0022-538X/85/020415-10$02.00/0
Copyright © 1985, American Society for Microbiology
Two
Separable
Functional Domains of Simian Virus 40 Large T
Antigen:
Carboxyl-Terminal Region of Simian Virus 40 Large
T
Antigen Is Required for Efficient Capsid Protein
Synthesis
JOANNE TORNOW,1t MARYELLEN POLVINO-BODNAR,'t GEORGESANTANGELO,1t ANDCHARLES N.
COLE',2*
DepartmentofHuman Genetics, Yale University School of Medicine, New Haven, Connecticut 065101; andDepartment of Biochemistry, Dartmouth Medical School, Hanover, New Hampshire 037562*
Received 12July1984/Accepted 18 October 1984
Thecarboxyl-terminal portion of simian virus 40 large T antigen is essential for productive infection of CV-1 andCV-lpgreenmonkey kidney cells. Mutant dlA2459, lacking 14 base pairs at 0.193mapunits,waspositive for viral DNA replication, but unable to form plaques in CV-lp cells (J. Tornow and C. N. Cole, J. Virol. 47:487-494, 1983).Inthisreport, thedefectof dlA2459 is further defined. Simian virus 40 late mRNAswere transcribed, polyadenylated, spliced,andtransportedindlA2459-infectedcells,but the level ofcapsid proteins produced in infected CV-1greenmonkeykidney cellswasextremely low. dlA2459 large T antigen lacks those residues knowntoberequired for adenovirus helper function, and the blocktoproductive infectionbydIA2459 occursatthesamestageof infectionasthe blocktoproductive adenovirus infection of CV-1 cells. Theseresults
suggest that the adenovirus helper function is required for productive infection by simian virus 40. Mutant
dlA2459 was able to grow on the Vero and BSC-1 lines of African green monkey kidney cells. Additional mutants affecting the carboxyl-terminal portion of large T were prepared. Mutant inv2408 contains an inversion of the DNA between the BamHI andBcll sites (0.144 to 0.189 map units). This inversion causes transposition of the carboxyl-terminal 26 amino acids of large T antigen and the carboxyl-terminal 18 amino acids of VP1. This mutant was viable, even though the essential information absent from dlA2459 large T antigen has been transferredtothecarboxyl terminus of VP1 of inv2408. TheVP1polypeptide carryingthis carboxyl-terminal portion of large T could overcome the defect of dlA2459. This indicates that thecarboxyl terminus oflarge T antigen isaseparateandseparable functional domain.
The simian virus 40(SV40) largeTantigenperformsmany
functions during lyticand transforming infections by SV40. Itisinvolvedintheinitiationof viral DNAsynthesis (10, 61), triggers host DNA replication (31), is involved in the
auto-regulation ofthe level of its own mRNA (2, 52, 63), and is required for boththe initiation and maintenance ofmalignant transformation ofnonpermissive cells (34, 35, 62). In addi-tion,itprovidesafunction thatpermitsthegrowthof human adenoviruses in monkey cells (14, 30, 50). The polypeptide contains both DNA-binding (32, 64) and ATPase (11, 28) activities.
Analysis ofthe biochemical properties of mutants with deletions in the A gene, encoding large Tantigen, indicates thatsequencesbetween0.50 and0.43mapunitsarerequired for specific binding to the SV40 origin of DNA replication (12, 48, 56), and sequencesin the vicinity of 0.30map units
(m.u.) (11) are associated with ATPase activity. Thus, the polypeptidecontainsmultiplefunctional domains. It islikely that both of these activitiesmust exist within each active T antigen polypeptide, since complementation between bind-ing-competent, ATPase-defective and ATPase-positive, binding-defective mutants has not been observed (65; J. Tornow, R. Clark, C. Cole, and R. Tjian, and C. N. Cole, manuscriptinpreparation), eventhougholigomeric forms of Tantigen arefound in virus-infected cells (8, 19, 20, 49).
*Correspondingauthor.
t Present address: Department ofBiologicalChemistry.
Califor-niaCollege of Medicine, University of California,Irvine, CA 92717.
tPresentaddress: DepartmentofPediatrics,SchoolofMedicine,
Yale University, NewHaven, CT 06510.
There is considerable homology between the large T antigens of SV40 and polyomavirus, but each contains a regionnotfound in the otherlargeTantigen (Fig. 1) (24, 27, 51, 58).Thecarboxyl-terminal113 amino acids ofSV40 large T(residues596through 708,encodedby0.25 through 0.174 m.u.) is the only portion of the polypeptide showing no homology to any portion of polyomavirus large T. This portion oflarge T is known to be involved in providingthe adenovirus helper function (23, 47). In addition, infected cells containa64-base RNA that iscomplementaryto early mRNA and that is encodedbysequences nearmapposition 0.21(1, 41). Many completelyviablemutantsofSV40donot
produce this small RNA (1, 47; M. Polvino-Bodnar, Ph.D. thesis, YaleUniversity,NewHaven,Conn., 1983),
suggest-ing that it does not play an important role during the lytic infection cycle. Finally, this portion of the early region contains an alternate open reading frame of 95 condons. Almost all of this alternativereadingframe is deleted inone oranother viable mutantgenome(15, 22,25,44,45, 47, 66), suggestingthat it also doesnotplayanimportantrole. There is noevidence that it is everused. The role played by this portion oflargeTantigen duringthenormal SV40infection cycle has remained amystery.
We describedpreviouslyasetofmutantswithdeletionsat DdeIsites in theearly regionofSV40 (65, 66). One of these
mutants, dlA2459, with a 14-base-pair(bp)deletionat0.193
m.u., isnonviable,but differs frommostnonviable Tantigen
mutants in its ability to catalyze the replication of SV40
DNA(66).Thismutantdefinesafunction that isprovided by SV40 large T and that is required after the onset of viral DNA replication. The function defective in dlA2459 can be
415
on November 10, 2019 by guest
http://jvi.asm.org/
Region Region Region
1 2 3
5
l
l
Region 4
N C SMALL-t
N - C MIDDLE-T
N
~~/4
Region Region
0 a
* 0 0
N C
N
a
to
Region 4
0
SMALL-t
FIG. 1. Comparison of the early regions c virus. The regions that show homology hav Unique regions (polyomavirus region 3 and different numbers. Homologywasdetermined ofpublished sequence information for SV40( 1 corresponds to the NH2-terminal regions s virustumorantigens. Region2corresponds tc smalltantigen; the polyomavirus middletant amino acid sequence. Region 3 encodes tI polyomavirus middle T antigen and, ina dit partof large T antigen.Region4encodesthat wherein map almost all of the tsA SV40 a
mutants. Region 5 encodes that portion of
which has nohomology to polyomavirus larg
provided by group A mutants, which c mutationsordeletions that aremultiples mutants able to express the carboxyl-I large Tare able to complement
dIA2451
Thisindicates that somecriticalfunction
portion
of large T antigen and that th performed by T-antigenmoleculesdefec gen properties.Inthisreport, thedefectofdlA2459 isi mutantwaspositive forthe synthesisan
bydigesting SV40 viral DNA tocompletion with BamHI and -I3' BcIl, yieldingfragmentsof237 and 5,006bp. Althoughthese tworestriction endonucleasesrecognize differentsequences, eachproduces 5'-GATC-3'terminiatitscleavagesite. These
POLYOMA
fragments
wereligated
withT4 DNAligase
ata DNAcon-P0LYO
MA
centration of 20,ug/ml.
