JOURNAL OFVIROLOGY,Apr. 1993,p. 2043-2054 0022-538X/93/042043-12$02.00/0
Copyright © 1993, AmericanSocietyfor Microbiology
Characterization of
a
Structurally
Tricistronic
Gene of
Human
Cytomegalovirus Composed of
Us18,
Us19,
and
Us20
YAW-WENGUO' ANDENG-SHANG HUANG'2,3,4*
Curiculumof Genetics, 1 Department of Medicine,2Departmentof Microbiology and Immunology,3 andLineberger Comprehensive Cancer Center,4 University of North Carolina
atChapel Hill, Chapel Hill, North Carolina 27599-7295 Received 22October 1992/Accepted 16 December 1992
Atricistronic genemapped between 0.91 and 0.93 mapunits within theEcoRI D fragment of the human cytomegalovirus uniqueshortregion
(Us)
has beencloned, sequenced,andexpressedinvitro.Cloned cDNAs of 2.3, 1.8, and 1.1 kb derived from this region were isolated from a Agtll cDNA library made from virus-infected fibroblastsand usedfor this study. Two majorclassesof 3'.coterminalmRNAs, 2.8 and 1.1 kb, weretranscribed fromthisregion. Sequence analysis of the cDNAs andtheupstreamgenomic DNA revealed threeopenreadingframes (ORFs),Us18, Us19,
andUs20,
anda common polyadenylation signal located15 bases upstream of thepoly(A) tail of both the2.85- and 1.1-kb mRNAs. Protein structureanalysespredicted theexistence ofmultiple hydrophobic moieties, suggestingthat theUs18, US19,
andUs20
polypeptideswere transmembrane proteins. The major transcription initiation site, determined by primer extension and S1 nucleasemapping,for the 2.85-kbtranscriptwaslocatedrightatthe firstinitiationcodon oftheUs20
ORF. Therewas notypicalTATA boxorCAAT boxupstreamof the 2.85-kb mRNAcapsiteexceptforaTATAAGAsequencethatwasfoundabout 210bpdownstream from themajorcapsite. The1.1-kbtranscriptwasinitiated 33 bpupstream of the
Us18
translationinitiationsite,andanatypicalTATAboxsequence (GATAAGA)wasfound 22bpupstream of the transcriptionstartsite. Differences intranscriptionkinetics and sensitivities to metabolicinhibitors suggestthattheywereregulated by differentmechanisms; the 2.85-kb mRNAbelongsto theearly
(13)
class oftranscripts, while the 1.1-kb mRNA isa late('y) message. Subgenomic DNAsegments derived from theUs18, Us19,
andUs20
ORFsweresubcloned andoverexpressedinEscherichiacoliasfusion proteinswithglutathione-s-transferase.Western immunoblotanalysiswithantibodiesagainst theUs18, Us19,
and
Us20
fusion proteins detectedvirus-specific polypeptides with molecular sizes of36, 32, and 43 kDa, respectively.Allthree antibodies alsoexhibitedapositiveimmunofluorescence reaction with human cytomeg-alovirus-infected cells harvested atlate stages ofinfection.Humancytomegalovirus (HCMV)is the fourth herpesvi-rustobesequencedand has thegreatestgenomic complexity
among the human viruses discovered to date(3). The viral
genome is a double-stranded DNA 230 kb in length. More than 200 openreading frames (ORFs) can be derived from this viral genome (3). Like that ofherpes simplex viruses, the HCMVgenomeiscomposedofaunique long (UL)anda unique short
(Us)
segment, each bounded by terminal re-peatsthatpermitULandUs
inversions to form four isomeric molecules (9,13, 14).TheHCMVUs
regionisverydifferent from that of other human herpesviruses. The 38-kbUs
regionof HCMV containsatleast six families ofhomologous ORFs, andexceptfor theGprotein-coupled receptor (GCR) family,noneof these showhomologytoother known human herpesviruses ORFs (25). Each family contains 2 to 13 membersorORFs whichoccureitherastandemarrays(the
Us2, Us6,
andUs12families) ordispersedin theUsregion(the
Us1, Us22,
and GCRfamilies).The
Us2
family, composed of theUs2
andUS3
ORFs, produces a group of abundant immediate-early (IE) tran-scripts encoding a group ofputative membrane-associated glycoproteins (24). The US6 family is composed of sixmembers,
Us6
to Us11, which encode a group of early transcripts(includingtwobicistronicmRNAs)foragroup ofvirion-associated envelope glycoprotein complexes, desig-natedgCII (gp47-52 [6]). TheUs22familyhas 12members,
*Correspondingauthor.
