Vol.66,No. 12 JOURNALOFVIROLOGY,Dec. 1992,p. 6922-6930
0022-538X/92/126922-09$02.00/0
Copyright© 1992, AmericanSociety for Microbiology
Assembly
of Viruslike
Particles
by
Recombinant Structural
Proteins of
Adeno-Associated Virus Type
2
in
Insect
Cells
MICHAELRUFFING,HANSWALTERZENTGRAF, ANDJURGEN A. KLEINSCHMIDT* DeutschesKrebsforschungszentrum, Forschungsschwerpunkt
Angewandte Tumorvirologie,
ImNeuenheimer Feld 242, D-6900
Heidelberg,
GennanyReceived29 June1992/Accepted24August1992
The three capsidproteinsVP1,VP2, and VP3 of the adeno-associated virus type 2 (AAV-2)areencodedby
overlapping sequences of thesameopenreading frame.Separate expressionof theseproteins byrecombinant baculovirusesininsect cellswasachievedbymutationof the internal translation initiation codons.
Coexpres-sion of VP1 andVP2, VP2 andVP3,and all threecapsid proteinsand theexpressionof VP2 alone inSf9cells resulted in theproduction of viruslike particlesresemblingempty capsids generated duringinfection ofHeLa cells withAAV-2 and adenovirus. These results suggest a requirementfor VP2 in theformation ofempty
capsids. Individual expression of the AAVcapsid proteinsin HeLacells showed that VP1 and VP2 accumulate in thecell nucleus and VP3 is distributed between nucleus andcytoplasm.Coexpressionof VP3 with the other structuralproteins also ledtonuclearlocalization ofVP3, indicatingthat the formation ofacomplexwithVP1
orVP2 is required for accumulation of VP3inthe nucleus.
The adeno-associated virus(AAV), ahumanparvovirus,
hasasingle-stranded DNA genomepackagedinan icosahe-dralcapsid with a diameter of 20to24nm(forareview,see
reference3).Thecapsid consistsof three structuralproteins, VP1,VP2, and VP3, with molecularweightsof90, 72,and 60 kDa, respectively. They are expressed from overlapping
sequencesof thesameopenreadingframebyusing alterna-tive initiation codons (1, 2, 28).A2.6-kb precursor RNA is encoded by the right half of the AAV type 2 (AAV-2)
genome (18, 19) and is spliced into two 2.3-kb mRNAs by usingthesamedonorsitebut different acceptor sites. In the presenceofadenovirus,mostof the2.6-kbRNAissplicedto
the acceptorsite atnucleotide 2228 (major
splice),
whereas theminorityof thetranscript isspliced to amore-proximalacceptorsiteatnucleotide 2201(minor splice; 1, 6, 45).VP1 isexpressed fromanAUG initiation codon atposition2203 which is present only in theminor-splice RNA, while VP2 and VP3 can be expressed from both differentially spliced
mRNAs.VP3isefficientlytranslated fromanAUGinitiation codon at nucleotide 2809, and VP2 is expressed about 10-fold less efficiently from an unusual initiation codon,
ACG, at position 2614. The relative accumulation of the
alternatively spliced mRNAs together with the reduced
translationinitiation frequency attheACG initiation codon leadstothesynthesis of VP1,VP2,and VP3 inrelative ratios of about 1:1:10, the mass ratio which is found in mature AAVvirusparticles (5).
Kinetic experiments allow us to distinguish two steps in theassemblyprocess ofinfectious AAVparticles: (i) rapid assembly of empty capsids and (ii) packaging of single-stranded DNA into preformed capsids (30)! While capsid assembly and association with the viral DNA are rapid processes,maturation to infectious particles requires several hours. It is not known which proteins compose the empty
capsids and whether additional proteins are required for DNApackaging. VP2 and VP3 are sufficient for accumula-tionorsequestration of single-stranded progeny DNA. VP1,
* Correspondingauthor.
however, is required for production of infectious particles (15, 37, 44).
Packaging signals have been postulated to be present in the inverted terminal repeats, since defective interfering particles contain at least one terminal repeat (9). This
assumption has been corroborated by the successful pack-agingof DNAwhich containedonlytheterminal repeats of the AAV genome into AAVparticles(35).
In order to study the requirements for the synthesis of
emptycapsids,weexpressedthe threecapsid proteins VP1, VP2, andVP3separatelyand in combination in
Spodoptera
frugiperda Sf9cellsbyusingbaculovirusexpressionvectors.
Ashas beenobservedwithautonomousparvoviruses (4, 20, 34), this resulted in formation of empty viruslike particles (VLPs). Unexpectedly, however,there seemedtobeastrict requirement for the minor capsid component, VP2, for detectable formation of VLPs in Sf9 cells. We failed to
observe capsid formation with the proteins VP1 and VP3 alone. Separate expression ofthe capsid proteins in HeLa cells showed that VP1 and VP2were abletoaccumulatein the cell nucleus, whereas VP3 was evenly distributed
be-tween nucleus and cytoplasm. Coexpression of VP3 with VP1and/or VP2, however,resulted inaccumulation of VP3 in the nucleus, too, suggesting that AAV capsid proteins need to form complexes for efficient nuclear accumulation andparticle assembly.
MATERIALSANDMETHODS
Mutagenesisandconstructionofrecombinantplasmids. For
oligonucleotide-directed
mutagenesis, a 2.9-kb fragmentcomprisingtheright half of the AAV-2 genome starting from nucleotide 1882 was isolated from the HindIII- and SstI-digestedpTAV-2plasmid (14)andcloned into theM13mpl8
vector. This DNA fragment contains the complete capsid
protein-coding sequences. For separate expression of the structuralproteins,the internal translation initiation codons
weremutatedasindicatedinFig. 1 essentially according to the methods described in reference 41 byusing an invitro
mutagenesiskit(Amersham,Braunschweig,Germany).
