0022-538X/82/110637-10$02.00/0
CopyrightC1982,American Society for Microbiology
Complete Nucleotide Sequences of Two Adjacent Early
Vaccinia
Virus
Genes
Located Within the Inverted Terminal
Repetition
SUNDARARAJANVENKATESAN,t ALANGERSHOWITZ, ANDBERNARDMOSS*
LaboratoryofBiology ofViruses,National InstituteofAllergyandInfectiousDiseases, National Institutes of
Health,Bethesda,
Maryland
20205Received28 May1982/Accepted9August 1982
Theproximal part ofthe 10,000-base pair (bp)inverted terminal repetition of vaccinia virus DNAencodesatleast threeearly mRNAs. A2,236-bpsegmentof the repetition was sequenced to characterize two of the genes. This task was
facilitated by constructing a series of recombinants containing overlapping deletions; oligonucleotidelinkers withsynthetic restriction sitesprovidedpoints forradioactive labelingbeforesequencing bythe chemicaldegradation method of Maxam and Gilbert (Methods Enzymol. 65:499-560, 1980). The ends of the transcripts were mapped by hybridizing labeled DNA fragments to early viral RNA and resolving nuclease Si-protected fragments in sequencing gels, by sequencing cDNA clones, and from the lengths of the RNAs. The nucleotide
sequencesforatleast 60 bpupstream ofbothtranscriptional initiation sites are more than 80% adenine * thymine rich and contain long runs of adenines and thymines with some homology to procaryotic and eucaryotic consensus se-quences. The gene transcribed in the rightward direction encodes an RNA of approximately 530 nucleotides with a single openreading frame of 420 nucleo-tides.PrecedingthefirstAUG,there isaheptanucleotidethatcanhybridizetothe 3' endof 18S rRNA with onlyonemismatch.Thederived amino acidsequenceof the proteinindicated amolecular weight of 15,500. The genetranscribed in the leftward direction encodesanRNA1,000to1,100nucleotides long withanopen
reading frame of 996nucleotidesandaleadersequenceofonly 5to6nucleotides. Thederivedamino acid sequenceof thisproteinindicated amolecularweightof 38,500. The 3' ends of the two transcripts were located within 100 bp of each other. Although there are adenine * thymine-rich clusters near the putative transcriptional termination sites, specific AATAAA polyadenylic acid signal sequences areabsent.
Poxviruses are large DNA viruses that
repli-catewithin thecytoplasm of infected cells. This extraordinary feat is accomplished partly by incorporating a complete viral transcriptional
system within the infectious particle (19, 23). Although apparentlynotspliced(12, 43, 45), the
mRNAs generated have typical eucaryotic
fea-tures including 5' terminal cap structures (39)
and 3' polyadenylic acid [poly(A)]tails (18, 24) enablingthem to usethe hosttranslational appa-ratus.Studies with vaccinia virus,the prototype
for thisgroup,revealed thathalfofthe genome
is transcribed early in infection (8, 14, 20, 26).
Since 100 or more early genes are scattered throughout thelength ofthe DNA (3, 10), they musthavecommonstructuralfeatures leadingto
tPresentaddress: Laboratoryof InfectiousDiseases,
Na-tional Institute of Allergy and Infectious Diseases, NaNa-tional Institutes ofHealth, Bethesda, MD 20205.
their selection by the viral multisubunit RNA polymerase (1,25, 32)orassociated factors. To identify such structural elements, we have
un-dertaken the sequencingof several earlygenes. Only recentlyhave technical advances made it possibletocharacterizeindividual vaccinia virus transcripts. Thus far, about a dozen early mRNAs mapping within and proximal to the invertedterminalrepetitionhave beenexamined (12, 43, 45). Theinverted terminal repetition of vaccinia virus is about 10,000 base pairs (bp) long (13, 42) and containsa novelflip-flop loop
at its distal end (2), followed by two sets of tandem 70-bp repeats (44) and then coding
re-gions for polypeptides of approximately 7,500 daltons(7.5K), 19K, and42K. The correspond-ing mRNAs are about 1,000, 600, and 1,050 nucleotides long, respectively, and are synthe-sized in vitrobydetergent-treatedvirusparticles (36)aswellasinvivo underconditions in which
637
on November 10, 2019 by guest
http://jvi.asm.org/
viral proteinorDNAsynthesis is prevented (11,
43).Furthermore,all three transcripts have mul-tiple 5' ends asjudged by the isolation ofcap structures containing adenine (A) and guanine (G)asthe penultimate nucleotide (36); the
reten-tionof the
P-phosphate
of GTP inmaturemRNA is evidence againstnucleolytic processing of 5' ends (36).Limited nucleotide sequence analysis of the 7.5K polypeptide gene revealed an extremely adenine * thymine (A * T)-rich cluster of nucleo-tides immediatleyupstreamof thecapsite anda
tandemly repeated hexanucleotide near the 3' end (35). In the present communication, we
provide the sequence fora2,236-bp segmentof theinverted terminal repetitionthatincludes the entire 19K and 42Kpolypeptidegenes aswellas
locations of putative transcriptional initiation andtermination sites.
MATERIALS AND METHODS
Construction of recombinantplasmids. Recombinant pVG3, a pBR322derivativecontaininga3.4-kilobase pair (kbp)SaIl-EcoRIfragmentfrom theleft end of the vaccinia virus genome,wasderived fromapreviously described recombinant, pAG1 (36). A setof in vitro deletionmutants wasconstructedbycleaving pVG3at
theuniqueKpnIsite (seeFig.1). Afterphenol
extrac-tion and ethanol precipitation, 10 ,ug of linearized plasmid was digested with 1.25 U of Bal 31 (15). Samples were removedat 1, 2,and 3 min, atwhich time thereactions werestopped byheatingat65°Cin the presenceof 0.25% sodiumdodecyl sulfate-0.012M ethyleneglycol-bis(3-aminoethyl ether)-N,N-tetraace-tic acid(EGTA)-0.02M EDTA. After phenol-chloro-formextractionand ethanol precipitation, theextent
ofexonucleolytic cleavagewasmonitoredbyagarose gelelectrophoresis. Frayedends remainingafter this stepwerefilled in withreversetranscriptase (2.5Uper FgofDNA) and 0.25 mM deoxynucleoside triphos-phate(dNTP)(each type)at37°Cfor 30 min. HindIII oligonucleotide linkers were labeled attheir 5' ends with[,y-32P]ATPbyanexchange reactioncatalyzed by polynucleotidekinase(5).Linkers,at amolar ratioof 100:1,wereligatedtotheplasmidDNAat4°Cfor4h, and theligatedmixwasrecoveredbyethanol precipi-tation after phenol extraction. The DNA was then digested with HindIII, phenol-chloroform extracted, and renderedfreeof linkersby gelfiltrationthrougha
Sepharose 4B column. The plasmid DNA was then incubatedat aconcentration of 10,ug/mlwith 7.5 Uof T4 DNAligaseat4°Cfor4h.Approximately0.2 ,ug of DNA was used to transform competent Escherichia coli K-12 HB101cells, andcolonieswere selectedby growthonampicillin plates (7). The number of
trans-formants obtained varied from5,000 for limited Bal31
digestions to 500forextensive digestions.
Recombi-nantplasmidswerepurifiedbyasmall-scale isolation procedure (6) and screenedbyrestriction
endonucle-asedigestions and agarose gelelectrophoresis.Atleast 75% of the tested recombinants hadaHindIIIsite,and themajorityof them had deletions that varied from100 to1,300bp. Fiverecombinants withoverlapping dele-tions(Dl,D3, D4, D5, andD6)wereselected forDNA
sequencing.
End labelingDNA.Restriction fragments were treat-ed with alkaline phosphatase and labeltreat-ed at the 5' end with [y-32P]ATP and T4 polynucleotide kinase (22). Avian myeloblastosis reverse transcriptase or the Klenowfragment of DNA polymerase was used to add
asingle complementary [a-32P]dNTPtothe recessed 3'end of restriction fragments (47). When the 3' end was protruding, labeling was accomplished with
[a-32P]cordycepin triphosphate and terminal deoxynu-cleotidyltransferase (34).