Subsequently, the DNA was digested with EcoRI and ligated with EcoRI-digested pBR322. The -C LARGE-T ligation reaction contained 10 ,ug of each DNA per ml. The HB101 strain ofEscherichia coliwas transfected with this DNA, andcolonies were selected on plates containing am-Region picillin (20,ug/ml). Minilysate DNA preparations were s screenedby
digestion
withEcoRI,
BamHI,
and Sau3AI. ,,1 3 inv2408 contained two sites recognized by EcoRI and a° SV40 single
BamHI
site andproducedthesame Sau3AIdigestion pattern as did wild-type SV40 cloned into the EcoRI site of pBR322.C dl(inv)2408E was constructed by combining the large LARGE-T ApaI-KpnI fragment of inv2408 (containing the entire early )f SV40 andpolyoma- region and including the inversion) with thesmall ApaI-KpnI
,e the same numbers. fragment of dlBC865 (containingmostofthelate region and
SV4o
region 5) have coveringthedeletionattheEcoRI
site) (Fig.3). Theligation 24,27,51f
58).Region mixture was then extracted twice with phenol-chloroform-shared by all pa ova- isoamyl alcohol (25:24:1) and four times with ether, ethanolsthe
uniqueportionof precipitated, and dissolved in 10 rmM Tris-chloride (pH igenalsocontains this 7.5)-i mM EDTA-10 mM NaCl. The DNA was digested he unique portion of with EcoRV andligated with anequimolarconcentration of fferent reading frame, EcoRV-digested pBR322 DNA, at a total DNA concentra-part of large T antigen tion of 20 p.g/ml. Insertion of DNA into the EcoRV site ofand
tsapolyomavirus
pBR322 inactivates the tetracycline resistance gene. After SV40 large Tantigen
transfection, colonies were selected on plates containing
ampicillin(20,ug/ml)andscreened toidentifythosecolonies sensitive to tetracycline(15 ,ug/ml).
dl(inv)2408L was constructed by combining the 946-bp
:ontain
either point ApaI-PstI fragment of inv2408 (0.091 to 0.271 m.u. and of3 bp (65). Thus, includingtheinversion), the2,004-bp PstI-KpnI fragment of terminal domain of dlA1209 (0.271 to 0.716 m.u. and covering the deletion 19 intracistronically. between 0.587and 0.650 m.u.)andthe6,326-bp KpnI-ApaIiisprovided by this fragment of pCC2, which contains the complete wild-type iis function can be SV40genome inserted into theEcoRI site of pBR322. For tive in other T-anti- the ligation, an equimolar concentration ofthe three frag-mentsand a totalDNAconcentration of25,ug/mlwereused. further defined.The Cells, viruses, and plasmids. The growth of CV-1 and Idprocessing oflate CV-lp cells has been described previously
(47).
The same mRNA, but the level ofcapsid proteins found in infectedCV-1 green monkey cells was drastically reduced. This defect resembles that of human adenoviruses in monkey cells (26, 37). Since dIA2459 large T antigen lacks those residues known to be required for helper function, these results suggest that
adenovirus
helperfunction is required forproductive infection by SV40. MutantdIA2459 isahost range mutant, sinceit is able toform plaqueson BSC-1 andVero
green monkeykidney monolayers.Additional mutations affectingthe carboxyl-terminal por-tion of large T are also described. In one, inv2408, the carboxyl terminal 18 amino acids of VP1 and the carboxyl-terminal 26aminoacids oflarge Tantigen wereexchanged. This mutant was viable. The VP1-Tantigen fusionproteinof this mutant can overcome the defect of dIA2459. This indicates that the portion of large T that provides the late function defined by dlA2459 is a separate and separable functional domain.
MATERIALS ANDMETHODS
Construction of SV40 mutants. The construction of mu-tants dlA2459, dlA2411, and
dIA2420
has been described previously(65, 66). Mutant inv2408 was constructed (Fig. 2)RI
BamHI(0143)
Bcl I(0188)
SV 40 - b
rO Bom HI
Bcl I
BamH1Hl 2
%BcII
V4 \,EcoRI
Ligase
2Heat
3EcoRI
B/BcI
Bcl/B
nv20
Bom HI Ligase
,-"-' 416aminoacids 2Heat
VP1 .. CCTGGG GAT CCAGAC....3BclI PRO GLY ASP PRO ASP 4Transfect
BcII
~~~~~~~~~RI
B/BcITag .OGTT CA T GAT+24 amino acidsCATAAT BcI/B
Tg..GTCATGAT CAT AAT..
VAL HIS ASP HIS ASN
[image:2.612.64.303.72.246.2]pBR322 pins2408 SV40
FIG. 2. Construction of SV40 mutant inv2408 and its insertion
intopBR322.Also shownis thenucleotidesequenceof thegenesfor
VP1 andlargeT antigenat theBamHIandBclI sites. SinceGAT
encodes aspartic acid in the reading frames ofboth polypeptides,
this inversion results intranspositionof thecarboxyl terminiof VP1
andlarge Tantigen.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.323.556.508.663.2]Produces an altered T antigen and an altered VP 1
Produces a defective VP 1
ond an altered T antigen
inv 2408 DNA dIA 1209 DNA
_~dlBC 865 DNA Wild type DNA
329bpOR
deleti
KpnI EcoRV
dl(inv) 2408L
EcoRX
PstI Bcl/Bom Apol
IBom/Oc
Produces no Tantigen
and an altered VP I
r---.Large Tantigen coding region
[image:3.612.163.452.70.519.2]vt,,,.VP1 coding region Large T intron
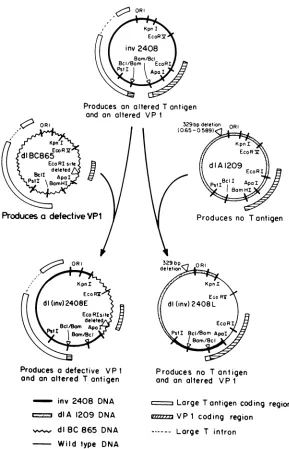
FIG. 3. Organization andoriginof SV40 sequences inmutantsdl(inv)2408Eanddl(inv)2408L.
protocols were used for growth of BSC-1 and Vero
cells,
which are alsoAfricangreen monkeykidney lines. Allwere
maintained in Dulbecco modified Eagle medium (DMEM) containing 5% fetal bovine serum
(FBS).
Ourwild-type
strain ofSV40 (WT830) was the small-plaque strain origi-nally characterizedby Takemotoetal. (60).
Wild-type
SV40 DNA was digested with EcoRI and cloned into theEcoRI site ofpBR322.All mutants were maintained as recombinant plasmids (inserted into pBR322) in the HB101 strain of E. coli. Previously describedmethods wereusedforbacterial
trans-formations (40)andfor thepreparation of
minilysates
(7)and purified plasmid DNA (13). All DNA preparations werepurified by equilibrium buoyant density centrifugation in gradientscontainingCsCland ethidiumbromide(300,ug/ml). MutantsdlBC865 (9),
dlA1209,
anddlE1226(15),and tsA1642 (16) were cloned in the BamHI site of pBR322. All other mutants exceptdl(inv)2408E were cloned in the EcoRI siteofpBR322; dl(inv)2408E was cloned in the EcoRV site of pBR322.
Transfection of eucaryotic cells. Plasmid DNA was first digested with either BamHI, EcoRI, or EcoRV to separate bacterial and viral sequences. This DNA was then ligatedat low concentration (5 ,ug/ml) with T4 DNAligaseto recircu-larize the viral DNA. Confluent monolayers of CV-1 cells wereincubated inTris-buffered saline (25mM Tris-chloride [pH 7.5], 137 mMNaCl, 5 mMKCI,0.6 mMNa2HPO4,0.05 mMMgCl2, 0.7 mM CaC12)containing1 p,g of DNA per ml and 500 ,ug of DEAE-dextran (5 x
105
daltons; Sigma Chemical Co., St. Louis, Mo.) per ml for 30 to 45 miri at 37°C. Plates 100 mm in diameter received 0.6to 1.0 mls of thismixture, 60-mm plates received 0.2 to 0.3 ml, and 35-mm plates received 0.1to0.2 ml. Cellswerethenwashed twice with Tris-buffered saline and incubated with DMEM con-taining 2%FBS and 100 ,uMchloroquine diphosphate(Sigma) for 4 to 6 h at 37°C. The treatment with chloroquinev, OR,
on November 10, 2019 by guest
http://jvi.asm.org/
A.