Mu-tants were identified byDNA sequencing according tothe
6922
on November 9, 2019 by guest
http://jvi.asm.org/
MV-2
genome organization
Pn4
0
I~~~~~~~~~II --I a~~~~~~~~
AAV-2 VP
coding region
jiO
f614r--U-I
ATQ A* ATG 1 iM T M
936 f814
I3
i"i
IATG
GCG CTGM A L
ATG CTG TM
M L
I
EmIIiiIT
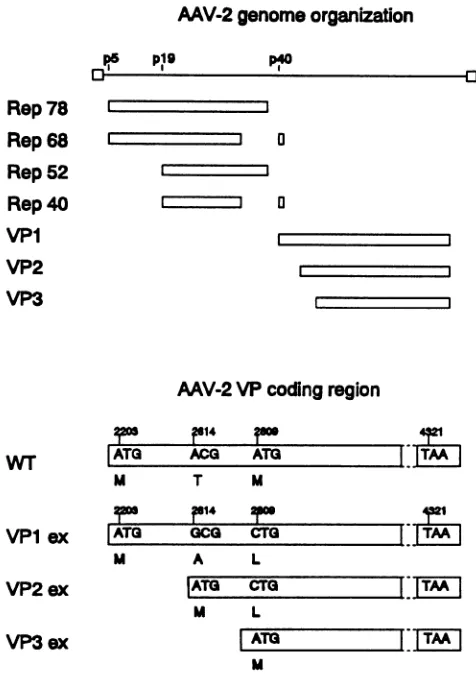
ATO | |TM |FIG. 1. AAV-2 genome organization and mutant VP coding
regions used for protein expression. (Top) AAV-2 genome, with inverted terminalrepeatsindicatedbyopenboxesattheends of the
genomeand the threepromotors(p)atmapunitpositions 5, 19,and 40.The coding regions for regulatory proteins Rep78, Rep68, Rep52, and Rep4O and structural proteins VP1,VP2,and VP3areshownby
openrectangular boxes. (Bottom)Overlappingopenreadingframes ofthe three capsid proteins with the translation initiation codons which code for the amino acids M, T, andM in thewild-type(WT)
genome.ForseparateexpressionofVP1, VP2,and VP3 in different expressionvectors, these initiation codonswere mutated as indi-cated in boxes VPlex, VP2ex, and VP3ex. Translationwas termi-nated in allconstructsatthesamestopcodonatposition4321.
method of Sanger et al. (36). In plasmid M13mpl8-VPlex,
the ACG translation initiation codon (nucleotides 2614 to 2616, according to reference 6)was changed to GCG, and ATG (nucleotides 2809 to 2811)was changed to CTG. In M13mpl8-VP2ex, ACG (nucleotides 2614 to 2616) was
changed to ATG, andanNdeI restriction site was
simulta-neously introducedby changingnucleotide 2611 from AtoC
and nucleotide 2613 from Gto T. ATG at nucleotide posi-tions2809to2811wasmutatedtoCTG. InM13mpl8-VP3ex
attheATG translationinitiation codon(nucleotides 2809 to 2811), anNdeI restriction sitewas introduced by mutating nucleotides 2806 to 2808 from ACG to CAT. The plasmid pVL-Vpl was constructed by insertion of a DraI-XbaI
fragment (DraIsite bluntended)fromM13mpl8-VPlexinto
theSmaI-XbaI-digestedbaculovirus vectorpVL1393, a
de-rivative of the plasmid pVL941 (27). Generation of the
plasmids pVL-Vp2andpVL-Vp3wasperformedby cloning NdeI-BalI fragments (nucleotides 2614 to 4552 and
nucleo-tides 2809 to 4552, respectively; NdeI site blunt ended) into
the SmaI site of pVL1393. The junction sequences were confirmed by dideoxy sequence analysis.
For expressionof the AAV capsid proteins in HeLa cells, thecorrespondingmutated viralcapsid protein (VP)-coding regions were inserted into the expression vector pKEX-2-XR(32). The plasmid pKEX-VP1 was generated by
liga-tionof a BamHI-XbaI(BamHI site blunt ended) fragment of pVL-VPl into pKEX-2-XR digested with XhoI and XbaI
(XhoIsite also blunt ended). The constructspKEX-VP2and pKEX-VP3 resulted from cloning the respective NdeI-XbaI fragments from M13mpl8-VP2ex and M13mpl8-VP3ex into the plasmid pUC131(kindlyprovided by H. Schaller, Zen-trum fur Molekulare Biologie, Heidelberg, Germany). The plasmids pUC-VP2 and pUC-VP3 were digested with ClaI andXbaI, respectively. The resulting 2.1- and 1.8-kb DNA fragments were inserted into pKEX-2-XR digested with the sameenzymes.
Cell culture andvirus stocks.S.
fnrgiperda
Sf9cells were grown as suspension or monolayer cultures in TNMFH medium(16)with10%fetal calf serum, 100 ,ug of gentamicin perml, and 2 mMglutamineat27°Cand95% humidity. For generation of recombinant baculovirusstocks,1x107
to5 x107 cells in suspension were infected for 1 h at 27°C at a
multiplicityof infection(MOI)of 2to5.Afterinfection,cells
were transferred into Spinner flasks and stirred for 5 to 6 daysat27°C. Cellswereremovedbycentrifugation, and the
supernatantwith the recombinant baculoviruseswasstored at 4°C. For expression of the capsid proteins,
106
Sf9 cells from a monolayer culture were infected by baculovirus recombinants for 1 h at 27°C at an MOI of 5 to 10. After infection, cells were incubated for 48 hat27°C.Mediumwasremoved, cellswerewashed with phosphate-bufferedsaline
(PBS; 140 mMNaCl, 6.5 mM
Na2HPO4,
2.5 mM KCI, 1.5mM
KH2PO4,
pH7.4), scraped off, andresuspended
in 150,ulofsample buffer(24) for protein
analysis by
electropho-resis.
HeLa cells were propagated in Dulbecco's modified Ea-gle's medium with 5% fetal calf serum, 100 ,ug each of
penicillin and streptomycin perml, and 2 mM
glutamine
at37°C and 5% CO2. For
generation
of AAV-2(strain H;
AmericanTypeCulture
Collection) stocks,
cellsweregrownto 40 to 60%
confluency
(5
x 104 cells percm2).
After themediumwasremoved,cellswerefirst infected for 1 hat
37°C
with AAV-2 (MOI = 10) and then with adenovirus(MOI
=2) under the same conditions. When morethan 90% of the HeLa cells showed a cytopathic
effect,
cellswere removed bycentrifugation. Supernatantwasincubated for 1 hat56°C
forinactivation ofadenovirus and then stored at-20°C.
Generation of recombinant baculoviruses. Recombinant
pVLplasmids wereused togenerate recombinant baculov-irusesaccordingtothe
protocol
ofSummers and Smith(40).
For transfection of Sf9 cells withrecombinantpVL
plasmids
and wild-typeAutographa califomica
nuclearpolyhedrosis
virusDNA,acalciumphosphate
precipitation
technique
was used. Recombinant baculoviruseswerepurified
by
aplaque
assayprocedure.