DNA sequence analysis.End-labeled DNAfragments werecleaved with appropriate restriction endonucle-ases, purified by agarose gel electrophoresis, and recoveredwith glass powder (37) or by electrophoresis
ontoDEAE paper (41).The limitedchemical degrada-tion procedureof Maxam and Gilbert (22) was used for sequencing. Reaction products were resolved on 0.4-mm-thick gels with either 4, 6, 8, or20%o polyacryl-amide (30 by 160 or 40 by 80 cm) in 8 M urea (31).
cDNA sequencing. Alibrary of cDNA recombinants was generously provided by B. Roberts (Harvard UniversitySchool ofMedicine).cDNA wasprepared using early RNA from cells infected with vaccinia virus (strain WR). Oligodeoxythymidylic acid [oli-go(dT)] served as a primer; hairpin 5' ends were removed with nuclease S1, and the double-stranded cDNAs were cloned in pBR322 by deoxycytidylate and deoxyguanylate tailing (B. Roberts, personal com-munication). We screened thelibrary by colony hy-bridization (33) witha3-kbpBamHI-EcoRlfragment (seeFig. 1) labeled with32Pby nick translation (29). A total of 35 colonies were selected for further restriction endonuclease analyses, and 8 were sequenced after labeling the 3' ends with [a-32P]cordycepin triphos-phate(34).
Mapping 5' and 3' ends by nuclease Si protection. poly(A)-containing RNA was purifiedfrom the cyto-plasmof cells 4 h after infection withvacciniavirus (15 PFU percell)in the presence of cycloheximide or was made in vitro by vaccinia virus particles (36). The RNAwashybridized to DNAfragments containing a labelatthe 5'or3' end on thecoding strand (35). After nuclease S1 digestion(35),hybrids were analyzed by electrophoresisonsequencing polyacrylamide gels.
Materials. Restriction endonucleases were pur-chased from NewEnglandBiolabs (Beverly, Mass.)or
BethesdaResearch Laboratories (Gaithersburg, Md.). Nucleases Bal 31 and S1 were supplied by New England Biolabs and Miles Laboratories, Inc., Elk-hart, Ind.,respectively. Bothterminal deoxynucleoti-dyltransferase and T4 polynucleotide kinase came from P-L Biochemicals, Milwaukee, Wis., whereas DNApolymerase holoenzyme and the Klenow
frag-ment camefrom BoehringerMannheim Corp. India-napolis, Ind. T4 DNA ligase was purchased from Bethesda Research Laboratories, and avian myelo-blastosis virus reversetranscriptase wassupplied by J. W. Beard of Life Sciences, Coral Gables, Fla. Oligodeoxynucleotideswereobtained from Collabora-tive Research, Inc., Waltham, Mass., [a-32P]dNTP
and[y-32P]ATPfrom AmershamSearle,Chicago, Ill., and [a-32P]cordycepin triphosphate from New En-glandNuclearCorp., Boston, Mass.
RESULTS
Sequencing strategy. Three mRNAsencoding polypeptides ofapproximately 7.5K, 19K, and
on November 10, 2019 by guest
http://jvi.asm.org/
VACCINIA VIRUS GENES
42K were mapped within the inverted terminal repetitionof the vaccinia virus genome(11, 42, 43) (Fig. 1). A strategy, avoiding either
exten-sive restriction endonuclease mapping or shot-gun methods, was used to determine the geno-mic sequence of the 19K and42Kpolypeptides. This deletion linkerapproach requiredthe clon-ing in pBR322 ofthe 3.4-kbp SalI-EcoRI frag-mentlocated 5.6to9.0kbpfrom the end of the viral DNA(Fig. 1). Therecombinantis referred to aspVG3. AuniqueKpnI site, located within the viral DNA segment, was cleaved, and the endsofthelinearizedplasmidweresubjectedto
exonucleasedigestionwith Bal 31(15).After the addition of synthetic oligonucleotide linkers containing a HindIII site, the plasmids were
recircularized and used to transform E. coli. Recombinantplasmidswerescreened, anda set
withoverlappingdeletions was selected(Fig.1).
A
TR
Sequencing of each recombinantwasfacilitated by use of the mobile HindlIl site for 5' and 3' labeling. Additional sequencing was carriedout
using therestriction sites andconventional strat-egyindicated at the bottom ofFig. 1.
A sequence of2,236 nucleotides encompass-ing genes for the 19K and 42K polypeptides is shown inFig. 2. As will be discussedlater,both genes have long open translational reading frames ofoppositepolarity.
Location of the 5' ends of transcripts. The mRNAs for the 19K and 42K polypeptides are transcribed fromopposite strands of the invert-edterminal repetition(Fig. 1). Approximate map positions of these mRNAs were determined by use of restriction fragments for hybridization selection and cell-free translation and for prob-ing blots of electrophoretically separated RNAs. To more precisely locatetheir 5' ends, a
modifi-CONSTRUCTION OF RECOMBINANT
vtASMIDS
TR
7.5K 19K 42K
,----go
1 2 3 4 5 6 7 8 91
Dl D4 D3 D5 D6
6.5 7.0 7.5 8.0 8.5 9.0
SEQUENCING STRATEGY
B 19K 42K
6.5 7.0 7.5 8.0 8.5 9.0
1T1~
t
it
T
t
T
Y
T
tt
ao
-_~
9 - m-Dl
D6 Dl - o- -D4
- 0 o D4 v
D5D3 ° _ a _D3
5 co
FIG. 1. Construction of recombinants and sequencing strategy. (A) Part of the 10-kbp inverted terminal repetitionof vaccinia virus is shown. The dark blocks labeled TR represent the two sets of70-bptandemrepeats. Arrows represent theapproximate lengths, map positions, and directions of transcriptionof three early mRNAs encoding polypeptides of approximately 7.5K, 19K, and 42K. TheBamHI-EcoRI segment of therecombinant plasmid pVG3 is expanded. Additional recombinants were constructed from pVG3 using Bal 31 to make deletionsattheKpnIsite. The extent of the deletions in recombinantsDl,D4,D3, D5, and D6 is shownby gaps in theheavy bar. (B) The BamHI-EcoRI segment of the inverted terminal repetition with the mRNAs for the 19K and 42Kpolypeptides is shown. Symbols for relevant restriction endonuclease sitesare:AvaIl,9;BamHI, l; EcoRI,4;HincII,6;Hinfl, I; HpaII,4; KpnI,*;TaqI,T;XbaI, .Filled and unfilled circles represent sites of 3' and 5'labeling, respectively. Solid arrows indicate the extent of sequence determination; interrupted parts of thearrowindicateregions where the sequence was not determined. ThedesignationsDltoD6refertotheuseof deletionrecombinants; otherwise, pVG3 or a related recombinant pVG1 wasused.
VOL.44,1982
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.491.83.414.292.574.2]J.VIROL.
