NOTES 281
occurred,
the supernatant fraction should show
an
enrichment for viral DNA
sequences.How-ever,
if the
presenceof viral DNA in the Hirt
supernatant
resulted from random
breakage
of
cell DNA,
therelative
amountof SV40 DNA in
the
supernatant fractionshould be
equal
to orless than
itsrelative
contentinthe
pellet
frac-tion.
4
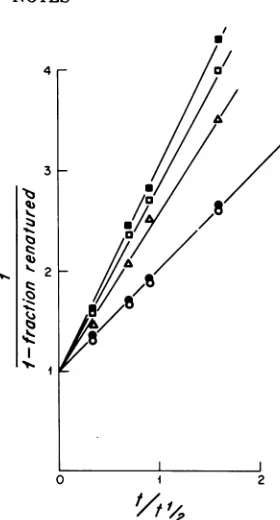
FIG. 2. Reassociationkineticsof 32P-labeled SV40
DNA inthepresenceof unlabeled total cellular DNA and fractionated high-molecular-weight cellular
DNAfrom clone E. Themixturesof 32P-labeled SV40 DNA and unlabeled DNAs were sonically treated,
heat-denatured, and incubated at 68C in medium
containing 0.001 M EDTA-0.4 M sodiumphosphate buffer(pH 6.8)-0.4% sodium dodecyl sulfate.Aliquots.
(0.2to0.3 ml) wereremovedatdifferenttime
inter-vals, and theproportion of reassociated 32P-labeled
SV40 DNA was determined by chromatographyon hydroxyapatite. The resultsareexpressedina
stand-ardsecond-orderplot (12).The abscissa(tlt1/2) is the, time of incubation of each mixture divided by the
time required for50% of 32P-labeled SV40DNA to reanneal in thepresenceof salmon spermDNA. In experiments shown,2 x 10-5optical densityat260
nm
(OD211)
unitsof32P-labeled SV40 DNA(specificactivity, 106 counts/min per pg) per ml was incu-bated withoneof the following: 0,36OD260u.nitsof
salmon sperm DNA per ml; 5 OD260 units of salmonspermDNAperml;A,36
OD26
unitsof totalcellular DNA (extracted from clone E cells by the methodof Marmur [8],withmodificationsdescribed inreference 6)perml; A,5.3
OD2b
unitsoffraction-ated high-molecular-weight cellular DNA (purified from bottomfractions ofalkalinesucrosegradients, asdescribed inFig.1)perml.
everyexperimentalgroup. Theamountof
acid-precipitable, labeled DNA was determined in
both the Hirtpellet andsupernatantfractions,
and theproportion of total cell DNA in the Hirt
supernatant fraction was calculated. Our
re-sultsareexpressed inanumberofways, oneof
whichindicates the amountsofSV40 DNAper
milligram of total cell DNA present in either
the supernatantor pellet fractions. If excision
[image:3.505.49.220.51.304.2]3
FIG. 3. Reassociation kinetics of 32P-labeled SV40 DNA incubated inthepresenceofunlabeled DNAs extractedfromHirtsupernatantandpellet fractions of mitomycin C-induced and control cells. Clone E cells were maintainedas described in thelegendto Fig.1.Whensubconfluent,amediumcontaining0.5
pgof mitomycin C(NutritionalBiochemicalCorp.) permlwasaddedtothe cultures(zerotime). A total
of10pgofaraC(cytosine-l-D-arabinofuranosyl hy-drochloride; Sigma)perml wasaddedto the
mito-mycin C-treated cellsat7hafterinitiationof
induc-tionoratthebeginning ofmitomycin Ctreatment.At theendoftheincubation withmitomycin C (24 h),
the cells were harvested and fractionated by the
methodofHirt(3).DNAfromthe Hirtsupernatant andpellet fractionswas extractedessentially bythe methodofMarmur (8),withmodifications described
in reference 6. Control cells were treated similarly with medium lacking mitomycin C andlor araC. Reassociationkineticsweredeterminedandthe data
wereexpressedasdescribed inthelegend toFig. 2.
3P-labeled SV40 DNA (2 x 10-1
OD26.
units/ml)wasincubated inthepresenceof the following unla-beledDNA samples: 0, 17
OD2,
units ofsalmonspermDNAperml; 0,10OD260unitsofDNAfrom
Hirtsupernatant ofcontrol cellsperml; A, 10 OD260
unitsofDNAfromHirtsupernatantofinduced cells
perml; 0,17OD260unitsofDNAfromHirtpellet of
control cellsperml; *,17OD260unitsofDNAfrom Hirtpellet ofinducedcellsperml.