Analysis
of protein expression. Foranalysis
ofprotein
expression bygel
electrophoresis,
2x106
pelleted cellswerelysed byheat at
100°C
for 5 min in 150 ,ul ofsample
buffer(24).Aftersonication
(10
s atlevel4;
BransonSonifier),
total cellularpolypeptides
wereanalyzed
by
electrophoresis
on a15% polyacrylamide
gel
in the presence ofsodiumdodecyl
sulfate
(SDS)
as described in reference 42. Proteins were visualized bystaining
with0.2% Coomassieblue. ForWest-ernblot
(immunoblot)
analysis,
proteins
wereelectrophoret-e5 p19
Rep
78
Rep
68
Rep
52
Rep
40VP1
VP2
VP3
WT
vpl
exVP2
exVP3
exon November 9, 2019 by guest
http://jvi.asm.org/
[image:2.612.62.300.76.415.2]6924 RUFFING ET AL.
ically transferred to nitrocellulose membranes (43). AAV
capsid proteinswere detected by incubations withan anti-VP3 antiserum andaphosphatase-coupled secondantibody (13).
Preparationof anti-VP3serum.The DNAfragment coding
for the AAV-2 VP3 capsid protein was isolated from the
M13mpl8-VP3ex plasmid by digestion withNdeI andBalI and cloned into the Escherichia coli expression vector pET3a (39).E.coliBL21(DE3) cells(39) transformedwith the recombinantplasmid pET-VP3 expressedthe VP3capsid protein.Cellswerelysed bysonication inlysisbuffer(50mM Tris [pH 8],5mMMgCl2, 1 mMEDTA,100 mMNaCl,2.5 mM dithiothreitol, 1 mg of lysozyme per ml), and the unsolubleproteinswere spundown at4,000 x gfor5min.
After thepellethad been heatedfor5minat100°Cinsample buffer, polypeptideswereseparated by SDS-polyacrylamide gel electrophoresis (PAGE) and visualizedby stainingwith 1% Coomassie blue inwater. The region corresponding to the VP3proteinwasexcised, andtheproteinwaseluted in 0.02% SDSovernight atroomtemperature. Gelpieceswere
spun down, and proteins in the supernatant were
precip-itatedbyacetone.VP3(100to200,ug)wassuspendedinPBS for each immunization, mixed with an equal volume of Freund'sadjuvant,andinjected subcutaneouslyintoarabbit
(chinchilla bastard). The rabbitwas immunized three times in3 months. One week after the thirdimmunization, serum wastestedbyan immunoblot analysis.
Preparation ofVLPs and electron microscopy. Sf9 cells
(2 x 108 to 5 x 108) were (co-)infected with recombinant baculoviruses at an MOI of 10 to 20. At 70 to 72 h
postinfection, cells were harvested by centrifugation and
suspended in 1 ml of a buffer containing 1% deoxycholate (DOC),0.1%SDS, 10 mM Tris(pH 8),0.1 mM EDTAand 2 mM phenylmethylsulfonyl fluoride. After sonication, cell debriswereremovedby centrifugation (12,000 x g, 5min),
and the supernatant was layered onto a double sucrose
cushion of 50 and 30% sucrose (wt/vol) in PBS. After
centrifugationat 100,000xgfor 2h,theparticle-containing
sediment was resuspended in PBS and centrifuged again (200,000 xg, 1 h).VLPswere resuspendedinPBS,stained
with 2% uranyl aqueous acetate, and examined in a Zeiss
EM10 electronmicroscope.Themagnificationwasroutinely
controlledby using a grating replica. In parallel, the same
fractionswere analyzed by SDS-PAGE and Western
blot-ting.
Purification ofAAV-2 virus particles. AAV-2 virus stock
was prepared as described above. A 12-ml portion of this
stockwasspundownfor 1 hat100,000x g.Thesedimented
viralparticlesweresuspendedin 200 ,ulof10mMTris (pH
8.0)-i mMEDTA (TE buffer) with 1% DOCfor30 min at 37°C anddiluted 1:10 with TE buffer. CsClwas addedto a
finaldensityof1.4g/cm3,andequilibrium centrifugationwas performed in a TST60-4 rotor (65 h, 35,000 rpm, 20°C; KontronT-1065ultracentrifuge). Fractions withadensity of
1.409 to 1.459 g/cm3 were collected, dialyzed against TE buffer, and analyzed byelectronmicroscopy.
Transfection of HeLacells andimmunofluorescence. Trans-fection ofHeLacellswasperformed by the protocol of Chen
andOkayama (7)inaslightly modified form. Briefly,the day beforetransfection,4 x 105cellswereseeded inapetri dish
(diameter, 9.4 cm) with 8 ml of medium and incubated at
37°C and 5% CO2. DNA(15 p,g) was mixed with 2x BBS buffer(7) (250 ,ul)towhichCaCl2 had been addedto afinal
concentration of175 mM andincubated for20minat room
temperature. The transfection solution was added to the cells,andcellswereincubated for18to22 hat35°Cand 3%
CO2. After the supernatant had been removed, cells were
washed twice with serum-free medium and then incubated in medium with fetal calfserumfor 24 hat37°C and 5% CO2. Forimmunofluorescence, cellsweregrown oncoverslips in tissue culture dishes and transfected as described above. After thecoverslipshad been washed inPBS, cellswerefirst fixed for 10 minat-20°C in methanol and then for 5minat -20°C in acetone. Cells were airdried, rehydrated in PBS for 5 minat roomtemperature, and then incubated with the anti-VP3 serum (diluted 1:50 in PBS with 1% bovine serum albumin [BSA]) for 15 minat roomtemperature. Cells were washed three times in PBS and then incubated with a
fluoresceinisothiocyanate-conjugated goat anti-rabbit
immu-noglobulinserum(Nordic, Talburg, Germany)diluted 1:50 in PBS with 1% BSA. Afterbeingwashed in PBS, coverslips
were embeddedin Elvanol (1 gof polyvinylalcohol mixed
with 8 ml of PBS and 4 ml of 86% glycerol, incubated
overnightat80°C,and storedat-20°C)and visualized under
aLeitz UVmicroscope.
RESULTS
Separate expression of individual AAV-2capsidproteins. In order toanalyze the contributions of the threeviral capsid proteins of AAV-2tothe assemblyofVLPs, we expressed VP1, VP2, and VP3 separately. For separate expression of VP1,westarted withasubfragmentofplasmid pTAV-2(14) which contained thecompleteopenreading framecodingfor all threecapsid proteinsandchangedtheinternal translation
start sites of VP2 and VP3at nucleotidepositions2614 and 2809toGCG andCTG, respectively, bysite-directed
muta-genesis(Fig. 1, VPlex; numbering accordingtoreference6).