10 20 30 40 50 60 70 80 90 100
TTTTTAACAG CAAACACATT CAATATTOTA TTGTTATTTT TATGTATTAT TTACACAATT AACAATATAT TATTAGTTTA TATTACTGAA TTAATAATAT AAAAATTGTC GTTTGTGTAA GTTATAACAT AACAATAAAA ATACATAATA AATGTGTTAA TTGTTATATA ATAATCAAAT ATAATGACTT AATTATTATA
'S 120 130 140 150 160 ...L 170 180 1 90 20 0
AAAATTCCCA ATCTTGTCAT AAACACACAC TGAGAAACAG CATAAACACA AAATCCATCA AAAATGTCGA TGAAATATCT GATGTTGTTG TTCGCTGCTA
TTTTAAGGGT TAGAACAGTA TTTGTGTGTG ACTCTTTGTC GTATTTGTGT TTTAGGTAGT TTTTACAGCT ACTTTATAGA CTACAACAAC AAGCGACGAT
210 220 250 240 250 260 270 280 290 300
TGATAATCAG ATCATTCGCC GATAGTGGTA ACGCTATCGA AACGACATCG CCAGAAATTA CAAACGCTAC AACAGATATT CCAGCTATCA GATTATGCGG ACTATTAGTC TAGTAAGCGG CTATCACCAT TGCGATAGCT TTGCTGTAGC GGTCTTTAAT GTTTGCGATG TTGTCTATAA GGTCGATAGT CTAATACGCC
510 320 330 340 350 360 370 380 390 400
TCCAGAGGGA GATGGATATT GT.TTACACGG TGACTGTATC CACGCTAGAG ATATTGACOG TATGTATTGT AGATGCTCTC ATGGTTATAC AGGCATTAGA AGGTCTCCCT CTACCTATAA CAAATGTGCC ACTGACATAG GTGCGATCTC TATAACTGCC ATACATAACA TCTACGAGAG TACCAATATG TCCGTAATCT
410 420 430 440 450 460 470 480 490 '500
TGTCAGCATG TAGTATTAGT AGACTATCAA CGTTCAGAAA ACCCAAACAC TACAACGTCA TATATCCCAT CTCCCGGTAT TATGCTTGTA TTAGTAGGCA
ACAGTCGTAC ATCATAATCA TCTGATAGTT GCAAGTCTTT TGGGTTTGTG ATGTTGCAGT ATATAGGGTA GAGGGCCATA ATACGAACAT AATCATCCGT
510 520 530 540 550 560 570 580 TL 590 600
TTATTATTAT TACGTGTTGT CTATTATCTO TTTATAGOTT CACTCOACGA ACTAAACTAC CTATACAAGA TATGGTTGTG CCATAATTTT TATAAATTTT AATAATAATA ATGCACAACA GATAATAGAC AAATATCCAA GTGAGCTGCT TGATTTGATG GATATGTTCT ATACCAACAC GGTATTAAAA ATATTTAAAA
610 620 630 13 650 660 6'70 680 690 700
TTTATGAGTA TTTTTACAAA AAAAATGTAT AAAGTOTATO TCTTATGTAT ATTTATAAAA ATGCTAAGTA TGCGATGTAT CTATGTTATT TGTATTTATC AAATACTCAT AAAAATGTTT TTTTTACATA TTTCACATAC AGAATACATA TAAATATTTT TACGATTCAT ACGCTACATA GATACAATAA ACATAAATAG
710 720 730 740 T1. 750 760 770 780 790 800
TAAACAATAC CTCTACCTCT AGATATTATA CAAAAATTTT TTATTTCGGC ATATTAAAGT AAAATCTAGT TACCTTGAAA ATGAATACAG TGGGTGGTTC
ATTTGTTATG GAGATGGAGA TCTATAATAT GTTTTTAAAA AATAAAGCCG TATAATTTCA TTTTAGATCA ATGGAACTTT TACTTATGTC ACCCACCAAG
810 820 830 840 850 860 870 880 890 900
CGTATCACCA GTAAGAACATAATAGTCGAA TACAGTATCC OATTGAGATT TTGCATACAA TACTAGTCTA OAAAGAAATT TGTAATCATC TTCTGTGACG GCATAGTGGT CATTCTTGTA TTATCAGCTT ATGTCATAGG CTAACTCTAA AACGTATGTT ATGATCAGAT CTTTCTTTAA ACATTAGTAG AAGACACTGC
910 920 930 940 950 960 970 980 990 1000
GGAGTCCATA TATCTGTATC ATCGTCTAGT TTATCAGTGT CCCATGCTAT ATTCCTGTTA TCATCATTAG TTAATGAAAA TAACTCTCGT GCTTCAGAAA
CCTCAGGTAT ATAGACATAG TAGCAGATCA AATAGTCACA GOGTACOATA TAAGGACAAT AGTAGTAATC AATTACTTTT ATTGAGAGCA CGAAGTCTTT
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
AGTCAAATAT TGTATCCATA CATACATCTC CAAAACTATC GCTTATACGT TTATCTTTAA CGATACCTAT ACCTAGATGG TTATTTACTA ACAGACATTT
TCAGTTTATA ACATAGGTAT GTATGTAGAG GTTTTGATAG CGAATATGCA AATAGAAATT GCTATGGATA TGGATCTACC AATAAATGAT TGTCTGTAAA
1110 1120 1130 1140 1150 1160 1170 1180 1190 1200
TCCAGATCTA TTGACTATAA CTCCTATAGT TTCCACATCA ACCAAGTAAT GATCATCTAT TGTTATATAA CAATAACATA ACTCTTTTCC ATTTTTATCA AGGTCTAGAT AACTGATATT GAGGATATCA AAGGTGTAGT TGGTTCATTA CTAGTAGATA ACAATATATT GTTATTGTAT TGAGAAAAGG TAAAAATAGT
1210 1220 1230 1240 1250 1260 1270 1280 1290 1300
GTATGTATAT CTATATCAAC OTCGTCGTTG TAGTGAATAG TAGTCATTOA TCTATTATAT GAAACGGATA TGTCTAGAAC GGCAATTGTT TTACGTCCAG CATACATATA GATATAGTTG CAGCAGCAAC ATCACTTATC ATCAGTAACT AGATAATATA CTTTGCCTAT ACAGATCTTG CCGTTAACAA AATGCAGGTC
1310 1320 1330 1340 1350 1360 1370 1380 1390 1400
TTAACACTTT CTTTGATTTA AAGTCTAGAG TCTTTGCAAA CATAATATCC TTATCCGACT TTATATTTCC TGTAGGGTGG TATAATTTTA TTTTGCCTCC
AATTGTGAAA GAAACTAAAT TTCAGATCTC AGAAACGTTT GTATTATAGG AATAGGCTGA AATATAAAGG ACATCCCACC ATATTAAAAT AAAACGGAGG
1410 1420 1430 1440 1450 1460 1470 1480 1490 0500
ACATATCGGT GTTTCCAAAT ATATTACTAG ACAATATTCC ATATAGTTAT TAGTTAAGGG TACCCAATTA GAACACGTAC GCTTATTATC ATCATTTGGA TGTATAGCCA CAAAGGTTTA TATAATGATC TGTTATAAGG TATATCAATA ATCAATTCCC ATGGGTTAAT CTTGTGCATG CGAATAATAG TAGTAAACCT
1510 1520 1530 1540 1550 1560 1570 1580 1590 1600
TCGTATTTCA TAAAAGTTAT TGTACTATCG ATGTCAACAC ATTCTACATT TTTTAATCGT CTATATAGTA TTTTTCTGAT ATTTTCTATA ATATCAGAAT
AGCATAAAGT ATTTTCAATA ACATGATAGC TACAGTTGTG TAAGATGTAA AAAATTAGCA GATATATCAT AAAAAGACTA TAAAAGATAT TATAGTCTTA
1610 1620 1630 1640 1650 1660 1670 1680 1690 1700
TGTCTTCCAT-CGGAAGTTGT ATACTATCGG AATCAGTTAC ATGTTTAAAT AATTCTCTGA TGTCATTCCT TATACAATCA AATTCATTAT TAAACAGTTT ACAGAAGGTA GCCTTCAACA TATGATAGCC TTAGTCAATG TACAAATTTA TTAAGAGACT ACAGTAAGGA ATATGTTAGT TTAAGTAATA ATTTGTCAAA
1710 1720 1730 -1 1750 1760 1770 1780 1790 1800
AATAGTCTGT AGACCTTTAT CGTCGTAAAT ATCCATTGTC TTATTAGTTA CGCTTATTTT TATGTGTTTT ACGTTGCTTT ATTATATTTT ATAAGAATGA
TTATCAGACA TCTGGAAATA GCAGCATTTA TAGGTAACAG AATAATCAAT GCGAATAAAA ATACACAAAA TGCAACGAAA TAATATAAAA TATTCTTACT
1810 1820 1830 1840 1850 1860 1870 1880 1890 1900
TTGTTTGACG AATCACGAGA ACTATTAAGACACATTATTA GGTATATATT ATAAAAAAGT TTTTGATTAC GATGTTATAA GAGGAAAGAG GACACATTAA AACAAACTGC TTAGTGCTCT TGATAATTCT GTGTAATAAT CCATATATAA TATTTTTTCA AAAACTAATG CTACAATATT CTCCTTTCTC CTGTGTAATT
1910 1920 1930 1940 1950 1960 1970 1980 1990 2000
CATCATACAT CAATTAACTA CATTCTTATA ACATCGTAAT CAAAAGAATT GCAATTTTGA TGTATAACAA CTGTCAATGG GTTATGAAAT TGTATATTAC
GTAGTATGTA GTTAATTGAT GTAAGAATAT TGTAGCATT4GTTTTCTTAA CGTTAAAACT ACATATTGTT GACAGTTACC CAATACTTTA ACATATAATG