M 1 2 3 4
19S-
1328-1264:J
945-
610-
237-B.1
2 3 4-1662
-1097
-615 -540
-435
16S-.
a
S-310-233 -170 -128
FIG. 4. Si nucleaseprotectionanalysis of latemRNAproduced indIA2459-infectedcells. Totalcytoplasmic RNAwasprepared72h after transfection. EcoRI-digested SV40 DNA was 3' end labeled (A) or 5'end labeled (B) asdescribed in the text. The DNA probes werehybridized to 25 ,ug of RNA under conditions of DNA excess at 47°C for 15 h. After S1 nuclease treatment, samples were analyzed by electrophoresis in 1.4% alkaline agarose (30 mM NaOH, 2.5 mM EDTA) gels. Afterelectrophoresis, gelswerefixed in7% trichloroacetic acid, dried, and exposed to KodakXAR-5film.
Lanes (A and B): 1, wild-type SV40; 2, dIA2411; 3, dIA2459; 4,
mock infected. In A, 5' end labeled, MboI-digested SV40 DNA servedfor size markers; in B,3'endlabeled,Ddel-digested plasmid containingSV40 inserted into theEcoRIsite of pBR322wasusedfor size markers.
diphosphate increases the percentage of cells expressing transfected DNAfrom 1 to2%to 40 to60% (39). Afterthe chloroquine diphosphatetreatment, the chloroquine-contain-ing mediumwas replaced with DMEM containing2% FBS, andcellswere incubatedat 37°C.
Plaque assays andgenetic complementation tests.Wild-type and mutant DNAs wereseparated from bacterialandplasmid sequences as described above. Confluent monolayers of CV-lp, BSC-1, or Vero cells were transfected with three differentconcentrations of each DNA(10, 1, and 0.1 ng per 60-mmplate) by usingDEAEdextran asdescribedbyMertz and Berg (42). The cells were incubated under an agar overlaycontaining DMEM, 1%Bacto-agar(Difco
Laborato-ries,
Detroit, Mich.),
4%FBS,
and antibiotics for 10 to 12 days at 37°C (or other temperatures when temperature sensitivity was being determined). Plaques were visualized bystaining
with neutral red. Plaques were counted 24 h later. To determine rates ofplaque enlargement, averages weredetermined for 50 plaques on adailybasis once plaques had appeared.Genetic
complementation
analysesweredone in thesame way, except cells were cotransfected with dilutions of the DNA tobe tested and 10ng of helperDNAs.RNA analysis. Cytoplasmic RNA was extracted from transfectedCV-1 cells 48 to 72 hpostinfection bythemethod ofWhiteetal. (68)andanalyzed bytheS1 nuclease method ofBerk and Sharp (6)asdescribed by Favaloro et al. (21). Probes were end labeled at the EcoRI site of SV40by using either
alpha[32P]deoxynucleoside
triphosphates and the Klenow fragment of DNA polymerase I (3' end-labeled probes) orgamma[32P]ATP
and polynucleotide kinase (5' end-labeled probes).Analysis of viral DNA replication. DNA replication was analyzed after transfection of cultures of CV-1 cells in 60-mmdishesasdescribedpreviously (47, 66).
Analysis of virus-specific proteins byimmunoprecipitation. At appropriate times after infection (indicated in figure legends), cells were washed twice with Tris-buffered saline
and incubated for 1 h in DMEM lacking methionine and containing2%dialyzed FBS.This medium was removed and replacedwith DMEM lacking methionine and containing2% dialyzed FBS and 300 ,uCi of[35S]methionine per ml (0.5 ml per100-mmplate, 0.25 ml per 35-mm plate). The durationof the labeling period was as indicated in the figure legends. After the labeling period, cells were washed twice with Tris-buffered saline, and protein extracts were prepared. Ice-cold lysing buffer (0.15 M NaCl, 0.02 M Tris-chloride [pH 8.0],1%NonidetP-40)wasaddedtothe cells(0.5ml per 100-mm plate, 0.25 ml per 35-mm plate). Cells were incu-bated in thelysing buffer for 30 min at 4°C. Cells and buffer werethen scraped from theplates into Eppendorf tubes with a rubberpoliceman. Tubes were spun in amicrocentrifuge to pelletthe cellulardebris, and the supernatant was removed to another tube and stored at -70°C.
A 0.1-ml sample of extract was immunoprecipitated as described previously (47). Either hamster antitumor serum (lot 8-OOX from the National Institutes of Health), rabbit antiserum against sodium dodecyl sulfate (SDS)-disrupted virions, or monoclonalantibody was used to precipitate T antigens or capsid proteins. Monoclonal antibody recogniz-ing determinants in thecarboxyl-terminal region of T antigen was a gift from S. Tevethia. Samples were analyzed by electrophoresis on SDS-polyacrylamide gels. After elec-trophoresis, gels were fixed in 50% methanol-10% acetic acid-10%trichloroacetic acid,fluorographed with En3Hance (New England Nuclear Corp., Boston, Mass.), dried, and exposed toKodak XAR-5 film for1 to5 daysat -70°C.
Enzymes and Chemicals. Restriction endonucleases were purchasedfrom NewEngland Biolabs(Beverly, Mass.), P-L Biochemicals (Milwaukee, Wis.), or BoehringerMannheim (Indianapolis, Ind.)and usedaccordingto theinstructions of thesupplier. T4DNAligasewaspurchased from Collabora-tive Research (Waltham, Mass.) or New England Biolabs, T4 polynucleotide kinase was from New England Biolabs, calf intestinal alkaline phosphatase was from Boehringer Mannheim, Si nuclease was from P-L Biochemicals or BoehringerMannheim, and proteinase K was from Beckman
30h pi. 54h pi
T k capsid T capsid
2 m 2 m2
w 4w o 4 w 0 4 wO 4
tM t c t
rC
5 tc 5M g k g k 9 k 9
92
69 * | X Tac
6
9
vp2
12 *
FIG. 5. Autoradiogram of[35S]methionine-labeled viralproteins immunoprecipitated from wild-type-, dIA2459-, or mock-infected
cell extracts. Extracts wereprepared30or54 h after transfection of
CV-1 cells. Each extract wasimmunoprecipitated with either
ham-ster antitumor serum or rabbit antiserum against SDS-disrupted SV40 virions.The numbersattheleft representthepositionsofsize
markers(sizes inkilodaltons).
30
-*
vp3
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.96.263.77.212.2] [image:4.612.356.521.480.662.2]TABLE 1. Complementation between dIA2459andtsA1642"
Plaquesformed (PFU/,ug) by comple-MutantMutant no.no. mutationLocation of
(m.u.)
mentation with:dIA2459 tsA1642 dIBC865 dIBC865 0 or1.0 1 x 105 1.3 x 105 <103 dlA1209 0.650 to 0.587 <103 <103 9 x 104 d1A2411b 0.497 9 X 104 <i0l 2.0 x 105 dlA2420b 0.497 <103 <103 1.2 x 105 d1A2416b 0.288 <103 <i0l 1.4 x 105 d1A2432b 0.288 6 x 104 <103 9.5 x 104
dIA24lOb
0.243 <103 <103 1.3 x 105dlA2433b 0.243 7 x 104 <10 1.4 x 105 dlA2459b 0.193 <103
1.1
X 105 1.9 X 105dl(inv)2408E' <103 9 X 104 <103
a Monolayers of CV-lp cells were exposedtodilutions ofmutantDNAand 10 ng ofcomplementing mutant DNA. Plateswere incubatedat 37°C,and plaques were counted 12 days afterinfection.
bThese mutantswere constructed and characterizedbyTornowand Cole
(5). MutantsdlA2411,dlA2432, anddIA2433produce nearly full-sizedlargeT
antigens, since amultiple of 3 bp is deleted. Mutants dlA2420, dIA2416,
dIA2410,anddIA2459produce truncatedlargeTantigens,since thedeletions
are notmultiples of 3 bpandthereforecause ashift inthetranslationreading frame.