These mutations introduceda change of aminoacid 138 of VP1 fromItoA and of aminoacid 202 fromMtoL. For separate expression of VP2, we mutated its unusual ACG translation initiation codon to ATG, thereby also changing the N-terminal amino acid I to M (Fig. 1, VP2ex). The
internalstartsiteof VP3 at 2809 was also mutated to CTG to prevent simultaneous expression of VP3. In addition, we introduced auniqueNdeI restriction site at the new trans-lation initiation codon ofVP2, which allowed the separate
subcloning of the VP2 open reading frame into different
expressionvectors. Similarly, the translation codon of VP3
was converted into a unique NdeI restriction site, which allowedexpressionof VP3withoutVP1and VP2 after VP3
was subcloned into a suitable expression vector (Fig. 1,
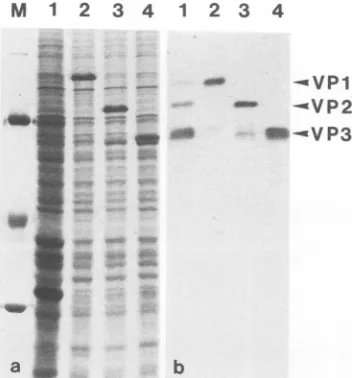
VP3ex). Thethree open reading frames were expressed in Sf9 cells under the control of the baculovirus polyhedrin promoter. Analysis of proteins of total cell extracts by SDS-PAGE shows that VP1, VP2, and VP3 were strongly
expressed
(Fig.
2a, lanes2, 3,and4),constituting up to 5% of total cellular protein compared with extracts of cellsexpressing only wild-typebaculovirus proteins (Fig. 2a, lane
1).Therelative molecular weights of the recombinant capsid
proteinscorrespondtothose of the viral capsid as shown by Western blot analysis (Fig. 2b). The kinetics of AAV-2
capsid protein expression inSf9 cells showed that levels of VP1andVP3 reachedamaximumabout72 h after infection, whereas the VP2 contentdid not increase significantly after 48 h(datanotshown).In extractsof cells expressing VP1 or
VP2, variable amounts of polypeptides in the molecular
weightrangeof VP3which reacted with our anti-VP3 serum
(Fig.
2b, lane 2,3)could be observed. These and productswith even lower molecular weights were more evident in VLPpreparationsfrom cells expressingVP1 and VP2(Fig. 3a) or VP2 alone (data not shown). In addition, in cells
J. VIROL.
on November 9, 2019 by guest
http://jvi.asm.org/
CAPSID ASSEMBLY OF AAV 6925
M 1 2 3 4 1 2 3 4
1
2
2'
3
3'
4
41
5
5'
X--
-VP1t_ -~~VP3 VP1
VP2
VP3;:.
[image:4.612.99.275.78.267.2]*1
aFIG. 2. Expressionof AAV-2 structuralproteinsV'Pl, VP2,and VP3ininsectcellsbyrecombinantbaculoviruses.(a)Totalproteins
of S.
frugiperda
Sf9 cells infected with A.califomnica
nuclearpolyhedrosisvirus(lane 1)or'VP1(lane2),VP2(lane 3),and'VT3
(lane 4)recombinant baculoviruswereseparated bySDS-PAGE and stained withCoomassie blue. Cellswereharvested 48 h
postinfec-tion. Molecularweightmarkers(lane M):
0-galactosidase
(115 kDa),BSA(68 kDa), egg albumin (45kDa), and carbonicanhydrase (29 kDa). (b)shows thecorrespondingimmunoblotanalysisofextracts of insect cells infected with recombinant baculovirus expressing
VP1(lane 2),VP2(lane 3),or'VP3 (lane 4)witharabbit antiserum raised against 'VP3. For comparison, an extract of HeLa cells
infected with AAV-2 and adenovirus-2 isanalyzed in lane 1.
.-b
expressing solely
IVT3,
the resultantoverexpressed
VP3protein
oftenappeared
as a doublepolypeptide
band onSDS-PAGE
(Fig.
2a, lane4). Although
the variable appear A. ances of thesepolypeptides
suggest thatthey
aredegrada-tion
products
ofVP1, VP2, or VP3,wecannot exclude thepossibility
thatthey
arise from usage ofalternative initiationcodons
(for example,
at nucleotidepositions
2833 to2835)
neartheauthentic translation initiation codon AUG for'VP3
~~~~~~~~~~~~~~~~~~a
Assembly
ofVLPs byrecombinantcapsid
proteins. Individually
expressed capsid proteins
accumulated as insolubleaggregates in Sf9 cells.
They
were recovered in alow-speed-centrifugation
sediment of cell extractsS;rprepared
bysonication of the cells in buffers with
physiological
salt concentrations and neutralpH.
Addition of low- andhigh-ionic-strength
buffers or low concentrations of differentC [image:4.612.325.563.87.712.2]detergents
ororganic
solvents did not solubilize thecapsid
FIG. 3. Analysisofcapsidformationbycoexpressionofdifferent structural proteins in insect cells. The insoluble fractions of soni-cated Sf9cells containingrecombinant AAV-2capsidproteinswere treated with 0.1% SDS and 1% DOC. Soluble(a,lanes2',3',4',and
5')and insoluble(a,lanes 3,4,and5)materialswereanalyzed
by;;
-in gunoblottingwithan anti-VP3 serum.Lane1,authenticAAV-2
capsid proteins produced in HeLa cells infected with AAV-2 and
3 and 3', cells coexpressing'V1 and 'VP3; lanes 4 and 4', cells j.'
coexpressingVP2and
VP3;
and lanes5and 5', cellscoexpressing
VP1,VP2andVP3.Electronmicrographs
areofnegatively
stainedempty-capsid-like
structures recovered from the solubilized frac- 'tionsof insectcellscoexpressingV V1andVP2(b) (cf. panela, lane
d
2'),VP2 and Vt3
(c)
(cf. panela,lane4'),andAVU1,
T2,andVP3(d)
(cf.
panela,lane5').Bars = 50nm.VOL. 66, 1992
on November 9, 2019 by guest
http://jvi.asm.org/
6926 RUFFING ET AL.
TABLE 1. Detection ofVLPs in insect cellsexpressing AAV-2capsid proteins
Capsidprotein(s) presentps
VP1(+ VP3)a*...
-VP2(+VP3)a... ... +
VP3.-VP1 +
VP3.-VP1+VP2(+VP3)a.+
VP2+VP3.+
VP1+ VP2 +VP3. +
aInthese cases, we observedpolypeptides in the molecularweightrange of VP3byWesternblotting, althoughcellswere notinfected withVP3
recom-binant baculoviruses.
proteins to asignificantextent. Highconcentrations ofurea
(8 M) or 1% SDS, which denatured the proteins, also
solubilizedthem. A combination of0.1%SDSand1%DOC, however, by which it was possible to solubilize VLPs of rotavirus from Sf9 cells expressing recombinant structural
proteins,
alsosolubilized the AAVcapsid proteinspresentascapsidlikestructures.