only4ofwhich,
Us22, Us23, Us24,
andUs26,
arelocated within theUs
region. This family encodes a group ofearly viralproteinswhichcontain N-linkedglycosylation sitesand C-terminal charged residues but no obvious hydrophobic transmembraneregion. HCMVproteinICP22wasidentified as a member of thisUs22
family. This protein is an early viralproteinpredominantlylocalized inthe nucleusbut also secreted into the culture medium from infected cells (15). The GCR family produces membrane-spanning glycopro-teins which havesequencehomologywith theopsinfamily of cellular receptors (3). All three members of the GCR family,Us27,
Us28,
andUs33,
have sevenpotential mem-brane-spanning regions. Members of this diverse family transduce differentsignalsinavarietyofsystemsthat have roles invision,olfaction, memory, learning, andregulation of thecirculatorysystem(16). Exceptfor data obtained from computer analysis of viral DNA sequences, there is little information about the remaining two large gene families,Usl
andUs12.
TheUs12
familyhas 10 members(Us12
toUs21),
whichareclustered andtandemly arrangedbetweenthe
Us6
andUs22
familiesin theUs
region.Members ofthis family aregrouped together bythe characteristic ofhaving multiple hydrophobic regionswithin their products, whichare tentatively classified as membrane-associated proteins with potential glycosylation sites. Each has up to seven highly hydrophobic putative transmembrane regions. The HCMV ORFs studied in this communicationbelongto this family.
Most virus genes are monocistronic, encoding only one
2043
Vol. 67, No. 4
on November 9, 2019 by guest
http://jvi.asm.org/
polypeptideinonegene.HCMV isanexceptionstothisrule,
as polystronic messages are not uncommon in this human
herpesvirus.For example, pp65 andpp7l (UL82 andUL83)
areencoded bybicistronic genes in HCMV(3,
18).
In thiscommunication, we show that the
unique
shortregion
of HCMV between 0.91 and 0.93 map units encodesastructur-ally
tricistronic genefortheUS18,
Us19,
andUs20
polypep-tides. Transcription of polycistronic genes in prokaryotic organismsis regulated in a
sequential
manner. In general,thecodingregion closestto the promoter
regulatory
regionproduces
moreproducts.Therefore,
atthebeginning
of thisstudy,weassumed that
regulation
of thetranscriptionof this HCMVtricistronic gene might be similar to that of apro-karyoticpolycistronicgene.Northern
(RNA
blot) hybridiza-tion, however, revealed twomajor
mRNAs with differenttranscription kinetics. Therefore, the transcriptional and translationalregulationsof these messageswere major
sub-jectsforthisstudy.
Sequence analysis
of these threenonoverlapping
ORFs(transcription
is leftward, fromUs20
toUs18)
predictedthree
polypeptides
of342,240,and 274 aminoacidresidues,
respectively. Toverifywhether these three ORFs are func-tional andtoconfirmthat this tricistronic gene is translated in HCMV-infected
cells,
subgenomic DNA fragments of these three ORFs wereengineered
and subcloned into a bacterialexpression
vectorcontaining
thecarboxy
terminus of theglutathione-S-transferase
(GST)
gene from Schisto-somajaponicum under the control ofa tac promoter, thepGEX-GST
system(21).
Thesebacterially expressed
fusionproteinswere used as
antigens
to preparepolyclonal
anti-bodies in rabbits forusein Westernimmunoblot andimmu-nofluoresence
analyses.
MATERUILSANDMETHODS
Virus, cells, andbacterial strains. Human
embryonic lung
(HEL)
fibroblasts(ATCC
HEL229;
passages15to24)
wereused for
propagation
of the virus and allbiochemicalstudies. HEL cells were cultured inEagle's
minimal essentialme-dium
supplemented
with 10% fetal calfserum. The Townestrain of HCMV
(passage
34to39)
waspropagated
in HEL cells at a lowmultiplicity
of infection(0.001 PFU/cell)
forseed
stocks,
andthe virustiterwasdeterminedbyaplaque
assayin HEL cells. Formost
experiments,
HELcellswere grownto nearconfluence inaculture flask and infected with Townestrain HCMVat amultiplicity
of infection ofapprox-imately
2PFU/cell.
Virus absorption was carried out in aCO2
incubator at37°C
for 2 h. Virus-infected cells were harvestedatthetimes indicated. Two bacterial strainswere used in theseexperiments.
Eschenchiacoli JM107wasused in thepropagation
ofplasmid
DNAandM13single-strandedphage
DNAtemplate.