2010 2020 2030 2040 2050 2060 2070 2080 2090 2100
ATATTATACG GTATGTTGGT AACGACAAAT ACCGATCGGT AATTGTCTGC CGGTGTACGA GAATTATATA TATCTATCTA TTACACCGGC TGAGTATGCA
TATAATATGC CATACAACCA TTGCTGTTTATGGCTAGCCA TTAACAGACG GCCACATGCT CTTAATATAT ATAGATAGAT AATGTGGCCG ACTCATACGT
2110 2120 2130 2140 2150 2160 2170 2180 2190 2200
TAATAATAAG TTGTGGTAGT ATGATCTCCA TATTTATAAT TTAGGACTTT GTATTCAGTA TTTTTGGAAT CATAAAAAAT AAAAAAAAGT TTTACTAATT ATTATTATTC AACACCATCA TACTAGAGGT ATAAATATTA AATCCTGAAA CATAAGTCAT AAAAACCTTA GTATTTTTTA TTTTTTTTCA AAATGATTAA
2210 2220 2230 TAAAATTTAA AAAGTATTTA CATTTTTTTC ACTGTT ATTTTAAATT TTTCATAAAT GTAAAAAAAG TGACAA
FIG. 2. Nucleotide sequence ofa2,236-bpsegment of DNA encompassingthegenesfor the 19K and 42K
polypeptides.Thenucleotide numbered 1 is approximately6,800nucleotides from the end of the genome, and nucleotide 2,236 is afew nucleotides before the first EcoRI site. The upper and lower lines represent the
noncodingstrandsfor mRNAsexpressingthe 19K and 42Kpolypeptides,respectively. Theputative
transcrip-tionalinitiation sites for thetwomRNAsareindicatedbyan arrowwith 5'overit. The 3'end(s)of the mRNA for the 19K polypeptide is shown by an arrow with 3' written over it. The 3' end of the mRNA for the 19K
polypeptideidentifiedbycDNAsequencingis shownby!.Iand Trefertothepresumptivetranslational initiation andterminationcodons,respectively, of the twomRNAs.
cationintroducedbyWeaver andWeissman(38) beled strand of this fragment was expected to of the nucleaseSi procedureof Berk andSharp containsequencescomplementary tothe 5' end
(4) was used. A HincII-AvaII fragment (6.6 to of the 19KpolypeptidemRNA. After hybridiza-7.1 kbpfrom the end of thegenome)with 5' 32P tion to early RNA, made in infected cellsor in label at the AvaII site was employed. The la- vitro by detergent-treated virus particles,
on November 10, 2019 by guest
http://jvi.asm.org/
maining single-strandedDNA wasdigested with
nuclease SI. The radioactively' labeled DNA segment that wasprotected from nuclease
diges-tion represented the distance from the RNA 5'
terminus to the 5' end of the DNAprobe. The
size ofthis segment was determined by
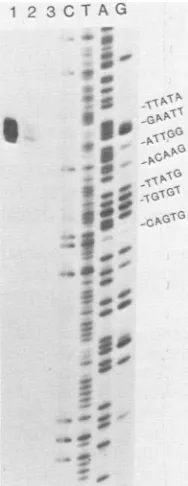
poly-acrylamide gelelectrophoresis under denaturing conditions. As shown in Fig. 3, at least six
closely spaced nuclease-resistant bands were resolved. By coelectrophoresis with the DNA sequence reaction products of the original
HinclI-Avall fragment, the nuclease-resistant bands werelinedup with a sequence ladder. As
pointed outpreviously (16), a 1.5-bp downward
displacementof the nuclease S1 bands is neces-sary for proper alignment. This placed the 5'
ends of the RNA complementary to the se-quence AGATT in Fig. 3. Since the
nuclease-protected bands were spaced one nucleotide
apart, the heterogeneity could result from a
1 2 3CTAG
:x0
A^ aw
Jar
S. ,
U?E,Fa
.
0.
_a
i-.p
ZS
_,.sv,P G>hv,t,l
-1G-GIM
[image:5.491.248.441.71.288.2]_rNG-T
FIG. 3. Map locations of the 5' end(s) of mRNA encoding the 19K polypeptide. Approximately 0.5 pmol of the AvaII-HincII fragment (7.0 to 6.64 kbp fromthe endof theDNA) 5' labeledatthe AvaIl site
wasdenatured and incubated under conditions
favor-ing RNA-DNA hybridization with 5 to 10 pmol of poly(A) selected early in vivo RNAorin vitroRNA. Afterhybridization, single-stranded DNAwas
digest-ed with nuclease S1 (750 U/ml). Resistant products after incubation with (1) in vivo RNA, (2) in vitro RNA, or(3)noRNAandsequencereaction products (C [cytosine]; Tis Tplus C; A is A plusG; G)were
resolved on an8%polyacrylamide gel (30by160cm) in8 Murea.
AJAA
;CA
rc04A
A
CTAG 1 2
.. _ w4
&-S
_ ,.2,d
@00
*a 1.
e~dP
4w
.Ae
S.
_p
a'#
sip
33 OTAGI 2
U, U'
..
'-r
!
S
"SE te
hA.v-'!
d- .1 -...!.ir Ut
A,%,cc-1'
FIG. 4. Map locations of the 5' ends of mRNA
encoding the 42K polypeptide. Approximately 0.5
pmol of(A) anXbaI-EcoRI fragment(8.1 to 9.0 kbp
from the end of the DNA) and (B) a TaqI-EcoRI fragment(8.3 to 9.0 kbp from the end of the DNA)5' labeledat theXbaIand TaqI sites, respectively, was
hybridizedtoapproximately 25 pmol of poly(A)
select-ed early cytoplasmic RNA. Nuclease Si-resistant products obtained with(1)in vivoRNA, (2) no RNA, or(3) in vitro RNA andsequence reactionproducts (C; Tis T plus C;AisAplus G; G) were resolved on an
8%polyacrylamide gel(30by160 cm)in 8 M urea.
tendency of nuclease Si to leave overhanging
ends or to nibble into a base-paired region.
Analysis ofthe capped 5' ends of this message
indicated that 80% ended in A and 20% in G, suggesting thatthe major endsare
complemen-tary to one orboth of the adjacent deoxythymi-dylate residues (positions110and111inFig. 2.). Themapposition of the 5' endwasconfirmed by usingas aprobe theHincII-HpaII fragment(6.6 to7.2 kbp from the end of the DNA) labeledat
the5' endatthe HpaII site(notshown). The 5' endsof themessageencodingthe42K
polypeptideweremappedin ananalogous man-ner. Two DNA fragments, XbaI-EcoRI (8.1 to
9.0 kbp from the end of the DNA) and
TaqI-EcoRI (8.3 to 9.0 kbp from the end of the DNA),
labeled attheXbaI and TaqI site, respectively,
were employed. The major nuclease-resistant bands lined up within a sequence TCTT(Fig.4).
Previous cap analysis indicated approximately equalamounts of G and A ends suggestingthat
they are complementary to the CT residues of the above sequence (numbered 1,740 and 1,741
in Fig. 2).