4
2
A
A
//
0 2
VOL. 19, 1976
3
13 Z., '!Zl
I%.
t, 91-Q%.,. C.- 2 4. 9,0 C-3
11
%.j
I
I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.505.285.420.156.388.2]Figure3shows the results ofarepresentative
experiment in whichclone E cellswereinduced
by mitomycin C. The rate of reassociation of
32Plabeled SV40 DNA was the same in the
presence ofsalmon sperm DNAasinthe
pres-enceofDNAextracted from the Hirt
superna-tantfraction of uninducedcells,
indicating
thatnovirus-specific DNA could be detected in the
latter. Incontrast, reassociation of32P-labeled
SV40 DNAwas accelerated by the addition of
DNAextracted from the Hirt supernatant
frac-tion ofmitomycinC-treatedcells. Theaddition
ofDNAs purified from Hirtpellet fractions of
both induced and uninduced cells accelerated
the rate of reannealing of SV40 DNA to the
samedegree. The amount ofSV40 DNAin the
Hirtfractionswascalculated from thedegreeof
acceleration of '2P-labeled SV40 DNA
reasso-ciation (seeTable 1, experiment4).
Several similar
experimentsaresummarized
in
Table
1.The
data
arepresented
asthe
num-ber
of SV40genomespresentpercell(column
7)
as well as the percentage of total viral DNA
recovered
ineach of the Hirtfractions
(columns
8
and 9). The table
also shows the amount ofSV40 DNA per-milligram of cellular DNA
pres-entinthe
Hirt fractions
(columns
4and5)
andthe changes
inthe
contentof SV40
DNA in the Hirtsupernatantfractions compared with
Hirtpellet
fractions(enrichment ratio,
column6).
The conditions of mitomycin C treatment
used
(0.5,ug/ml,
24 h ofexposure) were thosethat were previously shown to induce
maxi-mum virus yields (6). In all the experiments,
treatment with mitomycin
C resulted
in anenrichment
of the Hirt supernatant fractionwith SV40 DNA sequences (column 6).
[image:4.505.67.468.275.490.2]Treat-ment with mitomycin C alone (experiment 1)
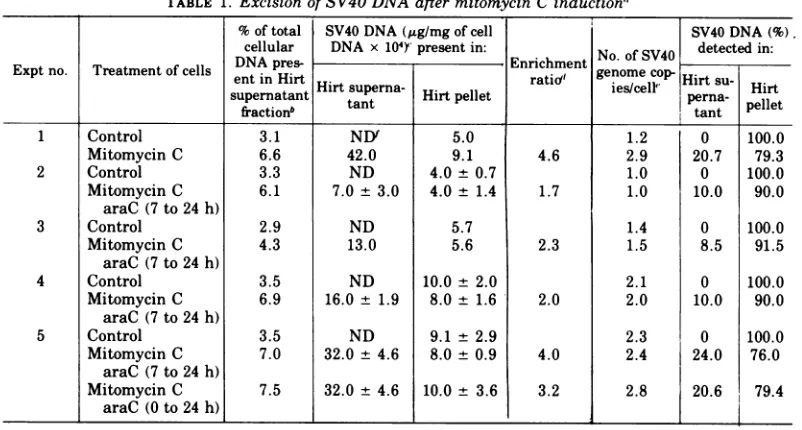
TABLE 1. Excision ofSV40DNAafter mitomycin C induction"
%of total SV40 DNA
(jg/mg
of cell SV40 DNA(%).
cellular DNA x104)'
present in: No. ofSV40
detected in: Expt no. Treatment of cells entin Hirt Enrchment genome cop Hirtsu-Hirtsuperna-
Hirt..