E. coliY1090wasused in thepropa-gationofAgtll cDNAclones(8).
Construction and screening ofcDNA libraries. Total and
cytoplasmic
RNAswereisolatedfromHCMV-infectedHELcells at 72 hpostinfection (p.i.) bytheguanidinium isothio-cyanatemethod(10).A cDNAlibrarywasconstructed from this RNAasdescribedpreviously (8). Essentially,
polyade-nylated
[poly(A)+] RNAswerepurified by usingoligo(dT)-cellulose column chromatography. Double-stranded cDNA was synthesized frompoly(A)+RNAby using oligo(dT) as
the
primer.
The cDNAsweremethylatedwith EcoRImeth-ylase
beforethe EcoRIlinkerwasadded. After fractionationthrough
aSepharose4Bcolumn, cDNAslarger than 0.5 kbwere inserted into the EcoRI site of
Xgtll
andpackaged invitrowithpackagingextractsobtained from Promega
(Mad-ison, Wis.)asdescribed before(8). Two cDNA libraries with 1.5 x 106 and 2 x 106 recombinants wereconstructed from total andcytoplasmic mRNA,respectively.
DNA labeling. The radioactive viral DNA probe was labeled by the random-primed DNA labeling method (Ran-dom PrimerLabeling SystemU1110;Promega). To label the 5' termini of an oligonucleotide probe, 20 pmol of dephos-phorylated oligonucleotides in an oligonucleotide kinase buffer with 0.1 mCi of [_y-32P]ATP (3,000 Ci/mmol; Amer-sham Corp.) and 4 U of T4 polynucleotide kinase was incubated at 37°C for 30min. After heat inactivation at 65°C for 10 min, the labeled oligonucleotides were purified by
three rounds of ethanolprecipitation.
Southernhybridization. Southern hybridizations were per-formed by standard methods (20). Prehybridization was carried out in a mixture containing 25 mM KPO4 (pH 7.4), 6x SSC(0.9 M NaCl, 0.09 M sodium citrate), 5xDenhardt's solution (0.1% Ficoll, 0.1%polyvinylpyrrolidone, 0.1% bo-vine serumalbumin), 50 ,ug of denatured salmon sperm DNA per ml, and0.1% sodium dodecyl sulfate (SDS) at 42°C for 6 h. After prehybridization, denatured 32P-labeled cDNA probe (5 x 105 cpm/ml)was added and incubated at 42°C overnight. Following hybridization, filters were washed twice in lx SSC-0.1% SDS and twice in 0.25x SSC-0.1% SDSat roomtemperature for 15 min each.
Northern hybridization. Total RNAs (8 ,g per well) pre-pared from mock- and HCMV-infected HEL cells by guani-diniumisothiocyanate cell lysis and cesium chloride equilib-rium centrifugation were subjected to electrophoresis in a 1.2% agarose gel containing 6% formaldehyde and then transferredtonitrocelluloseasdescribedbefore(10,20). The
resulting filterwasprehybridized at42°C for 18 h and then hybridized with a random-primed or nick-translated 32P-labeled cDNAprobe(5 x105
cpm/ml)
at42°Cfor 24 h. After being washed, the hybridized filter was autoradiographed overnight at -70°C. The same blot was stripped of the labeled probe by incubation at 90°C in TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA) for 15 min. The blot was then hybridized with the 5'-end _y-32P-labeled antisense oli-gonucleotides or a-32P-labeled cDNA probe for the yeast 28S rRNA gene. For the study of mRNA synthesis in the presenceof drugs, IE RNAswerepreparedfrom cyclohex-imide (50,ug/ml)-treated
HEL cells harvested at 10 h p.i. Early RNAswereprepared from phosphonoacetic acid (100,ug/ml)-treatedHEL cells harvestedat24 hp.i. Late RNAs wereharvestedat 60 hp.i.without drug treatment.
DNA sequencing. cDNA and genomic DNA sequencings were performedby the dideoxy chain termination method (19), with the Sequenase kit from U.S. Biochemical Corp.