VLPscould beprepared from cellscoexpressingVP1and
VP2, VP2 and VP3, or all three capsid proteins (Fig. 3, b
through d) by sedimentation of the solubilized material
throughadoublesucrosecushion and resedimentation of the
resuspended pellet. The same type of particles could be solubilized from cellsexpressingVP2onlybutnotfrom cells
expressingVP1onlyorVP3only (datanot
shown). Analysis
of protein expression by Western blotting confirmed the presence ofpolypeptides with the correctsize in theSDS-DOC-solubilized and -purified particles and in the corre-sponding insolublefraction, althoughwith different relative
proportions (Fig. 3a, lanes 2, 2', 4, 4', 5, and 5'). The detection of immunologically cross-reactive polypeptides comigrating with VP3 in extracts of cells whichwere not
infected withVP3 recombinantbaculoviruses (for example, Fig. 3a,lanes 2 and2') complicates theinterpretationwhen VP1 and VP2 or VP2 alone was expressed. Although the
origin andnatureof thesepolypeptidesareunclear,asstated
above,wecannotexclude thepossibilityofacontribution of these VP3-likemolecules toempty-capsid formation.
Coex-pression of VP1 and VP3, however, did not promote the formation ofempty-capsid-like structureswhich were solu-ble in0.1% SDS-1% DOC(Fig. 3a, lanes 3 and3'), which suggeststhatVP3is notsupportingcapsid formation. On the contrary, whenever VP2 is coexpressed with any other
capsid protein, we observed empty-capsid-like structures
(Table 1). A detailed comparison of the particles derived from recombinantproteins(Fig.4c, d,ande)with different
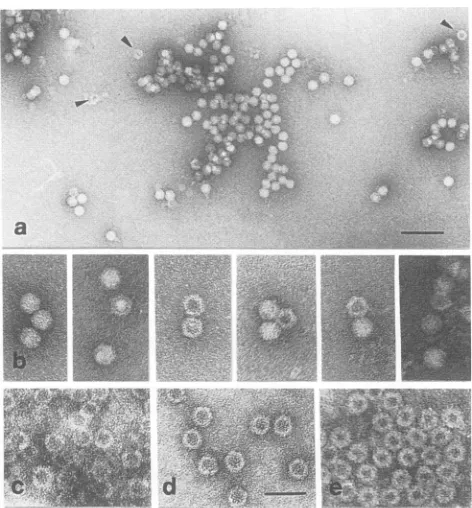
images obtained from AAV-2 virus preparations (Fig.4a and
b) shows that in addition to the full particles in the virus preparation, there are also empty capsids (arrowheads in
Fig. 4a and some examples in Fig. 4b) with morphologies verysimilartothose ofcapsids obtained in Sf9 cells.
Subceliular
localization of AAV capsid proteins. Since itwas difficult to determine the subcellular distribution of
individually expressed capsid proteins in Sf9 cells, we
ex-pressed themunder the control of the human
cytomegalovi-rus (HCMV) immediate-early promoter-enhancer in HeLa cells
(Fig.
5,lanes 2, 3, and4)and localized the proteins byindirect immunofluorescence. Besides the expressed capsid
proteinsofexpected sizes(Fig. 5, lanes 2 through 4,
arrow-heads), we observed in all cases lower-molecular-weight
products (Fig. 5, unlabeled bands), which are especially
a
FIG. 4. Comparison of reconstituted and wild-type capsid struc-tures of AAV-2. Different images of negatively stained AAV-2 capsid particlespresentin anAAV-2 virus preparationwere com-pared withemptycapsids formed in Sf9 cells after coinfection with recombinant baculoviruses expressingVP1 and VP2(c), VP2 and VP3 (d), andVP1, VP2, and VP3(e). Arrowsin panel a indicate ringlikestructureswhichprobablyrepresent emptycapsids,someof which areshownindetails inpanelb. Specimenswerenegatively stainedwithuranylacetate. Barscorrespond to100nminpanel a and40nminpanelsbthrough e.
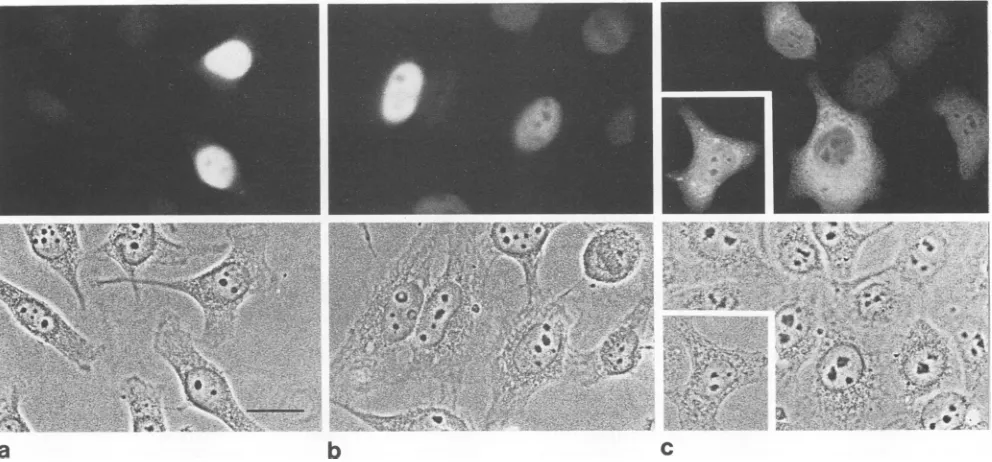
pronounced in thecaseof VP1 expression (Fig. 5, lane 2). Since these
expression products
didnot interfere with our conclusions drawn from the subcellular localizationdata,wedid not investigate their identities any further. While VP1 and VP2efficientlyaccumulated in the cell nucleus(Fig. 6a and b), VP3 was distributed throughout the nucleus and
cytoplasmand seemed insome cases evenpartiallyexcluded from the nucleus (Fig. 6c). In about 25% of VP3-positive cells,weobservedagranular stainingin thecytoplasm(Fig.
6c, inset). The nuclear staining of VP1 and VP2 is rather
homogeneousandnotin the clusters observed in HeLa cells infected with AAV-2 andadenovirus(17). In the presence of
coexpressedVP1orVP2 orboth, VP3 disappears from the
cytoplasm, suggestingthatit is accumulated in the nucleus
(Fig. 7). This assumption is supported by Western blot analysis,which shows that VP3 is notdegraded (Fig. 5, lanes
6, 7,and8). Especiallywhen VP1 and VP3arecoexpressed,
VP3 ismoreabundant in the cells thanVP1 (Fig. 5, lane 6);
thus, it should bepossibleto detectcytoplasmicstainingof VP3 if theprotein is not transported into the nucleus(Fig.