Location of the3'ends oftranscripts. Previous studies indicated that the mRNAs for the 19K
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.491.93.187.297.540.2]A B
G AC C;' A -T C
A.e
-~,
I~~~~~~~~I
4
FIG. 5. Map locations of the 3' ends of mRNA
encoding the 19K polypeptide by cDNA sequencing
and nuclease Si protection. (A)A recombinant
plas-mid (pVBR12) was selected from a vaccinia cDNA
library by its ability to hybridize specifically to a
vaccinia DNAfragment encodingmRNAfor the 19K
polypeptide. The recombinant DNA was cleaved at
Axv i t [ .2pt
the Pstl site and 3' labeled with
kx3Pcordycepin
triphosphate and terminal
deoxynucleotidyltransfer-ase. After cleavage with Hpall, the smaller of the labeled DNA fragments was purified, sequenced by
the chemical degradation method, and analyzed by electrophoresis on 8% polyacrylamide gel. The
se-quenceof thenoncoding (i.e., RNA)strandis indicat-ed. The 15 adenylate residues representthe proximal portionof thepoly(A)tail of the mRNA.(B) Approxi-mately0.5pmolofanHpall-Hinflfragment(7.2to7.6
kbpfrom the end of theDNA)labeledatthe 3' endwas
hybridizedto 10pmol ofpoly(A) selectedearly
cyto-plasmicRNA in80% formamideat380C.Nuclease
Si-resistant products obtained with (1) or without (2) RNAand sequencereactionproducts (G;Ais Aplus
G;Tis TplusC;C)wereresolvedon an8%
polyacryl-amide gel (30 by 160 cm) in 8 M urea. The coding
strand DNA sequence isindicated.
and 42Kpolypeptidesareapproximately580 and 1,050 nucleotides long, respectively (36, 43). Using the map locations determined above for the 5' ends, this wouldplace the3' ends of the oppositely oriented 19K and 42K polypeptide messagesclosetoeach othernearpositions690
and 740, respectively, of the genomic sequence in Fig. 2.
Attempts were made to map the 3' ends of
bothmRNAs moreprecisely.The first approach involved a procedure analogous to that used for
mapping5'termini.AHpaII-Hinfl fragment(7.2 to7.6 kbp from the end of the DNA) labeled at the 3' end at the HpaII site was used as a probe for the 19K polypeptide message. After hybrid-ization to early RNA, the nuclease-protected DNA segmentrepresented the distance fromthe 3' end of the RNA to the labeled restriction site.
The size of this segment was determined by
polyacrylamide gel electrophoresis, again using a sequence ladder prepared from the original hybridization probefor comparison. Two major and several minor bands were detected (Fig.
SB). After making the appropriate correction, they appeared to line up with the complemen-tary AA residues of the sequence GAAT. This
corresponds to the two thymine (T) residues
located at positions 643 and 644 of Fig. 2 and
places the 3'end within 50 bpof the site
predict-ed from the length ofthe RNA using northern
blotanalysis. This slightdeviation from its orig-inally reported size might reflect the length of
thepoly(A)tail.
Similarattempts to mapthe 3' end of the42K
polypeptide mRNA were unsatisfactory, possi-bly because of the low abundance of the mes-sage, the existence oftranscripts with overlap-ping 3' ends that might compete for
hybridization, andpossible heterogeneity ofthe 3' ends (36,43).
An alternative procedure was investigated to
confirm the mappositions of the 3' endsofthe
19K and 42K polypeptide mRNAs. For this
approach we needed recombinants containing cDNAs prepared using anoligo(dT) primer hy-bridized to the poly(A) tails of early vaccinia
virus mRNAs. By sequencing the recombinant cDNAs we hoped to identify the nucleotides
adjacenttotheoligo(dT) primeror
complemen-tary oligodeoxyadenylate [oligo(dA)]. Such a
cDNA recombinant library, generously provid-edbyB.Roberts(Harvard Medical School),was screened by colony hybridization. Selected re-combinants were further characterized by re-striction endonuclease analyses, andeightwere
sequenced fromtheir PstI sites. The sequenceof
one is shown in Fig. SA. After a cluster of
deoxycytidylate residues, derived from the
deoxyguanylate and deoxycytidylate tailing
usedforconstruction oftherecombinants,there were 15 deoxyadenylate (dA)residues andthen a sequence corresponding to the genome near
the 3' endofthe19Kpolypeptide. The deoxyth-ymidylateresidueadjacenttotheoligo(dA)isat
position 639 ofFig. 2, four nucleotides down-streamfromthe 3' end deducedby nuclease S1
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.491.80.223.79.365.2]SEQUENCES OF EARLY VACCINIA VIRUS GENES protection experiments. Further comparison of
the cDNAsequence with the genomicsequence
(position 678 to 638, Fig. 2) revealed twominor
discrepancies. There were seven dAs incDNA
in contrast toeightdAsin genomic DNA
(posi-tion 618, Fig. 2) and six dAs in cDNA
corre-spondingto seven dAs in the genome (position
603, Fig. 2). These differences might represent
slippage during reverse transcription or minor
variations inthe vaccinia virus WR isolatesused
for genome sequencing and cDNA cloning. Since these differencesoccurin the
nontranslat-edregion of thegene, theremightbe no
biologi-calconsequencesof such variations.
Unfortunately, the otherseven cDNA
recom-binants from thisregion of the genome as wellas mostfromasecondregion lackedtheoligo(dA)
sequence and therefore could not be used to
identify the precise 3'end ofthe message.
DISCUSSION
The majority ofDNAviruses engage cellular RNApolymerases forgene transcription within
the nucleus ofthe infected cell. Thus it is not
surprisingthattheirregulatory signalsresemble
those ofthe host (9). By contrast, poxviruses are
cytoplasmicandpossess aunique transcription-al systemmaking it likely that distinctive DNA sequencesinteract withthe viral RNA
polymer-ase. To elucidate structural features related to
the organization and expression ofthe vaccinia virus genome, several regions have been
se-quenced. These include the terminal loops that link thetwo DNAstrands together andadjacent
DNA (2) and portions of the most distal gene
coding for a 7.5Kpolypeptide (35). To these is
nowaddeda2,236-bpsegmentthat encodestwo
early polypeptides. Sequencingwasachievedby thechemical degradationmethodofMaxamand
Gilbert (22) using a strategy that involved the
preparation of a series of recombinants with
overlapping deletions and inserted restriction site linkers. This deletion linker approach
elimi-nated the need for extensive restriction site
mapping or shotgun sequencing.
A procedure of Weaver and Weissman (38)
was used to map the 5' ends of both mRNAs. After hybridization ofa DNAfragment labeled
atthe 5' endtoRNA, thenuclease S1-protected
segment and the nucleotide sequence reaction
productsof the originalfragmentwerecompared by electrophoresis on the same pol7yacrylamide gel. These data andourfinding ofm G(5')pppAm
and m7G(5')pppGm caps on both the 19K and
42Kpolypeptide mRNAs(36)placedtwomajor
5' ends at or near nucleotides 110 and 111 and
1,740 and1,741ofFig. 2, respectively.A
previ-ousanalysis of the5' ends of the 7.5K polypep-tide mRNA alsosuggestedthepresence of
multi-ple closely spacedpurine ends (35).
Since the cap structuresof both the 19Kand 42K mRNAs retain the ,B-phosphate of GTP
(36), the5'endsof their messagescorrespondto
sites oftranscriptional initiation. Accordingly, theadjacent DNA maycontainpromoter
recog-nition sequences. In Fig. 6, regionsupstreamof the 5' ends of the mRNAs coding for polypep-tides estimated to be 7.5K, 19K, and 42K are
shown. Foreach, the 60bppreceding the start
siteare atleast80%A * Trich with manyrunsof
A's and T's including some 14 to18nucleotides
long. Embeddedwithin the A * T-richregionare
possible equivalents of the Pribnow and Hog-ness-Goldberg boxes ofprocaryotesand eucary-otes,respectively (9, 30).Nevertheless,the sim-ilarity to the eucaryoticTATA sequence is not exact.Interestingly, thesequenceCGTAAAA is found starting at -28 and -29 ofthe 7.5K and
42Kpolypeptidegenes,respectively (Fig. 6). In
addition, each of the genes contains other ho-mologousA *T-rich clustersincludingCAAT40 to60 bpupstream ofthe capsites.