(Cleveland, Ohio)andSequentide(NEG-034;NewEngland
Nuclear, Boston, Mass.). Single- and double-stranded tem-plates were obtained from appropriate subclonings in M13mpl8 and mpl9. The enzymes used for the subcloning include PvuII, RsaI, PstI, andXhoI. All sequences were obtained andconfirmed bysequencing data taken from both strands. Eight synthetic oligonucleotideswere used as
se-quencing primers to resolve sequence gaps and to confirm sequenceambiguities.ORFanalysis was carried out with the DNA Inspector II (Textco, West Lebanon, N.H.). Two-dimensional protein structures (4) and hydrophobicity and hydrophilicity profiles of the deduced peptide were deter-minedbyusingasoftware package provided by the Genetics
Computer Group (UniversityofWisconsin, Madison). Primerextension.Primer extension experiments were per-formed by the method described by Kingston (11), with minor modification. Four 30-bp oligonucleotides, including J. VIROL.
on November 9, 2019 by guest
http://jvi.asm.org/
STRUCTURALLY TRICISTRONIC GENE OF HCMV 2045
primer 517 (5'-ACACAAGCGAGCGAGTGGGGCACGGT GACG-3') and primer 520 (5'-CCACTCGAGAGCCTCCAT
GCGGGAGAGCAG-3'), both complementary to the 2.85-kb message, and primer 130 (5'-TGTTCGGAAACCGAGGCG GTGTCGCCATGC-3') and primer 136 (5'-TCGGCCACC
AGCGCGTGGCTGCGATGGAGC-3'), both complemen-tary to the 1.1-kb message, were synthesized on an Applied Biosystems synthesizer and end labeled with
[,y-32P]ATP
by using T4polynucleotide kinase. Seven micrograms of total RNA and 104 cpm of labeled primers were mixed for 5min at 80°C and then precipitated with alcohol. Pellets were dis-solved in 20,1 of lx hybridization buffer [80% formamide, 40 mM piperazine-N,N'-bis(2-ethanesulfonic acid) (PIPES, pH6.4), 400 mM NaCl, 1 mM EDTA] and allowed to anneal at30°C overnight. After ethanol precipitation, reverse tran-scriptase extension reactions were carried out at42°C
for 90 min. These were done in a final volume of 25,u containing 18 U of avian myeloblastosis virus reverse transcriptase, 50 mMTris-HCl (pH 8.0), 5 mMMgCl2,5 mM dithiothreitol, 50 mM KCI, 50 U of RNasin, and 50,uM
each of the fourdeoxynucleosidetriphosphates. After RNasedigestion, phe-nol extraction, and ethanol precipitation, the pellets were dissolved in 4
RI
of TE and 4RI
of gel loading buffer, heated at 80°C for 3 min, and loaded on a 6% polyacrylamide sequencing gel containing 7 M urea. The extension products wereanalyzed by comparison with genomic DNA sequenc-ing reactions, which were run in parallel. The control genomic DNA was derived either from a 1,008-bp (bp 3 to1010)NcoI-PstI fragment upstream of the 2.85-kb mRNA or from a 602-bp (bp 1722 to 2333) PvuII-PvuII fragment upstream of the 1.1-kb mRNA.
S1 nuclease mapping. The four oligonucleotides used in primerextension were also used inS1 nuclease mapping (1). Primer 517 and primer 520 were used for the 5'-end mapping of the2.85-kb message. Primer 130 and primer 136 were used forthe 5'-end mapping of the 1.1-kb messages. The proce-dure used was as follows: 5'-end-labeled oligonucleotides wereannealed tosingle-stranded templates derived from the
correspondingNcoI-PstI region (for the 2.85-kb message) or PvuII-PvuII region (for the 1.1-kb mRNA), extended with Klenow enzyme, andrestrictedto generate uniform restric-tion fragments. A polyacrylamide gel electrophoresis-puri-fied probe (2x104 cpm) was annealed to 10,ugof total RNA in 30
RI
ofhybridization buffer (80% deionized formamide, 0.4 M NaCl, 10 mM PIPES [pH 6.5], 1 mM EDTA). Thisannealingmixture was placed in a55°Cwater bath for 5min
and then incubated at 30°C for 16 h. To the annealed mixture, 200jilofS1buffer (30 mM sodium citrate [pH 4.5],
0.25mMNaCl, 1mMZnSO4, 5% glycerol) and 100 U of S1 nuclease wereadded. The mixture was incubated at
16°C
for 1 h. TheS1 nuclease-resistant fragments were precipitated with ethanol, denatured in 4 jI of formamide sample buffer(90% formamide, 0.1% bromophenol blue, 0.1% xylene
cyanol), and electrophoresed in a 6% polyacrylamide
se-quencinggel with 7 M urea. The nuclease-resistant products were analyzedbycomparison with genomic DNA sequenc-ing reactions run in parallel.
Overexpressionof
US18,
US19,
andUs20
in the pGEX-GST system. The GST gene fusion system was used to express viral DNAsubclones as fusion proteins (21). To construct in-frame clones, DNA fragments were inserted into the BamHIandEcoRIsites of one of three expression plasmids,pGEX-1N, -2T, or -3X (21), with thereadingframematched in the sense orientation to yield recombinant plasmids
pGEX-lN-US18, pGEX-lN-US19, and pGEX-3X-US20.