ies/cellF Hirtsupernatant tant H pellet perna- pellet
fraction"
~~~~~~~~~~~~tant
1 Control 3.1 NDf 5.0 1.2 0 100.0
Mitomycin C 6.6 42.0 9.1 4.6 2.9 20.7 79.3
2 Control 3.3 ND 4.0 + 0.7 1.0 0 100.0
Mitomycin C 6.1 7.0 + 3.0 4.0 ± 1.4 1.7 1.0 10.0 90.0
araC (7 to24h)
3 Control 2.9 ND 5.7 1.4 0 100.0
Mitomycin C 4.3 13.0 5.6 2.3 1.5 8.5 91.5
araC (7to24h)
4 Control 3.5 ND 10.0 ± 2.0 2.1 0 100.0
Mitomycin C 6.9 16.0 + 1.9 8.0 ± 1.6 2.0 2.0 10.0 90.0
araC (7to24h)
5 Control 3.5 ND 9.1 ± 2.9 2.3 0 100.0
Mitomycin C 7.0 32.0 ± 4.6 8.0 ± 0.9 4.0 2.4 24.0 76.0 araC (7to 24h)
Mitomycin C 7.5 32.0 ± 4.6 10.0 ± 3.6 3.2 2.8 20.6 79.4
araC (0to24h)
l_I
I
I_I
"The
cells were maintained and treated as described in the legends to Fig. 1 and 3, respectively. In experiments2to5, araC (10,8g/ml)wasaddedtothe mitomycin C-treated cells at the indicated times after initiation ofinduction.bPartitioning of cellular DNA between Hirt supernatant andpellet fractions was determined by labeling
one to twoplates in eachexperimentalgroupwith[3Hlthymidine(0.5to1.0
,uCi/ml;
specificactivity, 20Ci/ mmol;NewEngland Nuclear Corp.) for24hprior to the induction treatment.After24h, the labeled medium wasremoved, and the cellswereinducedasdescribed above. At the end ofincubationwithmitomycin C, the cells were harvested and fractionated by the Hirt procedure, and the trichloroacetic acid-precipitable radioactivity in the Hirtsupernatantand pelletfractions was determined."The numbers shown represent the meanfromtwo tofourindependentdeterminations of SV40 DNA
content performed at different times with 32P-labeled DNA preparations of different specific activities. Wherethreetofour determinations wereperformed, standard deviation from the mean is shown.
dEnrichment ratio is thecontentof SV40 DNA in the Hirt supernatant DNA (micrograms permilligram)
dividedby the content ofSV40DNA in the HirtpelletDNA (micrograms permilligram), i.e.,valuesin
column4dividedbyvaluesincolumn5.
' Values shown as number of SV40 genomes per cell represent corrected values for the actual DNA
contentofclone E cells (6).
fND, None detected. No SV40 DNAwasdetected. This means that the content ofSV40 DNAintheHirt
supernatantfraction of the control cellswasless than itscontent inthe Hirtpelletfraction of control cells.
J. V
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.505.72.463.277.489.2]NOTES 283
led
to anincreaseinthetotal number of
SV40genome copies per
cell,
which indicates thatminimal replication of SV40
DNAoccursinthe
first 24 h oftreatment. To
inhibit viral
DNAreplication, cytosine
arabinoside
(araC, 10 ,ug/ml)was
included
inthe culturemedium
inall
subsequent
experiments.This
dose of araCwasfoundtocause>99% inhibition
of
incorporation
of
[3H]thymidine
into the DNAof clone E cellseither
exposed
orunexposed
tomitomycin
C. InthepresenceofaraC, noincrease intotal SV40
DNAcontentpercell was
observed.
Inexperi-ments 2 to 4, araC was added to the
induced
cells
7 hafter
theaddition
ofmitomycin
C.Experiment 5 shows that thesame
results
wereachieved whether araC was included in the
medium from
thebeginning of
mitomycin C treatment or added 7 h later. Movement ofSV40 DNA from the Hirtpellettothe
superna-tantfraction
could
notbedetected
inthepres-enceofaraC alone.
Since treatment with
mitomycin
Crequires
prolonged
exposure times(i.e.,
24h), theexci-sion event could not be
pinpointed
in time.Therefore, -y-irradiation,
whichcausesimmedi-ateanddirect
damage
toDNA,wasutilized inthe next series of
experiments.
Results of arepresentative experiment (no.1inTable2)are
shown in Fig. 4. Whereas addition of DNA
extracted from the Hirt supernatant fraction of
unirradiated cells did
notaccelerate
therateof
reassociationof
32P-labeled
SV40 DNA,indicat-ing that
this fraction did
notcontaindetectable
amounts
of
viral DNA,addition
of DNAex-tracted from the Hirt supernatant fraction of
irradiated
cells increased therate of reaction.When theamount ofSV40 DNApresentinthe
Hirtsupernatant
and
pellet fractionswascal-culated,
anenrichment
ofsupernatantDNAforviral
DNA sequences wasobserved
in theab-sence
of
anincrease inthetotal
number of SV40
viral genomes percell.
Table
2 presents results of several experi-ments in which we examined excisionsubse-quent to
y-irradiation of clone
Ecells.
Insepa-rate experiments, the cells were incubated for
different lengths of time after irradiation, both in the presence and in the absence of araC.