-100 -90 -80 -70 -60 -50 -40 -30 -20 -10 0
( !) AACTGATCACTAATTCCAAACC CAC C CGCTTTTTATAGTAAGTTTTTCACCCATAAATAATAAATACAATAATTAATTTCTCGTAAAMGTAGAAAATATATTCTAATTTA
... ... . ... ... . ... ... ...
(II) TTTTTAACAGCAAACACATTCAATATTGTATTGTTATTTTTATGTATTATTTACACAATTAACAATATATTATTAGTTTATATTACTGAATTAATAATATAAAATTCCCA
... ... ... ...
(111) TATAATATATACCTAATAATGTGTCTTAATAGTTCTCGTGATTCGTCAAACAATCATTCTTATAAAATATAATAAAGCAACGTAAAACACATAAAAATAAGCGTAACTAA
(I)~~~~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~~~... .. ... .. .
(IV)
(V) GGCCAATCTT
TTGACA TATAATG
TATAAAAT T
FIG. 6. Sequences upstream of three early transcriptional sites of vaccinia virus. I, II, and III refer
respectively to the upstream sequences of mRNAs encoding 7.5K, 19K, and 42K polypeptides. The most
proximal initiation site is indicated by 0. Lines IV and V represent the near upstream and far upstream sequences in procaryotes and eucaryotes, respectively.
VOL.44, 1982
on November 10, 2019 by guest
http://jvi.asm.org/
Examination ofmore geneswillbe necessary
to know whether to designate any of the
se-quences referred to above as specific "pox-boxes."Toassessthefunctional significance of
DNA structures, genetic experiments are re-quired. With this in mind, we have recently
mapped theselectablethymidinekinasegeneof
vacciniavirus (40). Ourplansareto mutatethe putativepromoter ofthe thymidinekinasegene withinplasmidorphage recombinants and then
to introduce the mutated DNA into infectious virus by transfection procedures. In this
man-ner, structure/function relationships can be ex-plored.
A sequenceof four or more nucleotides pre-ceding the translational initiation codon in pro-caryotic mRNA is complementary to the 3' terminal segment of 16S rRNA. This comple-mentarity is thought to facilitate the bindingof
mRNA to ribosomes. The situation in eucary-otes is less certain since the presence of a
sequence complementaryto 18SrRNA is
vari-able (17, 46). For this reason, itwas of interest
to examine the leader sequences ofearly vac-cinia virus mRNAs. Before the first ATG of
the19Kpolypeptidegene,thereisa
heptanucle-otide ATCCATC that has close sequence
complementarity to the 3' end of 18S rRNA (3'AUUACUAGGAAGGGCG,indicatedin
ital-ics; 17). Similarly, a complementary
tetranu-cleotide TCCTwasnoted(35)afewnucleotides
before the secondATG of the 7.5Kpolypeptide
gene. However, lesshomology to the 3' end of
18S rRNA is present in the 42K polypeptide
genewhichappearsto havealeader ofonlyfive to six nucleotides. Whetherthese differencesin
leadersequencesareresponsiblefor thegreater invitro synthesisof 19Kand 7.5Kpolypeptides
193 223 253
ATGTCG ATG AAA TAT CTG ATG TTG TTGTTC GCT GCT ATGATA ATC AGA TCA TTC GCC GAT AGT GGT AAC GCT ATC GAA ACGACA TCG CCA MET SER MET LYS TYR LEU MET LEU LEU PHE ALA ALA MET ILE ILE ARGSER PHE ALA ASP SER GLY ASN ALA ILEGLU THR THR SER PRO
283 313 343
GAA ATT ACA AAC GCT ACA ACA GAT ATT CCA GCT ATC AGATTA TGC GOT CCAGAG GGA GAT GGA TAT TGT TTA CAC GOT GAC TGTATC CAC GLU ILE THR ASH ALA THR THR ASP ILE PRO ALA ILEARG LEU CYS GLYPROGLU GLYASP GLY TYR CYS LEU HIS GLYASP CYS ILE HIS
373 403 433
GCT AGA GAT ATT GACGGT ATG TAT TGT AGA TGC TCT CAT GGT TAT ACA GGCATT AGA TGT CAG CAT GTA GTATTA OTA GAC TAT CAA CGT
ALA ARGASP ILE ASPGLYMET TYR CYS ARG CYS SER HIS GLYTYR THR GLY ILEARG CYS GLNHIS VAL VAL LEUVAL ASP TYRGLN ARG
463 493 523
TCA GAA AAC CCA AAC ACT ACA ACO TCA TAT ATC CCA TCT CCC GGTATT ATO CTTGTA TTA GTAGGCATT ATT ATT ATT ACG TGT TGTCTA SER GLU ASN PRO ASNTHR THR THR SER TYR ILE PRO SERPRO GLY ILEMET LEU VAL LEU VAL GLY ILE ILE ILE ILE THR CYSCYS LEU
553 583
TTA TCT GTT TAT AGG TTC ACT CGA CGAACT AAA CTACCT ATA CAA GAT ATG GTT GTGCCA TAA LEUSER VAL TYR ARG PHE THR ARG ARGTHR LYS LEU PRO ILE GLNASPMET VAL VAL PRO END
MOLECULAR WEIGHT - 15525
1707 1677 1647
ATGGAT ATT TAC GAC GAT AAA GGT CTA CAG ACT ATT AAA CTGTTT AAT AAT GAATTT GAT TOT ATA AGGAAT GAC ATC AGAGAA TTA TTT MET ASP ILE TYR ASP ASP LYS GLY LEU GLN THR ILE LYS LEU PHEASN ASNGLUPHE ASP CYS ILE ARG ASNASP ILE ARG GLU LEU PHE
1617 1587 1557
AAA CAT GTA ACT GAT TCC GAT AGTATA CAA CTT CCG ATG GAA GAC AAT TCT GAT ATT ATAGAA AAT ATC AGAAAA ATA CTA TAT AGA CGA
LYS HIS VAL THR ASP SER ASP SERILEOLN LEU PROMET GLUASP ASNSER ASP ILEILE GLU ASN ILE ARGLYS ILE LEU TYR ARG ARG
1527 1497 1467
TTA AAA AAT GTA GAA TGT GTT GACATC GAT AGT ACA ATA ACT TTT ATO AAA TAC GAT CCA AAT GAT GAT AAT AAG CGT ACO TGT TCT AAT LEU LYS ASN VAL GLU CYS VAL ASP ILEASP SERTHR ILETHRPHEMET LYS TYR ASP PRO ASH ASP ASPASN LYS ARG THRCYS SER ASN
1437 1407 1377
TGG GTA CCCTTA ACT AAT AAC TAT ATG GAA TAT TGT CTA GTA ATA TAT TTO GAA ACA CCOATA TGT GGA GGC AAA ATAAAA TTA TAC CAC TRPVAL PRO LEU THR ASN ASHTYR MET GLUTYR CYS LEU VAL ILETYR LEUGLU THRPRO ILE CYSGLY GLY LYS ILE LYS LEU TYR HIS
1347 1317 1287
CCTACA GGAAAT ATA AAG TCG GAT AAG GAT ATTATG TTT GCA AAGACT CTA GACTTT AAA TCA AAGAAA GTGTTA ACT GGA CGT AAA ACA PRO THR GLYASH ILE LYS SER ASP LYS ASP ILE MET PHE ALA LYS THR LEU ASP PHE LYS SER LYS LYS VAL LEUTHR GLY ARG LYS THR
1257 1227 1197
ATT GCC GTTCTA GAC ATA TCCGTT TCA TAT AAT AGA TCA ATG ACT ACT ATT CAC TAC AAC GACGACGTT GAT ATAGAT ATA CAT ACT GAT
ILE ALA VAL LEU ASP ILESER VAL SER TYRASN ARG SER MET THR THR ILE HIS TYR ASNASP ASP VAL ASP ILEASP ILE HIS THR ASP
1167 1137 1107
AAA AAT GGA AAA GAG TTA TGT TAT TGT TAT ATA ACA ATA GAT GAT CAT TAC TTG GTT GAT GTGGAA ACT ATA GGA GTT ATA GTC AAT AGA LYS ASH GLY LYS GLU LEU CYS TYR CYS TYR ILE THR ILEASP ASP HIS TYR LEU VAL ASP VAL GLU THR ILE GLY VAL ILE VAL ASN ARG
1077 1047 1017
TCT GGAAAA TGT CTG TTA GTAAAT AAC CAT CTA GGT ATA GGT ATCGTT AAA GAT AAA CGT ATA AGC GAT AGT TTT GGA GAT GTA TGT ATG
SER GLY LYS CYS LEU LEU VAL ASN ASNHIS LEU GLY ILE GLY ILE VAL LYS ASP LYSARG ILE SER ASP SER PHE GLYASP VAL CYS MET
987 957 927