'dl(inv)2408E contains an inversion ofthe BamHI-to-BclI fragment of
SV40 (0.144to0.189m.u.) andthe samedeletionattheEcoRI site(0or1.0
m.u.) asmutantdlBC865.
InstrumentsInc.(Fullerton,Calif.). Allradiochemicalswere
purchasedfrom AmershamCorp. (Arlington Heights, Ill.). RESULTS
Thedefect ofdlA2459 affectsalate stage ofinfection. SV40
mutant dIA2459 lacks 14 bp (nucleotides 2,785 through 2,798)at0.193 m.u.(66). Itdirectsthesynthesis ofalargeT
antigen that lacks the carboxyl-terminal 35 amino acids. Progeny viral DNA issynthesized in CV-1 cells transfected with
dIA2459
DNA (66). Todetermine whether the block to productive infection by dlA2459 was at the level of late mRNA transcription, the synthesis of late mRNA wasexamined (Fig. 4).
CV-1 cells were transfected with dlA2459, dIA2411 (a replication-defective group A mutant), or wild-type SV40 DNA. Total cytoplasmic RNA was isolated 72 h later and analyzed by the
Si
nuclease mapping procedure (6, 21). EcoRI-digested SV40 DNA was labeled at its 3' end. This probe will be protected by all late mRNAs that extend beyondthe EcoRIsitetothepolyadenylationsite for the late mRNAs. Analysis oftotal cytoplasmic RNA prepared 72 h aftertransfection demonstrated thatdlA2459(Fig. 4A, lane 3)producedasmuchlate mRNAasdidwild-typeSV40(Fig. 4A, lane 1). No late mRNAwasdetected in cellstransfected with dIA2411 DNA (Fig. 4A, lane 2) or mock-infectedcells (Fig. 4A,lane4). To determine whether the blocktodlA2459 infection was at thelevel ofmRNAprocessing, anotherS1 nucleaseprotectionexperimentwasperformed withaprobe thatwas5' end labeledat theEcoRI site. Thisprobewill be protected bythat partofthe late mRNAs thatisbetween the acceptorsplicejunctions and the EcoRI site. Both 16Sand 19S late mRNAs were produced in dlA2459-infected cells (Fig. 4B, lane 3) and the ratio of 16S/19S mRNAs was the same as in wild type-infected cells (Fig. 4B, lane 1). The probe was not protected by RNA prepared from either mock-infected(Fig.4B, lane 4)ordIA2411-infected
(Fig. 4B, lane 2) cells. Identical results were obtained when polyad-enylated mRNA was used (data not shown). We conclude that late mRNAs are synthesized, polyadenylated, spliced, and transported in dlA2459-infected cells, and that theabundance of these species is comparable to that of wild-type SV40 late mRNA. We have not yet examined the structures of the leaders and the locations of splice donor sites of dlA2459 late mRNAs.
Todetermine whether the blocktoproductiveinfection by dlA2459was at the levelofcapsid protein synthesis, CV-1 cellswereinfectedwith dlA2459orwild-type DNAormock infected and labeled 30 or 54 h later with [35S]methionine. Extracts were prepared, immunoprecipitated with rabbit capsid protein antiserum or hamster tumor anti-serum, and analyzed by electrophoresis on SDS-polyacrylamide gels (Fig. 5). Although dlA2459 produces almost as muchlargeTantigen as the wild type at both times (Fig. 5), the production of capsid protein VP1 is greatly reduced, especially at 54 h after infection. VP2 and VP3 couldnotbe detected inprotein extracts ofd1A2459-infected cells. We conclude that dIA2459 does not form plaques in CV-lpcells due to aninability to produce sufficient levels of capsid proteins.
One otherreplication-competent group A mutant of SV40 has been described (16). Mutant tsA1642 produces viral DNA,late mRNA, and capsid proteins at the nonpermissive temperature, but does not form plaques. It is blocked at some stage aftercapsid protein synthesis, perhaps in virion morphogenesis. To determine whether tsA1642 and dIA2459 affected different functions of SV40 large T antigen, a geneticcomplementation experiment wasperformed (Table 1).tsA1642 complemented the defect ofdlA2459, butnot the defect of dlA2411. Therefore, dIA2459 and tsA1642 affect different functions oflarge T antigen.
ANTISERUM:
TIME
NIH mcI anti mcI
anti--Tumor T SD6 T
cooh capsid cooh
30 30
2M 2M 2M 2 M
S 40S4 s 4 0S4 0
V 00cVO CV CVO C
8 KS8 KS 8 K S8 K
AD
t(ag
69->
a-30-
-*-VPi
4-VP2
-VP3
t ag
12X-FIG. 6. Autoradiogram of[35S]methionine-labeledviral proteins immunoprecipitatedfrom wildtypeinv2408-,andmock-infected cell
extracts. Extracts were prepared 30 or 50 h after transfection of
CV-1cells. Extractswereimmunoprecipitated with either hamster
antitumor serum, rabbit antiserum against SDS-disrupted SV40 virions, or tissue culture fluid containing a monoclonal antibody directed against the carboxyl terminus of large T antigen. The
numbersattheleft represent thepositions of size markers (sizesin
kilodaltons).
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.612.319.558.391.641.2]inv 2408
SV40
M 1 2 3 5 1 2 3 5 M
FIG. 7. Analysis of viral DNAreplication inCV-1 cellsinfected by wild-type SV40orinv2408. Confluent monolayersofCV-1 cells were transfected with wild-type SV40 or inv2408 DNA. Extracts
werepreparedat various times after infection (1,2, 3, or5 days).
Cells werewashed with Tris-buffered saline, and0.3 ml of
extrac-tion buffer (0.01 M Tris-chloride, [pH 7.5], 0.6% SDS, 0.01 M EDTA);0.1ml of 5 MNaCl, and 0.025 mlofasolution(10mg/ml)
ofproteinaseKwereaddedtoeachplate. Each plate alsoreceived
0.015 ml of3H-labeled M13 phagereplicative-form DNA
(approxi-mately 30,000 cpm). The plates wereincubated at 37°C overnight. Thenextday,lysateswerescrapedintoEppendorftubes andstored at4°C overnight.Thenextday,tubeswere spunfor50minat18,000
rpm in a refrigerated Sorvall centrifuge. Supernatants were
re-moved, phenolextractedonce,andprecipitated bythe additionof 2
volumes of ethanol. After centrifugation, DNA pellets were
dis-solvedin 0.1 mlof10mMTris-chloride (pH 7.5)-lmM EDTA-10 mM NaCl. A sample of each preparation was precipitated with
trichloroacetic acidtodetermine thepercentrecoveryofthe
3H-la-beled phage DNA. Samples containing equal numbers ofcounts
were subjected to electrophoresis in a 1% agarose gel in 89 mM
Tris-chloride (pH 8.2-89 mM boric acid-2.5 mM EDTA. After electrophoresis, DNAin thegelwasdepurinated by soakingthegel
in 0.25 M HCl fortwo 15-min periods, denaturedby washingwith
0.5 M NaOH and 1.5 M NaCl fortwo30-minperiods, and
neutral-izedby washingwith 0.5 M Tris-chloride(pH 7.5)-3MNaCl fortwo 30-min periods. The DNA was then transferred to nitrocellulose (BA85;Schleicher & Schuell Co.,Keene, N.H.) bythe method of
Southern(59).32P-labelled probe (specific activity,108cpm/,ug)was
prepared bynicktranslation ofaplasmid containingSV40 inserted
into the EcoRI site ofpBR322 (54). Thefilterwasprehybridized (8
h)and hybridized with probe (15 h)in standard bufferscontaining
50%formamide at42°C. The filterwaswashed twice with 2x SSC
(lx SSC is 0.15 M NaCl, 0.015 M sodium citrate)-0.1% SDS at
roomtemperaturefor 15minandtwice with 0.2x SSC-0.1% SDSat
55°C for 15min. The filterwasdried in air and exposedtoKodak
XAR-5 film. Themarker lanes (M)contained 20 ngofSV40DNA
includedtomonitor theevennessof transfertonitrocellulose.