7b).This result suggests thatcomplexing of VP1 and/or VP2 with themajor capsid protein VP3has to occur in order for VP3 to accumulate in the nucleus, the site of viral DNA
packaging.
DISCUSSION
Assembly
ofAAV-2 VLPs ininsect cells. The threecapsidproteins of AAV-2 are encoded by overlapping reading J.VIROL.
VP,
MC:
on November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.319.555.77.331.2]1 2 3 4
VP1o.- '
VP2
-
-ft _
VP3wm-5 6 7 8
I I
V '
[image:6.612.110.264.77.234.2]v
FIG. 5. Western blot analysis ofAAV-2capsid protein expres-sionin HeLacellsunder thecontrol of theHCMVimmediate-early promoter-enhancer. Lane 1showselectrophoretic positions ofVP1, VP2, and VP3 polypeptides present in an extract ofHeLa cells coinfected with AAV-2 and adenovirus-2 afterseparation by SDS-PAGE and detection withananti-VP3serum. VP1,VP2,andVP3 individually expressed inHeLacells underthecontrol ofthe HCMV immediate-early promoterareshown in lanes2, 3, and 4, respec-tively. Expressedcapsid proteins ofcorrectsizes areindicatedby arrowheads, whereas lower-molecular-weight products which are also recognizedby the antiserumare notmarked.Coexpression of VP1 andVP2(lane5),VP1andVP3(lane 6),VP2andVP3(lane7),
andVP1, VP2, and VP3(lane8)arealsoshown.
frames, resulting in obligatory coexpression of the three
proteins in the wild-type situation. In order to achieve separate expression of single capsid
proteins,
we mutated theinternaltranslation initiationsites of VP2 and VP3. This resulted in achangeof two amino acids in VP1 (I 138toA,.JK;a'.4:''
1Q.~~~~~ ~~~~~~~~or
~
~
~
~
:
7
a
b
andM202toL)and twoamino acidsin VP2(I1 toM,and M 64 toL). The amino acid sequence of VP3 was not altered. By choosing conservative substitutions, we tried to mini-mize theinfluence on the capsid protein structure andcapsid assembly. Using these mutant open reading frames, we expressed thethree capsid proteins in all possible combina-tions by recombinant baculoviruses. Unexpectedly, when-everVP2 was expressed, we observed formation of VLPs, whereas in cells expressing VP1 or VP3 or both, we did not detect any viruslike structures. There may be several rea-sons for these occurrences. It is possible that particles are formed by VP1 and/or VP3 but are not stable in the presence of the detergents used duringthe particle preparation proce-dure. Alternatively, the particles might not be solubilized by 0.1% SDS and 1% DOC. Our Western blot control does not distinguish between these two possibilities. A number of alternativeprocedures for solubilizing the expressed capsid proteins, including VP2, in anative state have failed so far (data not shown). Since both VP1 and VP3 assembled into VLPsin thepresence of VP2, we tend to favor the interpre-tation that VP2 somehow promotes capsid assembly, per-haps by preventing VP1 and VP3 from forming insoluble aggregates. However, we cannot completely rule out the
possibility that VP2 only facilitates the accessibility ofthe assembledviruslike structuresunder theconditions used in this study.
We also cannot definitively state that VP2 alone or VP1 and VP2aloneformedthecapsidlikestructures isolated from insect cells expressing these proteins. In all particle prepa-rations from cells which should not express VP3, we de-tected polypeptides in the molecular weight range ofVP3 that cross-reacted with our antiserum. Thepossibility of a
contributionofthesemolecules to particle formation cannot be excluded, because the presence of VP3 subspecies has also been observed in purified virions and empty capsids
__~~~~J
il F~~~*A
-
e,y
-#--w S k. ~~~~~~~~~~~~~~~~.J
C
iX
4h
FIG. 6. Subcellularlocalization ofindividually expressedAAV-2capsid proteinsinHeLacells. Plasmidsexpressing VP1,VP2,orVP3 (Fig. 1) under the control of the HCMVimmediate-early promoter-enhancerweretransfected into HeLacells.Thesubcellulardistributions ofVP1(a),VP2(b),andVP3(c)wereanalyzedbyindirectimmunofluorescencewithananti-VP3serum.The
corresponding phase-contrast
picturesareshownineachpanel. Bar inpanelacorresponds to20pm.
on November 9, 2019 by guest
http://jvi.asm.org/
[image:6.612.69.565.455.686.2]6928 RUFFING ET AL.
.
-.
r::< tt :.
S
,
,\..
jS, u.:...
.....o'
:,:'.?..'
*, ^ ;5:.. ,.w
.. /
et
w
.K
*,r's
'4
*'.,2:...'',...'-"'
..e..qif
'.. Z.,-"'.,.'
:
e
; :. .:
..
- ..
*...
1W..E
^
CX ,
b
I
_.,
d
FIG. 7. CoexpressionofVP1orVP2withVP3leadstonuclear accumulationof VP3. Variouscombinations ofplasmidsforindividual expression of VP1, VP2,orVP3 under the control of the HCMVimmediate-earlypromoter-enhancerwerecotransfected into HeLacells. Nuclear accumulation ofcapsidproteinswasanalyzed byindirect immunofluorescence withananti-VP3serum.(a) CoexpressionofVP1and VP2;(b)coexpression of VP1 andVP3;(c) coexpressionofVP2 andVP3;(d) coexpressionofVP1, VP2,and VP3. Bar inpanelacorresponds
to20
pum.
(28). The nature of these N- and/or C-terminal-truncated copy.
Nevertheless,
inspite
ofthepossible
differencesatthe VP1orVP2 molecules has notyetbeen clarified. molecular level, thestructures of VLPsand emptycapsids Preparations of VLPs from insect cells infected with found in AAVpreparationsarestrikinglysimilar. This result different combinations of recombinant baculoviruses alsoconfirms that the mutations introduced forsingle
capsid
showed different relative proportions of theexpressed pro- proteinexpression donotpreventcapsidassembly.
teins. In addition, there is acleardiscrepancy between the Expression of VP2, the major capsid protein of humanstoichiometry of the capsid proteins in VLPs from insect parvovirus B19 (4,
20)
and canine parvovirus(34),
led tocells and inpurifiedwild-typeAAV-2virions. Whereas the particleformation ininsect cells. Thiscontrastswith the fact
ratio of thecapsid proteins VP1, VP2,and VP3 is approxi- thatwecouldnotdetect AAVVLPsincellsexpressing VP3, mately 1:1:10 in infectious AAV particles (5), the relative the major capsidprotein of AAV-2. On the otherhand, it is
amountofVP3 isalways reduced in the VLP preparations. alsotheexpression ofVP2,themiddle-sizedcapsid protein, Inprinciple, this difference may be explained by the lack of which allows formation ofempty-capsid-like structures of DNA in the VLP preparations. It is more plausible, how- AAV-2,although it represents a minor structural component ever, that the capsid protein stoichiometry is controlled by in this virus.
the supply ofcapsid proteins for the assembly reaction. It Subcellular localization of capsid proteins.