GAT ACA ATA TTT GAC TTT TCT GAA GCA CGA GAGTTA TTT TCA TTA ACT AAT GAT GAT AAC AGGAAT ATA GCA TGGGAC ACT GAT AAA CTA ASP THR ILE PHE ASP PHE SERGLU ALA ARGGLU LEU PHE SER LEU THR ASN ASP ASP ASN ARGASH ILE ALA TRPASP THRASP LYS LEU
897 867 837
GACGAT GAT ACA GAT ATA TGG ACT CCCGTC ACA GAA GAT GAT TAC AAA TTT CTT TCT AGA CTAGTA TTGTAT GCA AAA TCT CAA TCG GAT
ASP ASP ASP THRASP ILE TRP THR PRO VAL THR GLU ASP ASP TYR LYS PHE LEU SER ARGLEU VAL LEU TYR ALA LYS SER GLN SER ASP
807 777 747
ACT GTA TTC GAC TAT TAT GTT CTT ACT GGT GAT ACG GAA CCA CCC ACT GTA TTC ATT TTCAAG GTA ACT AGA TTT TAC TTT AAT ATG CCG
THR VAL PHEASP TYR TYR VAL LEU THR GLY ASP THR GLU PRO PRO THR VAL PHE ILE PHE LYS VAL THR ARG PHE TYR PHE ASH MET PRO AAA TAA
LYS END
MOLECULAR WEIGHT = 38508
FIG. 7. Derived amino acidsequencesofthetwoearlypolypeptides. Open readingframesareshown with theirpredicted molecularweights.
on November 10, 2019 by guest
http://jvi.asm.org/
relativetothe 42Kpolypeptide(11) isnot under-stood.
Nuclease Si protection experimentswerealso
used to map the major 3' ends of the 19K
polypeptide mRNA at or near nucleotides 643 and 644 (Fig. 2). We considered that a more preciseidentificationof thesequenceadjacentto
thepoly(A)tail of themessagecould beobtained bysequencingcDNArecombinants. Indeed,aT residue atposition 639 wasfound adjacent toa
nongenomic oligo(dA) cluster. Thesequence of additional cDNA recombinants is necessary to
determine theextent of 3' heterogeneity. How-ever, as mentioned above, most of the cDNA recombinants did not have the oligo(dA) * oli-go(dT) terminus because thesecondpolymerase reaction didnotgo tocompletion orthis struc-ture was nibbled away during the nuclease Si step usedto remove the 5' hairpin. An alterna-tive method of cDNA cloning that avoids this
stepwouldseempreferable(21). Foravariety of
possiblereasons, we could notprecisely locate the 3' end of the 42K polypeptide message. However, fromthelocation of the 5' end and the length of the RNA, itmustoccurbetween nucle-otides 640 and 740.Thus, the3' ends of thetwo oppositely oriented mRNAsmustbeveryclose
toeach other.
Examination of the genomic sequence near
the 3' ends of thetwo mRNAs does notreveal the tandem CTATTC that was found with the 7.5Kpolypeptide'message (35). Evidently, this repeated structure is not a general termination
sequence. Just before the end of the 19K poly-peptide mRNA there is a possibly related se-quence CTTATG. The significance of the
tan-demly repeated sequence TAGAGGTAGAGG
beyond the coding regions of the 42K polypep-tide mRNA is questionable (Fig. 2). Although A * T-rich sequences are present, a precise
AATAAApoly(A)signal sequence (27)was not
found. It is not known whether the 3' ends of vaccinia virus mRNAs represent termination
sitesor sites ofRNAprocessing.
Thegenomicsequencein Fig. 2wasanalyzed with the aid of a computer program (28) for amino acid and stop codons in both directions and all three reading frames. The first ATG occurs at position 164, about 50 nucleotides downstream from the start site for the 19K
polypeptide mRNA. A second ATG occurs in-phase, six nucleotides further downstream.
Be-tweenthefirst ATG and the TAA stopcodonat
position 584, there is a 420-nucleotide open
reading frame sufficient to code for 140 amino acids (Fig. 2). Additional inphase stop codons occurbefore the RNAterminates. In both other reading frames, there are many stop codons throughout thegene.
Ontheopposite DNA strand, anATGoccurs
at position 1,736, only five to six nucleotides downstreamfromthe5'endof the 42K polypep-tide mRNA. A 993-nucleotide open reading
frame continues until the TAA at position 743 neartheendof the message. Thisregionwould code forapolypeptide of 331 amino acids (Fig. 2). Both of the other reading frames have stop
codons throughout.
Based on the predicted amino acid sequence (Fig. 7), the molecular weights of the two poly-peptides would be 15.5K and 38.5K. These
numbersare somewhat lower thanthe 19Kand
42K values determined by polyacrylamide gel electrophoresis (11). It seems likely that the latter values were overestimated since in that studytheendogenousreticulocyte43K polypep-tide appearedto be 48K.
Theamino acid sequences of thetwo polypep-tidesareunremarkable. Neither hasaclusterof hydrophobic amino acids indicative of a mem-brane transportfunction. A * T-rich codons are
usedprimarily, reflecting the 60%A * T content
of the genes. As yet, there is no information regarding the functions ofthese two early
pro-teins.
LITERATURE CITED
1. Baroudy, B. M., and B. Moss. 1980. Purification and characterization ofa DNA-dependentRNA polymerase from vacciniavirions.J. Biol.Chem. 255:4372-4380.
2. Baroudy, B. M., S. Venkatesan, and B. Moss. 1982. Incompletelybase-pairedflip-flopterminal loopslink the two DNAstrandsof the vaccinia virus genome intoone
uninterruptedpolynucleotidechain. Cell28:315-324. 3. BelleIsle, H., S. Venkatesan,and B. Moss.1981.Cell-free
translation of early and late mRNAs selected by hybrid-ization to cloned DNA fragmentsderivedfrom the left 14 million to 72 million daltons of the vaccinia virus genome. Virology 112:306-317.
4. Berk,A.J.,and P. A.Sharp. 1977.Sizingandmappingof early adenovirus mRNA's by gel electrophoresis ofSi
endonuclease-digested hybrids.Cell 12:721-732.
5.Berkner, K. L., and W. R. Folk. 1977. Polynucleotide kinase exchange reaction. Quantitativeassayfor restric-tion endonuclease-generated 5-phosphoryl termini in DNAs. J.Biol. Chem.252:3176-3184.
6. Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extractionprocedure forscreening recombinant plasmid DNA.Nucleic AcidsRes. 7:1513-1523.
7. Bolivar, F.,R.L.Rodriguez,P.J. Greene,M.E.Betlach,
H. L.Heyneker,and H. W.Boyer. 1977.Constructionand characterizationofnewcloningvehicles. II. A multipur-posecloningsystem. Gene 2:95-112.
8. Boone, R. F., and B. Moss. 1978. Sequencecomplexity and relative abundance ofvaccinia virus mRNA's synthe-sizedinvivoand in vitro. J. Virol.26:554-569.