Viability is retained when the information absent from dlA2459large T antigen is transposedtothecarboxyl terminus of VP1. inv2408 is a viable SV40 mutant in which the carboxyl terminal 26 amino acids of large T antigen are transferred toVP1 and thecarboxylterminal 18 amino acids ofVP1 are transferred tolarge Tantigen. This mutant was constructed by inverting the small (237-bp) BamHI-to-BclI fragment of SV40. That these domains have actually been exchanged was confirmed by immunoprecipitation of [35S]methionine-labeled proteinextractsfrom wild type- and inv2408-infected cells with appropriate antisera or
mono-clonalantibodies (Fig. 6). When antitumorserum wasused,
wild-type large Tantigen wasimmunoprecipitated and both large T and VP1 were immunoprecipitated from inv2408-infected cells, indicating that inv2408 VP1 contains large T-antigen information. When a monoclonal antibody
di-rected against thecarboxyl-terminal portion of large T was used, large T was immunoprecipitated from wild type-in-fected cells, but not from inv2408-intype-in-fected cells. Instead, inv2408 VP1 was immunoprecipitated, indicating that it carried information from the carboxyl terminus of large T antigen. When antiserum prepared against SDS-disrupted SV40 virions was used, the three capsid proteins, VP1, VP2, andVP3, wereimmunoprecipitated from wild type-infected cell extracts. The same three polypeptides and large T antigen were immunoprecipitated from inv2408-infected cells, indicating that the inv2408 large T antigen carried capsid protein antigenic information.
Plaques formed in CV-lp cells transfected with inv2408 DNAwiththe same efficiency (PFU per microgram) as with wild-type SV40. However, the plaques appeared later and enlargedmore slowly than did wild-type SV40 plaques. The kinetics of progenyDNA synthesis in inv2408-infected cells was indistinguishable from that of wild-type SV40 (Fig. 7). This suggests that the slower rate of plaque enlargement reflects either aslower rate ofassembly of inv2408 progeny, production of a smaller number of inv2408 virions, or reduced specific infectivity of inv2408 virions.
Construction andanalysis of SV40 mutantsexpressing only one of the two fusion proteins expressed by inv2408. In inv2408, that portion of large T antigen missing from the large T of dlA2459 was fused to the carboxyl terminus of VP1. Since inv2408 was aviable mutant, this suggested that this information was able tofunction evenwhen separated from the remainder of large T antigen. Two derivatives of inv2408 wereconstructed whichexpressed onlyone of two fusion polypeptides produced by inv2408 (large T with the carboxylterminusof VP1 and VP1 with the carboxyl termi-nusoflargeT). These derivatives were constructed (Fig. 3) by replacing portions of the inv2408 genome with DNA fragments derived from other SV40 mutants.
dlBC865 isanonviableSV40mutantwith adeletion at the EcoRI site (9), which is located near the beginning of the coding regionfor VP1. This mutantproduces an 8-kilodalton
TABLE 2. Complementation properties ofdl(inv)2408E and
dl(inv)2408L"
Plaques formed(PFU/pLg)by
complemen-Complementing tation with:
mutant
dl(inv)2408E dl(inv)2408L
dIA1209 <103 <103
dlBC865 <103 3.5 x 104
dIE1226 2 x 105 2.5 x 105
dlA2459 <103 6.5 x 104
dlA24115 2.5 x 104 <103
dlA24205 <103 <103
dl(inv)2408E <103 8.5 x 104
Controls
SV40 3.0 x 105
inv2408 1.7 x 105
dlA2459 <103
dlE1226 2.2 x 105
dl(inv)2408E <103
dl(inv)2408L <103
aMonolayersofCV-lpcells weretransfectedwith dilutions ofdl(in')2408E ordi(inv)2408LDNA and 10 ngof complementing mutant DNAper 60-mm
plate.
bMutantdIA2411lacks 12bpat0.497 m.u.; mutantdlA2420lacks 20bpat 0.497 m.u.ThelargeTantigenofdlA2411containsthecarboxyterminus of
large T,whereasdIA2420directsthesynthesis ofa18-kilodaltonfragment of largeT(65).
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.612.99.266.80.217.2] [image:6.612.319.557.495.668.2]fragment of VP1 (Cole and Goff, unpublished results). A portion of the inv2408genomeincluding theEcoRI sitewas replaced with an analogous fragment of dlBC865, as de-scribed above, to yield mutant dl(inv)2408E, which can expressthe early region fusionpolypeptide (largeT contain-ing thecarboxylterminus ofVP1)ofinv2408, but not the late region fusion polypeptide. dlA1209 is another nonviable SV40mutantwitha329-bpdeletionbetween 0.650and0.587 m.u. (15; Polvino-Bodnar and Cole, unpublished results). This deletion removes the early mRNA capsite, the initia-tion site for largeTantigen, andthedonorsplice junction for large T mRNA. No large T-related polypeptides have been detected in dlA1209-infected cells (Cole, unpublished re-sults). Aportion ofthe inv2408genome,includingtheregion between0.650and0.587 m.u., wasreplaced byananalogous fragment ofdIA1209toyieldmutantdl(inv)2408L, whichcan express the late region fusion polypeptide (VP1
containing
thecarboxyl terminus of large Tantigen) of inv2408, but not theearly region fusion polypeptide.
These two mutants were examined for their
genetic
com-plementation behavior (Table 2). Neither dl(inv)2408E nor
dl(inv)2408Lwasabletoformplaques inCV-lp cells alone. They were, however, able to complement one another to
permit plaque formation. Furthermore,
dl(inv)2408L
wasabletocomplement
dIA2459,
indicatingthat theinformation missing from dlA2459 large T antigen can besupplied
in trans when this formation is attached to VP1. All of the control complementations agreed withpredicted
results: dl(inv)2408E and dl(inv)2408Lwerebothabletocomplement
dlE1226, which produces a defective VP2
(15),
indicating
thatVP2expression is normal in bothmutants;dl(inv)2408L was unable to complement dlA1209 because it could not expresslargeTantigen, butwasable tocomplement dlBC865 since the VP1-T antigen fusion
polypeptide
couldperform
the functions of VP1;
dl(inv)2408E
was unable tocomple-ment dlBC865, since it could not express VP1, and was unable tocomplement dlA1209or
dlA2411,
since itcouldnotproduce a large T antigen providing all of thefunctions of large T. We conclude that the
carboxyl-terminal
26 amino acids, transferredto VP1 in inv2408 anddl(inv)2408L,
con-stitute a separable functional domain of SV40 largeT anti-gen.
Mutant dlA2459 can grow in some African green monkey kidney cell lines. Confluentmonolayers of
CV-lp, BSC-1,
or VerocellsweretransfectedwithdIA2459orwild-typeDNA. Wefound that dlA2459was able to formplaques
on BSC-1 andVerocells; wild-type SV40 formed plaques onall three celllines. Theplaques formed in BSC-1 and Verocellswereapproximately half the size of
wild-type plaques
formed in thosecells. Noplaques appeared inCV-lpcells transfected withdlA2459
DNA, evenafter40days.DISCUSSION
The large Tantigen of SV40 is a multifunctional protein and may therefore contain multiple functional domains. Studies with mutants and monoclonal antibodies demon-strate that someof the propertiesoflarge Tantigen
require
different regions of the protein. The ATPase activity of T antigencanbe inhibitedspecifically bymonoclonal antibod-iesthatrecognizesequences encoded between 0.33 and 0.27 m.u. (11). The portionof T antigen encoded by sequences between 0.50 and 0.43 m.u. has been implicated in the
binding of large T antigen to the SV40 origin of DNA replication (12, 48, 56). The carboxyl-terminal 35 amino acids oflargeTaresufficient fornormal levelsofadenovirus
helper
function (14,23, 47).Weand othershaveisolatedand characterized many SV40 mutants with deletions in the A gene,encoding large
Tantigen (12, 14, 15, 46, 65, 66).