Overnight cultures ofplasmid-transformed E. coli JM107
were diluted 1:10 with fresh medium and grown at
37°C
for 1.5 h. They were subsequently induced with isopropyl-3-D-thiogalactopyranoside (IPTG) at a final concentration of 0.1 mM. After 1 to 6 h of growth, cells were pelleted and lysed in 0.25 volume of SDS sample buffer (0.006 MTris-HCl
[pH 6.8], 4% SDS, 40% glycerol, 3% dithiothreitol, 0.005% bromophenol blue). The samples were boiled and subjected to 12% polyacrylamide gel electrophoresis.The majority of the US18-GST,Us19-GST, and
US20-GST
fusion proteins were expressed as insoluble inclusion bodies. The inclusion bodies were prepared as described by Harlow and Lane (7) with modification. In brief, the induced plas-mid-transformed E. coliJM107
cultures(pGEX-1N-US18,
pGEX-lN-US19,
andpGEX-3X-US20,
respectively) werepelleted by centrifugation at 8,000 x g for 5
min.
The cell pellets were washed twice and resuspended at 1/10 their original volume in a washing buffer containing 100 mM NaCl, 1 mM EDTA, and 50 mMTris-HCl
(pH 8.0). Ly-sozyme was added to the suspension at a concentration of 1mg/ml, and the suspension was incubated at room tempera-ture for 20min. The lysate was then sonicated for three 30-s cycles at full power with a Branson Sonifier (model 200 cell disrupter) equipped with a microtip and centrifuged at 5,000 x g for 10
min.
The
supernatant
containing soluble fusion protein was further purified on a glutathione-Sepharose column. The pellets containing inclusion bodies were washed with an ice-cold washing buffer (100 mM NaCl, 1 mM EDTA, 50 mMTris-HCl
[pH 8.0]) with 0.1% SDS, then with washing buffer containing 1% Nonidet P-40, and finally with washing buffer alone. The inclusion bodies were then further purified by 10 to 50% sucrose gradient centrifugation. Fractions containing inclusion bodies were pelleted and resuspended in phosphate-buffered saline (PBS), analyzed for purity, and used as antigens for antibody production in rabbits.Antibody preparation. Antisera against the US18-GST,
Us19-GST, and
US20-GST
fusion proteins were made by individually immunizing New Zealand White rabbits. Inclu-sions with fusion protein aggregates were denatured in the presence of 7 M guanidineHCI
solution and dialyzed against PBS with several changes of buffer solution to remove the guanidineHCl.
A mixture of 100 jig of the fusion proteins with an equal volume of Freund's complete adjuvant (Cal-biochem Corp.) was injected into the rabbits subdermally at multiple sites near the hind legs. Booster injections with fusion proteins in an equal volume of Freund's incomplete adjuvant (Calbiochem Corp.) were given intradermally to the rabbits three times at three-week intervals. Ten days after the final injection, rabbits were bled, and the sera were tested for the presence of HCMV-specific antibodies by Western blot analysis and immunofluorescence staining.Western blot and indirect immunofluorescence
analysis.
HEL cells were infected with
HCMV
as described above. At the indicated times, cells were washed twice with ice-cold PBS and then lysed with 2x SDS sample buffer (0.125 mM Tris [pH 6.8], 20% glycerol, 0.2% SDS, 0.28 M 3-mercap-toethanol, 10 jig of bromophenol blue). The lysates were sheared by passage through a 20-gauge needle four times and then boiled for 5 min. After electrophoresis, proteins were transferred to nitrocellulose paper in transfer buffer (20 mMTris-HCl [pH
8.0],
150 mM glycine, 20% methanol). The blot was washed three times with blocking buffer (2% dry milk in PBS) at room temperature for 10min
each and then probed with antiserum against the fusion protein (diluted 1:100 in blocking buffer) at room temperature for 1 h. The antiserum was removed, and the blot was rinsed three times with aVOL.67, 1993
on November 9, 2019 by guest
http://jvi.asm.org/
2046 GUO HUANG
A. iCcV 0.0
gsne
IL
0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0
I I I I I I I I I.
B EoRI LT M C VOKUT E J N G R A S B LZXH I U/S F D WST
map II I 1111I I I I IIIII1 I I _I_
C Transcript
Xho lndE Hind RO
ago
lindl
.i I *I I I I I I I
E-OM
n i I
_
5Transcdfte
D. pbe
Hhid ag.
a-.A, I|I
T..AAA
T..AAA
4-=M
1.1Kb mRNA
E cDNAmd 5 en 3'..A -..