9. Breathnach, R.,and P.Chambon.1981.Organizationand expression of eukaryoticsplit genes codingforproteins.
Annu.Rev. Biochem.50:349-383.
10. Cabrera, C. V.,M.Esteban,R.McCarron,W. T.
McAllis-ter,andJ.A.Holowczak.1978.Vaccinia virus transcrip-tion: hybridization ofmRNA torestrictionfragments of vaccinia DNA. Virology 86:102-114.
11. Cooper, J. A.,R.Wittek,and B. Moss.1981.
Hybridiza-tion selection and cell-free translation of mRNA's en-coded within the invertedterminalrepetitionofthe
vac-ciniavirusgenome. J. Virol. 37:284-294.
12. Cooper, J. A.,R.Wittek,andB.Moss.1981.Extensionof
VOL.
on November 10, 2019 by guest
http://jvi.asm.org/
thetranscriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J. Virol. 39:733-745.
13. Garon, C. F., E. Barbosa, and B. Moss. 1978. Visualiza-tion of an inverted terminal repetiVisualiza-tion in vaccinia virus DNA. Proc. Natl. Acad. Sci. U.S.A. 75:4863-4867. 14. Grady, L. J., and E. Paoletti. 1977.Molecular complexity
ofvaccinia DNA and the presence of reiterated sequences inthe genome.Virology 79:337-341.
15. Gray, H. B., Jr., D. A. Ostrander, J. L. Hodnett, R. J. Legerski, and D. L. Robberson. 1975. Extracellular nu-cleases ofpseudomonas Bal 31. Characterization of single strand-specificdeoxyriboendonuclease and doublestrand deoxyriboexonuclease activities. Nucleic Acids Res.
2:1459-1492.
16. Green, M. R., and R. G. Roeder. 1980. Definition of a novel promoter for the majoradenovirus-associated virus mRNA. Cell22:231-242.
17. Hagenbuchle,O., M. Santer, J. A. Steitz, and R. J. Mons. 1978.Conservation of the primary structure at the 3' end of18S mRNA from eukaryotic cells. Cell 13:551-563. 18. Kates, J. R., and J. Beeson. 1970. Ribonucleic acid
synthesis in vaccinia virus. II. Synthesis of polyriboade-nylic acid. J. Mol. Biol. 50:19-33.
19. Kates, J. R., and B. R. McAuslan. 1967. Poxvirus DNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 58:134-141.
20. Kaverin, N. V., N. L. Varich, V. V. Surgay, and V. I. Chernos. 1975. A quantitative estimation of poxvirus genome fraction transcribed as "early" and "late" mRNA.Virology 65:112-119.
21. Land, H., M. Grez, H. Hauser, W.Lindenmaier, and G. Schutz. 1981.5'-terminal sequences ofeukaryoticmRNA
can be cloned withhigh efficiency. Nucleic Acids Res. 9:2251-2266.
22. Maxam, A., and W. Gilbert. 1980.Sequencing end labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560.
23. Munyon, W., E. Paoletti, and J.T.Grace, Jr. 1967.RNA
polymeraseactivity inpurified infectious vacciniavirus.
Proc. Natl.Acad. Sci. U.S.A.58:2280-2287.
24. Nevins, J. R., and W. K. Joklik.1975.Poly(A) sequences of vaccinia virus messenger RNA: nature, mode of addi-tion and funcaddi-tion during translaaddi-tion in vitro and invivo.
Virology 63:1-14.
25. Nevins, J. R., and W. K. Joklik. 1977. Isolation and properties of the vaccinia virus DNA-dependent RNA
polymerase. J. Biol. Chem. 252:6930-6938.
26. Oda, K., and W. K. Joklik. 1967. Hybridization and
sedimentation studies on "early" and "late" vaccinia messenger RNA. J. Mol. Biol. 27:395-419.
27. Proudfoot, N. J., and G. G. Brownlee. 1976. 3'noncoding region sequences in eukaryotic mRNA. Nature(London) 263:211-214.
28. Queen, C. L., and L. J. Korn. 1980.Computeranalysis of nucleic acids and proteins. Methods Enzymol.
65:595-609.
29. Rigby, P.W.J., M.Dieckmann, C. Rhodes, andP.Berg.
1977. Labeling deoxyribonucleic acid to high specific activityinvitroby nick translation withDNApolymerase I.J.Mol. Biol. 113:237-251.
30. Rosenberg, M.,and D.Court.1980.Regulatory sequences
involved in thepromotion and termination of RNA
tran-scription.Annu. Rev.Genet. 13:319-353.
31. Sanger, F.,andR. Coulson. 1978.Theuseof thin acryl-amidegelsfor DNAsequencing. FEBS Lett.87:107-110.
32. Spencer, E.,S.Shuman,andJ. Hurwitz.1980.Purification and properties of vaccinia virus DNA-dependent RNA polymerase.J. Biol.Chem. 255:5388-5395.
33. Thayer, R. E. 1979. An improved method fordetecting foreign DNA in plasmids of Escherichia coli. Anal. Bio-chem. 98:60-63.
34. Tu,C.-P.,andS. N.Cohen.1980. 3'-endlabeling of DNA with [a-32P] cordycepin triphosphate. Gene 10:177-183. 35. Venkatesan, S., B. M. Baroudy, and B. Moss. 1981.
Distinctive nucleotide sequencesadjacent to multiple ini-tiation and termination sites of an early vaccinia virus gene. Cell 25:805-813.
36. Venkatesan,S.,andB. Moss.1981.In vitrotranscription of the inverted terminal repetition of the vaccinia virus genome: correspondence ofinitiation and cap sites. J. Virol. 37:738-747.
37. Vogelstein, B., andD. Gillespie. 1979. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. U.S.A. 76:615-619.
38. Weaver,R.F.,andC.Weissman.1979.Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of P-globin mRNA have identical map
coordi-nates. Nucleic AcidsRes. 7:1175-1193.
39. Wei, C.-M.,and B.Moss. 1975.Methylated nucleotides block5'-terminus of vaccinia virus messenger RNA. Proc. Nati. Acad. Sci. U.S.A. 72:318-322.
40. Weir, J. P., G. Bajsiar, and B. Moss. 1982. Mapping of the vaccinia virus thymidine kinasegene by markerrescue
andby cell-free translation of selected mRNA. Proc. Natl. Acad. Sci. U.S.A.79:1210-1214.
41. Winberg, G.,andM. L.Hammarskjold. 1980. Isolation of DNA fromagarose gelsusing DEAE-paper. Application
torestriction sitemapping of adenovirus type 16. Nucleic Acids Res. 8:253-264.
42. Wittek, R., E. Barbosa, J.A. Cooper, C. F. Garon, H. Chan, and B. Moss. 1980. The inverted terminalrepetition in vaccinia virus DNA encodes early mRNAs. Nature (London) 285:21-25.
43. Wittek, R., J.A.Cooper, E. Barbosa,andB. Moss. 1980. Expression of the vaccinia virus genome: analysis and mapping of mRNAs encoded within the inverted terminal repetition. Cell 21:487-493.
44. Wittek, R., and B. Moss.1980. Tandem repeats within the inverted terminalrepetition of vaccinia virus DNA. Cell 21:277-284.
45. Wittek, R.,andB. Moss.1982.ColinearityofRNAswith the vacciniavirusgenome;anomalieswithtwo comple-mentaryearly and late RNAs result fromasmalldeletion
orrearrangementwithin the inverted terminalrepetition. J. Virol. 42:447-455.
46. Yamaguchi, K., S. Hikaka, and K.-I. Miura. 1982. Rela-tionship betweenstructureofthe5' noncoding region of viral mRNA andefficiency intheinitiationstepofprotein synthesis in aeukaryotic system. Proc. Natl. Acad. Sci.
U.S.A. 79:1012-1016.
47. Yang, R. C. A., and R. Wu. 1979. BK virus DNA sequencecodingfortheamino-terminusof theT-antigen. Virology 92:340-352.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/