Immunolocal-cannotbe deduced from our experiments that the stoichiom- ization of individually expressed capsid proteins clearly etryofcapsid proteinsin theparticle preparations detected shows that VP1 and VP2 efficiently accumulate in the cell
by Western blotting
corresponds
to the stoichiometry of nucleus, while VP3 equilibrates only between nucleus andcapsid proteins incorporated into empty capsids. Itis diffi- cytoplasm.
This
result is remarkable, because VP3 is too culttocorrelateaquantitativelyrepresentative protein anal- large topassively diffuse through nuclear pores(10). On theysis of a fraction by SDS-PAGE with a rather selective otherhand, VP3 hasnobasic sequenceswhich could allow
visualizationof structures in this fraction by electron micros- selective entry into the nucleus. In about 25% of the cells J. VIROL.
11
OP '.
1. .".
0'. ...
fi .i J
0 '.
I I
I
on November 9, 2019 by guest
http://jvi.asm.org/
transfected by VP3, we observed a granular appearance of VP3 in thecytoplasmthatindicatesaggregationwithitself or withcytoplasmic structures.
Coexpression ofVP1 orVP2with VP3 led tothe disap-pearance of VP3 from thecytoplasmandalso ofthe granular structures. Since VP3 is not degraded in coexpression
ex-periments, one has to assume that itis accumulated in the nucleus either by cotransport or by trapping within the
nucleus due to complex formation with VP1 or VP2.
Cotransport ofpolypeptides lacking a nuclear localization
signal by complex formation with polypeptides which
ac-tivelyaccumulated in thenucleus wasimplicatedin several reports(29, 46, 48).
The AAV-2capsid proteinsVP1 andVP2contain several basic amino acid stretches with some similarity to the nuclear localization signal of the simian virus 40 large-T antigen (PKKKRKV; 12, 21, 22, 25, 26). However, theyare shorter and separated from each other by sequences of different
lengths.
VP1, forexample, contains the sequence AKKRV(positions
121 to125),
and the overlapping openreadingframe of VP1 andVP2 containsthe motif GKKRP
(positions
141to145)
and the motif ARKRL(positions167 to171).For several viral andcellular nuclearproteins,bipartite
nuclear localization
signals
withsuchshort basicstretchesofamino acids have been described(23, 31, 33, 47).
Inarecentpublication,Hunter and Samulski(17)showed that the AAV structural proteins concentrate in nuclear
subcompartments
whencellsarecoinfected with AAV-2 andadenovirus 2. This additional
compartmentalization
obvi-ously requires
further protein-protein and/or protein-DNAinteractions whichare notprovidedin uninfected cells.
ACKNOWLEDGMENTS
WethankH.zurHausenfor continuoussupportandV. Boschfor critical readingof themanuscript. We aregrateful toB. Hub,A. Kameda,and0. Muller for excellent technical assistance.We also thankR. Webler forpreparingthefigures.
M. Ruffing was supported in partby grant KL 516/1-2 of the DeutscheForschungsgemeinschaft.
REFERENCES
1. Becerra, S. P., F. Koczot, P. Fabisch, and J.A. Rose. 1988.
Synthesisof adeno-associated virus structuralproteins requires
bothalternativemRNAsplicingandalternative initiations from asingletranscript.J.Virol. 62:2745-2754.
2. Becerra,S. P., J.A. Rose,M.Hardy, B.Baroudy,and C. W. Anderson.1985. Directmappingof adeno-associated virus cap-sidproteins B and C: a possible ACG initiationcodon. Proc. Natl. Acad. Sci. USA 82:7919-7923.
3. Berns, K. I., and R. A. Bohenzky. 1987. Adeno-associated viruses:anupdate.Adv.Virus Res. 32:243-306.
4. Brown, C. S., J.W. M.van Lent, J.M.VIak,and W.J.M. Spaan. 1991. Assemblyofemptycapsids by usingbaculovirus recombinantsexpressinghumanparvovirusB19 structural pro-teins. J.Virol. 65:2702-2706.
5. Buller, R. M. L., and J. A. Rose. 1978. Characterization of adeno-associated virus-induced polypeptides in KB cells. J. Virol. 25:331-338.
6. Cassinotti, P.,M.Weitz,andJ.-D.Tratschin.1988.Organization of the adeno-associatedvirus (AAV) capsidgene: mappingof minorsplicedmRNAcodingfor viruscapsid protein1.Virology
167:176-184.
7. Chen, C.,and H. Okayama. 1987. High-efficiency transforma-tion of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752.
8. Clever, J.,and H. Kasamatsu.1991.Simian virus 40VP2/3 small structural proteins harbor theirown nuclear transport signal.
Virology 181:78-90.
9. de la Maza, L. M., and B.J. Carter. 1978. DNA structure of incomplete adeno-associated virus particles, p. 193-204. In D. C.Ward and P. Tattersall(ed.), Replication of mammalian parvoviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor,N.Y.
10. Dingwall, C.,and R. A. Laskey. 1986. Proteinimport into the cell nucleus. Annu. Rev. Cell Biol. 2:367-390.
11. Gharakhanian, E.,and H. Kasamatsu. 1990. Two independent signals, a nuclear localization signal and a VP1-interactive
signal,residewithin thecarboxyl-35 amino acids ofSV40 VP3. Virology178:62-71.
12. Goldfarb,D.S., J. Gariepy,G.Schoolnik,and R. D. Kornberg. 1986. Synthetic peptidesasnuclearlocalizationsignals. Nature (London)322:641-644.
13. Harlow,E.,and D.Lane.1988.Immunoblotting,p.471-510. In E. HarlowandD. Lane(ed.),Antibodies.Alaboratorymanual. ColdSpringHarborLaboratory,ColdSpring Harbor, N.Y. 14. Heilbronn, R.,A.Burkle, S. Stephan,and H. zurHausen. 1990.
Theadeno-associated virusrep gene suppressesherpessimplex virus-inducedDNAamplification.J. Virol. 64:3012-3018. 15. Hermonat,P.L.,M.A.Labow,R.Wright, K. I.Berns, and N.
Muzyczka. 1984. Geneticsof adeno-associated virus: isolation andpreliminarycharacterization of adeno-associated virustype 2mutants.J.Virol. 51:329-339.
16. Hink, N. F. 1970. Established insect cell line from cabbage
looper,Trichoplasiani.Nature(London) 226:466-467. 17. Hunter, L. A., and R. J. Samulski. 1992. Colocalization of
adeno-associatedvirusRep and capsid proteins in the nuclei of infected cells. J.Virol. 66:317-324.