2.2KbCDNA
V...Am
10 Kb
g.o
5
primer 517 & 520
prime 130& 136
Is2bP !
SIbp
1.1 KbCDNA Om
F. Arrangementofputative geneproducts USi8
3'..AAA W
822 bp
5 end CAP ste
i CAPaft
USi 9 IH 720bp
US20
14 1026bp
.1
s.HWLF5 HWLF 4 HWLf 3
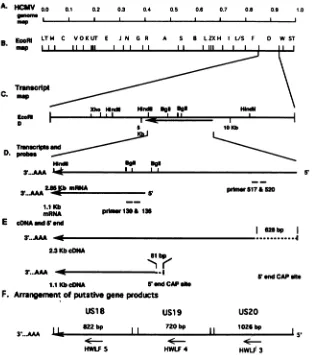
FIG. 1. PhysicalmapofHCMVTowne strain DNA and relativelocations of theEcoRI Dfragment, cDNAs,and varioustranscriptional
products derived fromthis tricistronic gene.(A) Fractionalmapof theHCMVDNA genome.(B)Schematicrepresentationof theHCMV genome,with the restrictionmapfor EcoRI.(C)EcoRIDfragmentof theHCMVgenome,designatedpHD4(5).Thetranscriptionorientation ofthe 2.3-kbcDNAisindicatedbyanarrow,and themagnifiedmapis shown underneath.(D) Transcripts andrelative locations ofDNA primers used forprimer extension and S1mapping.(E)Schematicdiagram showinglocationsof the2.85-kb and 1.1-kbmRNAsandcDNAs and their locationsrelativetotranscription initiation sites.A628-bpsequencewasmissinginthe 2.3-kbcDNAandan81-bpsequencewas
missingin the1.1-kb cDNAcompared withtheirmRNAs.(F)Thelocation and arrangement ofputativegeneproducts.Datawereobtained from theexperiments showninFig.2through8.
blocking buffer. Bound antigen-antibody
complexes
were detected by incubating the filter in buffer solution withprotein A-peroxidase-labeled secondary antibody (Boehr-inger Mannheim Co.) at a 1:500 dilution for 2 h. After
incubation, the filterwaswashedasabove and reactedwith substrate(10mgof diaminobenzidine in 100 ml ofPBSwith 10 ,ul of30% H202)for 30 min.
For theindirectimmunofluorescencetest,antibodies were preabsorbed with HEL cellular lysates at 37°C for 1 h. HCMV-infected HEL cells grown on eight-well chamber slides (Nunc, Naperville, Ill.) were harvested at the indi-catedtimes p.i. The cells were washed with PBS three times andfixed with ice-chilled methanol-acetone (1:1) at -20°C for 15 min. The fixed cells were then blocked with 20%
normalgoat serum(JacksonImmuno Research Laboratory
Inc.)inPBS for 30 minatroomtemperature.Cells were then reacted withdiluted rabbit antiserum (1:50 in PBS) for 1 h. After PBS washes, biotin-labeled goat anti-rabbit
immuno-globulinGsecondary antibodies (VectorLaboratories Inc.,
Burlingame, Calif.) at a 1:200 dilution were added to the
cells and incubatedat roomtemperaturefor 1 h.AfteraPBS
wash,fluorescein-conjugated avidinD(VectorLaboratories
Inc.)was added at a concentration of 12.5 ,ug/100ml. The slidewasmounted with50%glycerolinPBS and examined underafluoresent microscope.
Nucleotide sequence accession number. The GenBank ac-cession number for the sequencereportedinthis
communi-cationis L04998.
RESULTS
cDNA cloning. In our first attempt to isolate EcoRI D
fragment-related sequences, a 2.3-kb cDNA was isolated from our Xgtll cDNA library made from HCMV-infected fibroblasts. This 2.3-kb cDNAwasmappedin the EcoRI D
fragment.Themappingwasdoneby Southern hybridization and further confirmed by DNA sequence analysis at both ends of the 2.3-kb cDNA, as shown below (Fig. 1). In subsequent screenings with the 2.3-kb cDNA clone as the
probe,80positivecloneswereisolated from 4 x 104
recom-I
on November 9, 2019 by guest
http://jvi.asm.org/
[image:4.612.157.472.72.427.2]STRUCTURALLY TRICISTRONIC GENE OF HCMV 2047
A.
B.