These includemutantslacking
ATPaseactivity,
butretaining
bind-ing
to theSV40origin,
mutants unable tobind totheSV40
origin,
butretaining
awild-type
level ofATPaseactivity,
and mutants with
drastically
reduced levels of adenovirushelper
function,
butwhicharestillcompletely
viable.There-fore,
at least three separateproperties
of Tantigen
have beenlocalizedtodifferentregions
oftheprotein.
We described
previously (66)
amutant ofSV40, dlA2459,
lacking
14bp
at 0.193m.u. Thelarge
Tantigen
encodedby
dlA2459 lacks the
carboxyl-terminal
35 amino acids and contains 3 new amino acids in theirplace.
This mutant iscapable
of viral DNAreplication,
but is unable to form plaques onCV-lp
monolayers.
These results demonstrate that a latefunction,
required
after the onset of viral DNAreplication,
isperformed by large
Tantigen.
In this report, we have further
investigated
the defect of dlA2459. Our results indicate that the block toproductive
infection
by
dlA2459 occurred aftertranscription
andbefore translation of late mRNA. The defect ofdlA2459was very similarto the defect ofhuman adenoviruses whenthey
aregrownin CV-1 cells. Adenoviruslate mRNAsare
produced,
though
at a levelapproximately
10% of that present in adenovirus-infected human cells(5);
production
of somecapsid
proteins (e.g.,
fiber)
is reducedby
as much as100-fold. Sincethedeletion in dlA2459affectsthat
portion
oflarge
Tantigen
whichprovides
adenovirushelper
function (14,23, 47)
and the defect of dIA2459 resembles that of adenovirusesinmonkey
cells,
we concludethatproductive
infection of CV-1 cells
by
SV40requires
thehelper
function. Thisfunctioncanbeprovided by
intracistroniccomplemen-tation
by
anySV40
mutant whoselarge
Tantigen
contains the informationmissing
from dlA2459large
Tantigen (65)
and
by
mutantdl(inv)2408L,
which contains thisinformationtransposed
to thecarboxyl
terminus ofVP1. This indicates that thehelper
function information oflarge
Tisaseparable
functional domain.
A
large
numberof viable mutantswith deletions nearthe 3' end oftheSV40
early
region
have beendescribed(1,
15,
22,
25,
44,45, 66).
In all but one of these mutants, the deletion is located upstream from the deletion ofdIA2459 andisamultiple
of3bp. Thus,
thesemutantlarge
Tantigens
contain the residues
missing
from dlA2459large
Tantigen.
Many
of these mutants show normal levels of adenovirushelper
function.Theothermutant,d11265,
lacksthecarboxyl-terminal 9 amino acids and contains 4 different residues in their
place (67).
Thismutant shows asubstantially
reduced butmeasurable level(7%)
ofhelper
function(14).
Therefore,
productive
infection of CV-1 cellsby SV40
requires only
alow level of adenovirus
helper
function. Of mutants with lesions in thisregion, only
dlA2459 iscompletely
defective for adenovirushelper
function(66).
In this paper, we have shown thatasufficient amountofthefunctionwasprovided
when
only
the 26carboxyl-terminal
residues weretrans-posed
to VP1. Sinceproductive
infectionby SV40
requires
only
alow level ofhelper function,
we cannot tell whether the 26carboxyl-terminal
residuesprovided wild-type
levels ofhelper
function. Mutant inv2408provided
little or nohelper
functiontocoinfecting
adenovirus(Cole,
unpublished
results). However,
ininv2408, helper
function becomes alate SV40 function.
Since,
adenoviruseseffectively
inhibit both cellular andSV40
lateprotein
synthesis (4,
29,
55),
thisprobably
accountsfor ourinability
to measurehelper
func-tionactivity
in inv2408-infected cells.on November 10, 2019 by guest
http://jvi.asm.org/
The genetic analysis of the altered genes in inv2408 supports theidea that some orallof the residues deleted in dlA2459largeTdefineaseparatedomain. Althoughinv2408 was viable, it grew more slowly than the wild type. To determine thegenetic potentials ofthetwoalteredproteins, the altered genes were separated from one another. The results ofcomplementation analyses indicated that the al-tered VP1 of inv2408 was afunctionalcapsid protein, since dl(inv)2408L could complement dlBC865, a mutant which produces a defective VP1 (Table 2). However, the plaques enlarged at a significantly slowerrate than those generated bycontrolcomplementation [plaques formed by dl(inv)2408L and dlBC865 were not visible until 10 to 12 days after infection; plaques formed by coinfection with d1BC865 and
dIA1209
(which encodes awildtype VP1)werevisible7days afterinfection].
This demonstrated that the altered VP1,although
not completely defective, was impaired. Since inv2408 DNA replication proceeded with the same kineticsas wild-type SV40, this suggests that the slower growth
rateofinv2408wascausedprimarily by apartially impaired VP'.
It is not known whether oligomerization of T antigen is requiredfor it to perform all of its functions. It is possible thatoligomerization occurs as aconsequenceofthebinding of Tantigen monomers toDNA and thatmutant monomers
unabletoremain
oligomerized
afterdissociation fromDNAmight
still possess full activity. The monomerfraction ofTantigen displays
both ATPaseandspecific DNAbinding (8, 20). Our finding that the helper function of large T can beprovided by
aVP1polypeptide carryingthe carboxyl-termi-nal 26residues oflarge Tantigen suggeststhat oligomeriza-tion of large T is not required for this function. However, since VP1 does oligomerize in the formation of the viralcapsid,
the possibility that the helper activity of inv2408requires
the formation of VP1 oligomers cannot be elimi-nated.SV40
large
T antigen and the adenovirus 72-kilodalton DNA-binding protein show important similarities. Bothareinvolved in DNA replicationand in regulation of transcrip-tion (17, 33, 43). Although adenoviruses cannot grow
pro-ductively
in monkey cells, adenovirus mutants capable of productive infection ofmonkey cellscanbe isolated(3, 36, 38). The lesions in these host range mutants map to the 72-kilodaltonDNA-binding proteinandare locatedat asite in thepolypeptidedistant from themutationsitesofmutantsaffecting
thereplication
andtranscriptional regulation
activ-ities of the polypeptide (53). Thus, for both SV40 and the human adenoviruses, a separate domain ofa majorDNA-binding
and regulatorypolypeptide
is involved inpermitting
the virus to grow
productively
in monkey cells. In both cases, sites ofphosphorylation
are located in thehelper
function
region
ofthepolypeptide.
It will beinteresting
to determine whetherexpression
of the adenovirus host rangemutant 72-kilodalton
DNA-binding protein
willpermit
thegrowth
ofSV40mutants suchas dlA2459in CV-1orCV-lp
cells in a manner analogous to the role played by the
carboxyl
terminus oflarge T antigen inpermitting
produc-tive adenovirus infection of CV-1 cells.The helperfunction activityoflargeT is provided by the
onlypartoflargeTshowingnohomologywithpolyomavirus
(Fig.
1). This suggests that thisinformation entered the SV40 genome after the evolutionary separation of SV40 andpolyomavirus.