Movement
ofviral
DNA from the Hirtpellettothe
supernatant fraction ininduced cells
wasobserved 2.5 h after induction.
Preliminary
evi-denceindicates thatnomovement
of viral
DNAcouldbedetectedasearlyas30 min
postirradia-tion.
Upto10hpostirradiation, therewaslittledifferencebetween cells thatwere or were not
incubated
inthepresenceof araC
afterirradia-tion.By 10 h,anincreaseinthe number of viral
genomes present per
cell
wasobserved
incul-tureslacking
araC,
indicating thatviral
DNAreplication
occurred
in induced cells. By 16 h,TABLE 2. Excision of SV40 DNA aftery-irradiation"
%of total cel- SV40DNA ( g/mgof cell
SV40
DNA(%) lular DNA DNA x10Y'
present in: No.of SV40 detected in:Expt T o c Enrichment
no. reatment
OraCelS
presentin tio genome pHirtHirtsuperna- Hirt
superna-
Hirt
pellet ies/cell' pena Hirttant
fractionb
tant
perna- pellet1 Control 2.1 NDf 10.0 ± 2.4 2.3 0 100.0
2.5hpostirradiation 4.5 24.0 + 5.3 8.8 ± 0.8 2.7 2.2 9.0 91.0
2 Control 2.0 ND 9.0 2.1 0 100.0
4hpostirradiation 2.8 28.0 6.0 4.6 1.8 16.6 83.4
4 hr postirradiation 3.8 49.0 5.5 8.8 1.9 26.3 73.7
araC (Oto 4h postirradiation)
3 Control 5.1 ND 10.4 ± 0.9 2.6 0 100.0
10 h postirradiation 9.5 160.0 ± 18.8 7.2 ± 0.6 22.0 5.8 70.0 30.0
10 h postirradiation 11.2 35.0 ± 7.9 7.8 + 0.5 4.5 2.5 30.0 70.0 araC (Oto 10h
postirradiation)
4 Control 1.7 ND 7.2 1.8 0 100.0
16 h postirradiation 5.6 40.0 19.0 2.1 4.8 9.5 90.5
16 h postirradiation 3.0 40.0 8.0 5.0 2.2 13.6 86.4
araC (0-16 h postirradiation)
aThe cells were treated asdescribed in the legend to Fig. 4. Whereindicated, araC in a final
concentra-tion of 10 ,ug/ml was added to the cultures immediately after irradiation. The cells were harvested at different times postirradiation and fractionated by the method of Hirt. The amount ofSV40 DNA was determined as described in thelegends to Fig. 2 and 4.
b.e.d.ef See corresponding footnotes to Table 1. VOL.
19,
1976on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.505.54.450.390.611.2]NOTES
00
.S1~~~~~~~~~'
FIG. 4. Reassociation kinetics
of
32P-labeled SV40 DNA incubated in thepresenceof
unlabeled DNAfrom
Hirtfractions of Sy-irradiated
and control cells. Clone E cells were maintained as described in thelegend
toFig.
1.Whensubconfluent,the cultureswereexposed
toirradiation(25,000rads;dose rate,10,900n2ds/min; 6OCo source) and were harvested at2.5 h
postirradiation.
Control cells were not irradiated.Harvesting of
thecells,partitioningof
the host cell DNA between Hirtsupernatantandpellet
fractions, extraction of DNA,C(,j analysis,
and calculationswere
performed
asdescribedinthelegends
toFig.2and 3. 32P-labeled SV40 DNA(3 x10-5OD260unitsl ml) was incubated with the
following
unlabeled DNAsamples:
O,30OD2+;,0
unitsof
salmon spermDNAperml; *,10
OD161,
units ofDNAfromHirt supernatantofcontrolcellsperml;A~,
10OD260,units
ofDNAfromHirtsupernatantofirradiated cellsper
ml; El, 29
OD26.
units ofDNA from Hirtpellet
of irradiated cells perml; *, 28OD261)
units ofDNAfr"omHirt
pellet of
control cellsperml.much of theviral DNA in cultures
lacking
araCwas
present
inthe Hirtpellet
fraction. Whetherthis DNA is associated with the
high-molecu-lar-weight
host DNA and the nature of anysuch associationarenotknownat
present,
but-nvestigations
arebeing
carried out toclarify
the issue.
Thus,
underourexperimental
condi-tions,
excisionoccurredearly
afterirradiation,
butviral DNA
replication
wasnotdetectedun-til
approximately
10hpostirradiation.