C
Mock Cyto Total Nuci Mock Cyto Total Nuci Mock Cyto Nuci Total
MW(Kb)
9.49-_
7.486
4.40 -_
2.37 -- 2.85
[image:5.612.100.529.77.285.2]1.8 1.35 -_
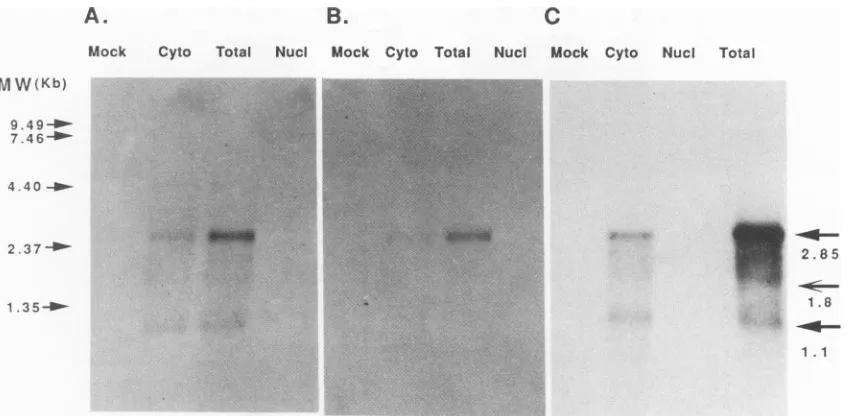
FIG. 2. Transcription analysis with the 2.3-kb cDNA and strand-specific oligonucleotide probes. Total RNA (8pLgperwell) was prepared from mock-and HCMV-infected HEL cells. Cytoplasmic, nuclear, and total RNAs were purified from HEL cells at 60 h p.i.Cytoplasmic and nuclear RNAs were furtherfractionated byoligo(dT)-cellulose column chromatography to obtainpoly(A)'mRNA. Sizes are indicated on the left, andarrowsonthe right indicate themessages detected and their sizes (in kilobases). (A) Probed with astrand-specific probe (primer 136) close tothe 3' end. Twomessages, 2.85 and 1.1 kb, were observed. (B) Probed with astrand-specific probe (primer 520) close to the 5' end of the 2.85-kbmRNA.Thisprobedetected only one message of 2.85 kb (see Fig. 1forrelative locations of oligonucleotide probes used in hybridization studies). (C) Probed with labeled 2.3-kb cDNA. This probe detected 2.8-kb, 1.1-kb, and a minor 1.8-kb message.
binant phages. Twenty positive clones were analyzed by restriction enzyme mapping and DNA sequencing. The clones were categorized into three groups, with a cDNA insert of 2.3, 1.8, and 1.1 kb, respectively. Three cDNA clones were further cloned into M13mpl8 for 3'-end and 5'-end analysis. Theresults of 3'-end analysis reveal that the 2.3-kb, 1.8-kb, and 1.1-kb cDNAs have an identical poly-adenylation signal (AATAAA) and a 14-bp downstream
sequence(GCTTT1TT1-1CACGCTl) and are3' coterminal.
Transcription pattern. To analyze the transcripts corre-spondingtothe2.3-kbcDNA, RNA samples isolatedat60 h after infection were probed with the nick-translated and random-primed labeled 2.3-kb cDNA fragment. Two major
groupsofmRNAs of2.85and 1.1kbweredetected (Fig. 2C).
Thedetection ofaminorband of 1.8kbwas notconsistent andvaried from batchto batchofRNA preparations. Hy-bridization withanantisense oligonucleotide probe (primer 520)derived fromthe 5'endof the 2.3-kbcDNA detected the 2.85-kb message but not the 1.1-kb transcript (Fig. 2B). When an antisense oligonucleotide probe (primer 136) de-rived fromthe 3' end of the 2.3-kb cDNAwasusedasthe probe, both the 2.85-kb and the 1.1-kb messages were
detected (Fig. 2A). This result suggests that the 1.1-kb transcript wastranscribed within the 3'-end portion of the 2.85-kbmessage. The DNA sequencebetween thepoly(A)
signal sequence and the 5' end of the 1.1-kb cDNAwas
equivalent tothe length detected in the 1.1-kb RNA tran-script. These data suggest that the 2.85-kb and 1.1-kb mRNAs are 3' coterminal and that the 1.1-kb transcript is unspliced. These3'-coterminal transcriptswerefurther con-firmedby3'-endsequencingofthe cDNAaswellasbythe methodoftherapid amplificationof thecDNA ends(RACE) (datanotshown).