Although the large Tantigen
of the human papovavirus, BKV,showshomologytothisportionofSV40 large T (57), the homology is much less than that between the remainderof SV40 and BK largeTantigens.The function defined bydlA2459 is essential only in some greenmonkey kidney cell lines. Although dlA2459 could not form plaques in CV-lp cells, plaques were formed at wild-type efficiency in both BSC-1 and Vero cells. Therefore, dlA2459 is a host range mutant. CV-lp, Vero, and BSC-1 cells are independent lines all derived from kidneys of Cercopithecus aethiops, but may have considerably dif-ferent cellular biology. Human adenoviruses are able to grow in Vero cells without an SV40 helper (18). We found thatadenovirus type 2 was also able to grow in BSC-1 cells (Cole, unpublished results). Eron et al. (18) reported that adenoviruses could not grow in BSC-1 cells. This discrep-ancy could reflect substantial differences between the BSC-1 line they used and the one we currently use. Another SV40 mutant, isolated by Pipas et al. (46), lacks 80 bp between 0.196and 0.181 m.u.; itforms plaques in BSC-1 cells, but not in CV-lp cells (Pipas, personal communication). This sug-gests that the host range behavior of these mutants is a general property of mutants whose large T antigens lack carboxyl terminal information. Atpresent, we do not under-stand the molecular basisfor the host range behavior, but it may indicate that the carboxyl-terminal domain oflarge T antigen is involved in determining the tissue or species growth range (or both) of SV40.
ACKNOWLEDGMENTS
We aregrateful to Katherine Ehlert and Deborah Klaus for
ex-cellent technical assistance. We thank Satvir Tevethia foragift of monoclonalantibodythatrecognizes the carboxyl terminal portion ofSV40 largeTantigen.
This workwas supported by National Science Foundation grant PCM-8340188.
LITERATURECITED
1. Alwine, J. C., and G. Khoury. 1980.Simian virus 40-associated
small RNA: mapping on the simian virus 40 genome and
characterization ofitssynthesis. J. Virol.36:701-708.
2. Alwine, J.C., S. I. Reed, and G. R. Stark. 1977.
Characteriza-tion of the autoregulation of simian virus40 gene A. J. Virol. 24:22-27.
3. Anderson, C. W. 1981.Spontaneousmutantsofthe
adenovirus-simian virus 40 hybrid Ad2+ND3 that grow efficiently in monkey cells. Virology. 111:263-269.
4. Anderson, C. W., P. R. Baum, and R. F. Gesteland. 1973.
Processing of adenovirus 2-induced proteins. J. Virol. 12: 214-252.
5. Anderson, K. P., and D. F.Klessig. 1983. Post-transcriptional
block to synthesis of a human adenovirus capsid protein in
abortively infected monkey cells. J. Mol. Appl. Gen. 2:31-43.
6. Berk,A.J.,andP. A.Sharp.1977.Sizingandmappingofearly
adenovirusmRNAsby gel electrophoresis ofS1 endonuclease-digested hybrids. Cell 12:721-732.
7. Birnboim, H. C., and J. Doly. 1979. Arapidalkaline extraction
procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1413-1423.
8. Bradley, M. K., J. D. Griffin, and D. M. Livingston. 1982.
Relationshipofoligomerizationtoenzymaticand DNAbinding propertiesof theSV40largeTantigen. Cell 28:125-134.
9. Carbon, J., T. E. Shenk, and P. Berg. 1975. Biochemical
procedure forproduction ofsmalldeletions insimianvirus 40
DNA. Proc. Natl. Acad. Sci. U.S.A. 72:1392-1396.
10. Chou, J. Y.,and R.G. Martin. 1974.Complementation analysis
of simian virus 40mutants.J. Virol. 13:1101-1109.
11. Clark, R., D. P. Lane, and R. Tjian. 1981. Use ofmonoclonal
antibodies as probes of simian virus 40 T antigen ATPase activity.J. Biol. Chem.256:11854-11858.
12. Clark, R., K. Peden, J. M. Pipas, D. Nathans, and R. Tjian.
1983. Biochemical activities ofT antigen proteinsencodedby
simian virus 40 A gene deletion mutants. Mol. Cell. Biol.
on November 10, 2019 by guest
http://jvi.asm.org/
3:220-228.
13. Clewell, D. B., and D. R. Helinski. 1970. A membrane filter techniquefor the detection ofcomplementary DNA. Biochem-istry9:4428 4440.
14. Cole,C.N.,L. V.Crawford,and P.Berg. 1979. Simian virus40
mutants with deletions at the 3' end ofthe early region are
defectiveinadenovirushelperfunction.J. Virol. 30:683-691. 15. Cole,C.N.,T.Landers,S.P.Goff,S.Manteuil-Brutlag,and P.
Berg. 1977. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J. Virol. 24:277-294.
16. Cosman,D.J.,and M.J. Tevethia. 1981.Characterization ofa
temperature-sensitive, DNA-positive nontransforming mutant
ofsimian virus40.Virology112:605-624.
17. Ensinger, M. J., and H. S. Ginsberg. 1972. Selection and
preliminary characterization oftemperature-sensitive mutants
of type5adenovirus. J. Virol. 10:328-339.
18. Eron, L., H. Westphal, and G. Khoury. 1975. Posttranscripti-onal restriction of human adenovirus expression in monkey cells. J. Virol. 15:1256-1261.
19. Fanning, E., B. Nowak, and C. Burger. 1981. Detection and characterization of multiple forms of simian virus 40 large T
antigen.J. Virol. 37:92-102.
20. Fanning, E.,K.-H. Westphal, D. Brauer, and D. Corlin. 1982. Subclasses of simian virus 40largeT antigenin freeform and boundtoDNA. J. Mol. Biol. 148:347-353.
21. Favaloro, J.,R. Treisman,and R. Kamen. 1981.Transcription
maps ofpolyoma virus-specificRNA: analysisby two-dimen-sional nucleaseS1gelmapping.MethodsEnzymol.65:718-749. 22. Feunteun, J.,G.Carmichael, J.C.Nicolas,and M. Kress.1981.
Mutantcarryingdeletions in thetwosimian virus40earlygenes.
J. Virol. 40:625-634.
23. Fey, G., J. B. Lewis, T. Grodzicker, and A. Bothwell. 1979. Characterization ofafusedproteinspecifiedbythe adenovirus type 2-simian virus 40 hybrid AD2+ND1 dp2. J. Virol 40: 201-217.
24. Fiers, W., R. Contreras, G. Haegeman, R. Rogiers, A.van de
Voorde, H.vanHeuverswyn, J. vanHerreweghe,G. Volckaert,
andM. Ysebaert. 1978. Completenucleotidesequenceof SV40
DNA. Nature(London)273:113-120.
25. Fitzgerald, M.,and T.Shenk.1981. Thesequence 5'-AAUAAA-3' forms part of therecognition site forpolyadenylationoflate
SV40mRNAs.Cell24:251-260.
26. Friedman, M. P., M. J. Lyons, and H. S. Ginsberg. 1970. Biochemical consequences of type 2 adenovirus and simian virus40doubleinfections of Africangreenmonkeykidneycells. J. Virol.5:86-97.
27. Friedmann, T.,A.Esty,P.LaPorte,and P.Deininger.1979. The nucleotidesequence and genome organizationof the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell 17:715-724.
28. Giacherio, D., and L. P. Hager. 1979. A poly dT stimulated
ATPaseactivityassociated with SV40largeTantigen. J.Biol.
Chem.254:8113-8116.
29. Ginsberg,H.S., L.J.Bello,and A.J.Levine. 1967. Control of
biosynthesis
of host macromolecules in cells infected withadenovirus,p.547-573.InJ. S.Colter andW.Paranchych (ed.), The molecularbiologyofviruses. AcademicPress, Inc., New York.
30. Grodzicker, T.,J.B.Lewis,andC. W. Anderson. 1976. Condi-tional lethal mutants of adenovirus type 2-simian virus 40
hybrids. II.Ad2+ND1host-rangemutantsthatsynthesize
frag-mentsof the Ad2+ND130Kprotein.J. Virol. 19:559-571. 31. Hatanaka, M., and R. Dulbecco. 1966. Induction of DNA
synthesisby SV40. Proc. Natl. Acad. Sci. U.S.A.72:673-677. 32. Jessel, D.,J.Hudson,T.Landau,D.Tenen,and D.M.