We have demonstrated
that,
early
afterthe induction of
virogenic
SV40-transformed
hamster
cells,
aportion
of the viral DNAmoves from its association with
high-molecular-weight
cellular
DNA to apool of
relatively
low-molecular-weight DNA. We
proposethat the
observed enrichment of
Hirt supernatantfrac-tions
for viral DNA
ininduced cells
isdue
toanexcision
of
viral DNA from
itsintegrated
state.It
isunlikely that this effect
issimply due
toreplication of
SV40 DNA
since it occurs tothe
same extent
in the
presenceand absence
ofaraC
atearly
timeperiods postirradiation. In
addition,
at the timeof
excision, no increaseoccurred
inthetotal number of SV40
genomecopies per
cell.
It is
difficult
atthe present time tocalculate
the
exactproportionof the cells
inthe
popula-tion in
which
excision occurs. Inexperiments
described
herein,approximately
10 to 30%of
the
total viral DNA moved from the
Hirtpellet
to
the
Hirt supernatantfractions
uponinduc-tion. We suspect that excision occurs in a
greater proportion
of
cells than those which
synthesize
infectious
virus(2
to6%)
by
infec-tious center assay
(6).
The
excision event that wehave
described
inSV40-transformed cells
mayalso
occurafter
induction of polyoma virus-transformed cells
since
the
polyoma viral DNA
isintegrated
intohost
cell
DNA (7). Itwould be of
interest todetermine whether induction of RNA
virusesfrom either
untransformed
orRNA
virus-trans-formed cells leads
to excisionof
proviral
DNA.
This investigation was supported by Public Health Serv-ice grant CA-10126-07 from theNational Cancer Institute. Joan C. Kaplan is the recipient of a Cancer Research Scholar Award from theAmerican CancerSociety (Massa-chusetts division).
Wewould like to thank LawrenceF. Kleinman for his excellent technical assistance and John B. Little for provid-ingradiation facilities for these experiments.
LITERATURE CITED
1. Black, P. H. 1968. The oncogenic DNA viruses: a re-view of in vitrotransformation studies. Annu. Rev. Microbiol. 22:391-426.
2. Gelb, L. D., D. E. Kohne, and M. A. Martin. 1971. Quantitationof simian virus 40 sequences in African greenmonkey, mouseandvirus-transformed cell ge-nomes. J. Mol. Biol. 57:129-145.
3. Hirt, B. 1967. Selective extraction of polyoma DNA
from infected mouse cell cultures. J. Mol. Biol. 26:365-369.
4. Kaplan, J. C.,P. H. Black, andH. Rothschild. 1971.
Induction of virogenic mammalian cells with
chemi-cal andphysical agents, p. 143-158. In R. W. Begg (ed.), Proceedings, Canadian Cancer Conference 1971, vol. IX.University of TorontoPress, Toronto.
5. Kaplan, J. C.,L. F.Kleinman, andP. H.Black.1975.
Cell cycledependenceofSV40virusinductionfrom
transformed hamster cellsby UV-irradiation. Virol-ogy68:215-220.
6. Kaplan, J. C., S. M. Wilbert, J. J. Collins, T. Rakusa-nova,G. B. Zamansky, andP. H.Black.1975.
Isola-tion ofSV40 transformed inbred hamster cell lines J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.505.92.232.60.320.2]VOL. 19, 1976 NOTES 285 heterogeneous for virus induction by chemicals or
radiation. Virology68:200-214.
7. Manor, H.,M.Fogel, andL.Sachs.1973.Integrationof viral intochromosomal deoxyribonucleic acid inan
inducible line ofpolyoma-transformedcells.Virology 53:174-185.
8. Marmur, J. 1961. Aprocedure for the isolation of deoxy-ribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218.
9. Ozanne, B., P. A.Sharp,and J. Sambrook. 1973. Tran-scription of simian virus40.II.Hybridizationof RNA
extracted from different lines of transformed cells to the separated strands of simian virus 40 DNA. J. Virol. 12:90-98.
10. Sambrook, J., H. Westphal, P. R. Srinivasan, and R. Dulbecco. 1968. The integrated state ofSV40 DNA in transformed cells. Proc. Natl. Acad. Sci. U.S.A. 60:1288-1295.
11. Signer, E. R. 1968. Lysogeny: the integration problem. Annu. Rev. Microbiol. 22:451-488.
12. Wetmur, J. G., and N. Davidson. 1968. Kinetics of renaturation of DNA. J. Mol. Biol. 31:349-370.