18. Janik, J. E., M. M. Houston, and J. A. Rose. 1984. Adeno-associatedvirusproteinsinthe nuclei of infected cells.J.Virol. 52:591-597.
19. Jay,F.T., C.A. Laughlin,and B.J. Carter. 1981. Eucaryotic translationalcontrol:adeno-associated virusprotein synthesis is affectedby mutation in the adenovirus DNAbinding protein. Proc. Natl.Acad. Sci. USA 78:2927-2931.
20. Kajigaya, S.,H.Fujii,A.Field, S. Anderson,S.Rosenfeld,L.J. Anderson,T.Shimada,and N. S.Young. 1991. Self-assembled B19parvovirus capsids,producedinabaculovirussystem, are antigenically and immunogenically similar to native virions. Proc. Natl. Acad. Sci. USA 88:4646-4650.
21. Kalderon, D., W. D.Richardson, A. F.Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40large-T antigen. Nature(London)311:33-38. 22. Kalderon, D.,B. L.Roberts,W. D.Richardson,and A. E.Smith.
1984. A short amino acid sequence able to specify nuclear location. Cell39:499-509.
23. Kleinschmidt, J. A., and A. Seiter. 1988. Identification of do-mains involved in nuclearuptakeand histonebinding of protein N1ofXenopuslaevis. EMBO J. 9:1309-1318.
24. Laemmli,U. K.1970.Cleavageofstructuralproteinsduring the
assembly of the head ofbacteriophage T4. Nature (London) 227:680-685.
25. Lanford,R.E.,andJ.Butel.1984.Constructionand character-ization of an SV40mutantdefective in nuclear transportofT
antigen.Cell 37:801-813.
26. Lanford,R.E.,P.Kanda,andR.C.Kennedy.1986. Induction of nuclear transport withasynthetic peptide homologoustothe
SV40Tantigentransportsignal. Cell46:575-582.
27. Luckow,V.A.,andM. D.Summers. 1989.Highlevelexpression
of nonfusedforeigngeneswithAutographacalifornicanuclear
polyhedrosisvirusexpressionvectors.Virology170:31-39. 28. McPherson,R.A.,andJ.A.Rose.1983. Structuralproteinsof
adeno-associated virus: subspecies and their relatedness. J. Virol.46:523-529.
29. Moreland,R.B., G. L.Langevin,R. H.Singer,R. L.Garcea, and L.M. Hereford.1987. Amino acid sequences thatdetermine the nuclear localization of yeast histone 2B. Mol. Cell. Biol. 7:4048-4057.
30. Myers, M.W., and B. J. Carter. 1980. Assembly of adeno-associatedvirus.Virology102:71-82.
31. Nath, S.T.,and D. P.
Nayak.
1990. Function oftwo discreteon November 9, 2019 by guest
http://jvi.asm.org/
6930 RUFFING ET AL.
regions required for nuclear localization of polymerase basic protein1ofA/WSN/33 influenzavirus(H1N1). Mol.Cell. Biol. 10:4139-4145.
32. Rittner, K, H. Stoppler, M. Pawlita, and G. Sczakiel. 1991. Versatile eucaryotic vectors for strong and constitutive tran-sient and stable gene expression. Methods Mol. Cell. Biol. 2:176-181.
33. Robbins, J., S. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targetingsequence: identification ofaclassofbipartitenuclear targetingsequence.Cell 64:615-623.
34. Saliki, J. T., B. Mizak, H. P.Flore,R. R.Gettig, J. P. Burand, L. E.Carmichael, H. A. Wood, and C. R.Parrish. 1992.Canine parvovirusemptycapsidsproduced by expression ina baculo-virusvector: use in analysis of viral properties and immuniza-tion of dogs. J. Gen. Virol. 73:369-374.
35. Samulski, R. J., L.-S. Chang,and T.Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal inte-gration doesnotrequire viral geneexpression.J.Virol. 63:3822-3828.
36. Sanger, F.,S. Nicklen, and A. R. Coulson. 1977. DNA sequenc-ing with chain-terminatsequenc-ing inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467.
37. Smuda, J. W., and B. J. Carter. 1991.Adeno-associated virus having nonsense mutations in the capsid genes: growth in mammalian cells containing an inducible amber suppressor. Virology 184:310-318.
38. Stamatos, N. M., S. Chakraburt, B. Moss, and J. D. Hare. 1987. Expression of polyomavirus virion proteinsby avacciniavirus vector:association ofVP1and VP2 with the nuclear framework. J.Virol. 61:516-525.
39. Studier,W.F., A. H. Rosenberg, and J. J. Dunn. 1990. Use of theT7 RNApolymerasetodirect expressionof cloned genes.
MethodsEnzymol.185:60-89.
40. Summers, M. D., and G. E. Smith. 1987. A manualof methods for baculovirusvectorsand insect cellcultureprocedures.Tex. Agric. Exp. Stn.Bull. 1555:1-56.
41. Taylor, J. W., J.Ott,andF.Eckstein.1985. Therapid genera-tion of oligonucleotide-directed mutations at high frequency using phosphothioate-modified DNA. Nucleic Acids Res. 13: 8765-8785.
42. Thomas, J. O., and R. D. Kornberg. 1975. An octamer of histones in chromatin and free in solution. Proc. Natl. Acad. Sci. USA 72:2626-2630.
43. Towbin, H., T. Staehelin, and J. Gordon. 1979.Electrophoretic transfer ofproteinsfrompolyacrylamide gelstonitrocellulose sheets:procedureand someapplications.Proc.Natl. Acad. Sci. USA76:4350-4354.
44. Tratschin, J.-D.,I. L. Miller, and B. J. Carter. 1984. Genetic analysis of adeno-associated virus: properties of deletion mu-tantsconstructedin vitro andevidence foranadeno-associated virusreplication function.J. Virol. 51:611-619.
45. Trempe, J. P., and B. J. Carter. 1988.AlternatemRNAsplicing isrequired forsynthesis of adeno-associated virusVP1 capsid protein. J.Virol.62:3356-3363.
46. Wychowski,C.,D. Benichou, and M.Girard. 1987. The intra-nuclearlocation of simian virus 40 polypeptidesVP2andVP3
dependson a specificamino acidsequence. J. Virol. 61:3862-3869.
47. Xia, Y.-P., C.-T. Yeh, J.-H. Ou, and M. M. C. Lai. 1992. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol. 66:914-921.
48. Zhao,L.-J., and R. Padmanabhan. 1988. Nucleartransportof adenovirus DNApolymerase is facilitated by interaction with preterminal protein. Cell55:1005-1015.
J. VIROL.