Living-ston. 1975. Interaction ofpartially purified simian virus 40 T
antigen with circularDNA molecules. Proc. Natl. Acad. Sci. U.S.A. 72:1960-1964.
33. Kaplan, M., H. Ariga, J. Hurwitz, and M. S. Horwitz. 1979.
Complementationofthe temperaturesensitive defectinH5ts125 adenovirus DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 76:5534-5538.
34. Kimura,, G., and R. Dulbecco. 1973. A temperature-sensitive
mutant of simian virus 40affecting transforming ability.
Virol-ogy 52:529-534.
35. Kimura, G.,and A.Itagaki. 1975. Initiation and maintenance of cell transformationby simian virus 40: a viral genetic property. Proc. Natl. Acad. Sci. U.S.A.72:673-677.
36. Klessig,D. F. 1977.Isolation ofavariant of human adenovirus serotype 2 thatmultiplies efficientlyonmonkey cells. J. Virol. 16:1650-1672.
37. Klessig,D.F.,andC. W.Anderson. 1975. Blockto multiplica-tion of adenovirus serotype 2 in monkey cells. J. Virol. 16:1650-1678.
38. Klessig, D. F., and T. Grodzicker. 1979. Mutations that allow human Ad2 andAdStoexpress late genes inmonkey cells map in the viral geneencoding the 72K DNA binding protein. Cell 17:957-966.
39. Luthman, H.,andG.Magnusson.1983.High efficiency polyoma DNAtransfection ofchloroquine treated cells. Nucleic Acids Res. 11:1295-1308.
40. Mandel, M.,and A. Higa. 1970. Calcium-dependent bacterio-phageDNAinteraction. J.Mol. Biol. 53:159-162.
41. Mark, D.,and P. Berg. 1979. A third splice site in SV40early
mRNA.ColdSpring Harbor Symp. Quant. Biol. 44:55-62.
42. Mertz, J. E., and P. Berg. 1974. Defective SV40 genomes: isolation andgrowth of individual clones. Virology 62:112-124. 43. Nevins, J. R., and J. J. Jensen-Winkler. 1980. Regulation of early adenovirus transcription:aprotein product of earlyregion 2specifically represses region 4 transcription. Proc. Natl. Acad. Sci. U.S.A. 77:1893-1897.
44. Pintel, D., N. Bouck,and G. di Mayorca. 1981. Separationof
lyticandtransforming functions of the simian virus 40Aregion:
two mutantswhicharetemperature sensitive forlytic functions
haveoppositeeffectsontransformation. J.Virol. 38:518-528. 45. Pintel, D., N. Bouck, G. di Mayorca, B. Thimmappaya, B.
Swerdlow, and T. Shenk. 1979. SV40 mutanttsA1499is heat-sensitive forlytic growth but generates cold-sensitive rat cell transformants. Cold Spring Harbor Symp. Quant. Biol. 44:305-309.
46. Pipas, J. M.,K. W.C.Peden,and D.Nathans. 1983. Mutational
analysisof simian virus 40Tantigen: isolation and characteri-zation ofmutants with deletions in the T antigen gene. Mol. Cell. Biol.3:203-213.
47. Polvino-Bodnar, M., and C. N. Cole. 1982. Construction and characterization of viable deletion mutants of simian virus 40 lackingsequencesnearthe 3' endof theearly region. J. Virol. 43:489-502.
48. Prives, C.,B.Barnet,A.Scheller,G.Khoury,and G.Jay.1982. Discreteregionsofsimian virus40largeTantigenarerequired for non-specific and origin-specific DNA binding. J. Virol. 43:73-82.
49. Prives, C., Y. Beck, D. Gidoni,M. Oren, and H. Shure. 1980. DNAbindingandsedimentationproperties of SV40Tantigens synthesized in vivo and in vitro. Cold Spring Harbor Symp. Quant. Biol. 44:123-130.
50. Rabson,A.S.,G. T. O'Conor, I. K.Berezesky,and F.J.Paul. 1964. Enhancement of adenovirus growth in African green monkey kidney cell cultures by SV40. Proc. Soc. Exp. Biol. Med. 116:187-190.
51. Reddy,V.B.,B.Thimmappaya,R.Dhar, K. N.Subramanian, B.S.Zain, J. Pan,P.K.Ghosh,M.Celma,andS.M.Weissman.
1978. Thegenomeof simianvirus 40.Science200:494-502.
52. Reed,S.I.,G. R.Stark,andJ. C.Alwine.1976.Autoregulation
of simian virus40geneAbyTantigen. Proc. Natl. Acad.Sci.
U.S.A. 73:3083-3087.
53. Rice,S.A.,and D. F.Klessig.1984.Thefunction(s) provided by the adenovirus-specified, DNA-binding protein required for viral late gene expression is independent of the role of the proteininviral DNAreplication. J. Virol.49:35-49.
54. Rigby,P. W.J.,M.Dieckmann,C.Rhodes,and P.Berg.1977. Labellingofdeoxyribonucleic acid to high specific activity in
vitroby nicktranslation with DNApolymerase I. J. Mol. Biol. 113:237-251.
55. Russell, W. C., and J. J. Skehel. 1972. The polypeptides of
on November 10, 2019 by guest
http://jvi.asm.org/
adenovirus-infected cells. J. Gen.Virol. 15:45-52.
56. Shortle, D.,R. F.Margoiskee, andD. Nathans. 1979.Mutational
analysis of the simian virus 40 replicon: pseudorevertants of mutants with a defectivereplication origin. Proc. Natl. Acad.
Sci. U.S.A. 76:6128-6131.
57. Sief, I., G. Khoury, and R. Dhar. 1979. Thegenomeof human
papovavirus BKV. Cell 18:963-977.
58. Soeda, E., J. R. Arrand, N. Smolar, J. E. Walsh, and B. E.
Griffin. 1980. Coding potential and regulatory signals of the
polyoma virusgenome. Nature(London) 283:445-453.
59. Southern, E. M. 1975. Detection of specific sequences among
DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517.
60. Takemoto, K. K., R. L. Kirschstein, and K. Habel. 1966.
Mutants of simian virus 40differing in plaque size, oncogenicity,
and heatsensitivity. J. Bacteriol. 92:990-994.
61. Tegtmeyer, P. 1972. Simian virus 40 deoxyribonucleic acid
synthesis: the viral replicon. J. Virol. 10:591-598.
62. Tegtmeyer, P. 1975. Function of simian virus 40 gene A in
transforming infection. J. Virol. 15:613-618.
63. Tegtmeyer, P., M. Schwartz, J. K. Collins, and K. Rundell.1975.
Regulation of tumor antigen synthesis by simian virus 40 A
gene.J. Virol. 16:168-178.
64. Tjian, R. 1978. The binding siteonSV40DNA foraTantigen
relatedprotein. Cell 13:165-179.
65. Tornow, J., and C. N. Cole. 1983. Intracistronic
complementa-tionin the simian virus 40 Agene.Proc.Natl. Acad. Sci. U.S.A.
80:6312-6316.
66. Tornow,J.,and C; N. Cole.1983. Nonviablemutantsofsimian
virus 40 with deletions near the 3' end of gene A define a
function for large T antigen required afteronsetof viral DNA
replication.J. Virol. 47:487-494.
67. van Heuverswyn, H., C. Cole, P. Berg, and W. Fiers. 1979.
Nucleotide sequence analysis oftwo simian virus 40mutants
withdeletions in the region coding for the carboxyl terminus of
the T antigen. J. Virol. 30:936-941.
68. White, R. T., P. Berg, and L. P. Villarreal. 1982. Simian virus
40-rabbitB-globin recombinants lacking late mRNA splice sites
express cytoplasmic RNAs with altered structures. J. Virol. 42:262-274.