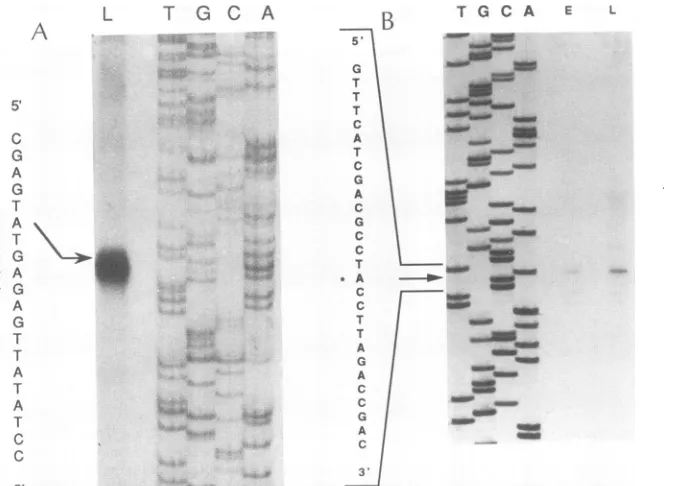
5'-endmapping by primerextensionand Si nuclease
map-ping.Twoantisenseoligonucleotides (primer517 andprimer 520), complementaryto twoareasclosetothe5' end of the
2.85-kb mRNA, were used in the primer extension studies
(datanot shown). Both of these primers showed the same
major transcription initiation site right at the translation
initiation codon of the
Us20
ORF, even though these twoprimers are 80 bp apart. These two oligonucleotides were also used in
Si
nuclease mapping. Themajor transcription initiation site for the 2.85-kb mRNA was similar to theprimer extensionsite (Fig. 3A).However,whenprimer 517 was used for S1 nuclease mapping, a minor band also
appeared about243bpdownstreamfrom the major initiation site indicated above, which is approximately 26 bp
down-streamfromaputativeTATAboxandapproximately21 bp
upstreamofapotential second initiation codon for the
Us20
ORF(datanotshown).
Anothersetofantisenseoligonucleotides
(primer
130and primer 136), complementary to the 5'-end region of the 1.1-kbmRNA, wereusedin the primer extensionstudiesto detect thetranscription initiation site of the 1.1-kb mRNA. This set ofprimers picked up four transcription initiationsites with equal intensity within a 7-base range
(data
notshown). S1 nuclease mapping withthesameoligonucleotides
alsodetectedamajortranscriptioninitiationsiteinthesame
region (Fig. 3B). This sitewas 33bpupstreamof the
Us18
translation start site. The 5' end of this transcript has a
TATA-likebox, GATAAGA
(bp
2115to2121), 22to 28bp
upstream of the transcription initiation site. A CAAT-like
box, CGTCAATCA (bp 2085 to 2092), was 50 to 58 bp
upstream of thetranscription initiation site. The
transcrip-tioninitiationsitewas 33bpupstream of the
Us18
transla-tioninitiationcodon. TheschematicdiagramsinFig.
1E and F outline the transcription initiation sites found and their relative locations in the cDNAs. The detailsareindicated inFig. 6.
Attempts to map the 5' end of the minor 1.8-kb mRNA were not successful when
primer
156(5'-GTAGCAGCAG
GAACA-3')
wasused forprimer
extensionandS1
nucleaseVOL. 67,1993
on November 9, 2019 by guest
http://jvi.asm.org/
2048 GUO AND HUANG
L T G C A
i.. 5.1
G T
T G C A E
B
T T
c A T
c
G A c G
c
c
T
* A
_ _
_-I
- _A
- a
___w
so=
wrwm
_ _ mo
4
__ _ _w
c , i_
T I
T A G A
C J:
C I
c
G |
A c
- a
-p
-u
-a
"o
G-FIG. 3. Si nucleasemappingof the2.85-kb and 1.1-kbmessages.The5'-end-labeledoligonucleotides (primer520 for the2.85-kbmRNA, and primer 136 for the1.1-kbmRNA)wereannealedtocorrespondingsensesingle-strandedDNAtemplatesfor thesynthesisof32P-labeled antisenseprobes.Thepreparationof radioactivesingle-strandedDNAfragmentsand theprocedureforS1mappingaredescribed in Materials
andMethods. Thedideoxynucleotides (T, G, C,andA)used in the reactions areshown above the lanes.The DNAsequencingdata from
parallel experimentsareshowntotheleft of eachpanel;possible transcriptioninitiation sitesaremarked with asterisks(*).(A) Si mapping
ofthe 2.85-kb mRNA. Late RNA(L)isolatedat60 hp.i.wasused in thisanalysis. (B)Si mappingofthe 1.1-kb mRNA. RNAs isolatedat
24(E) and 60 (L)hp.i.wereused in thisstudy.Thetranscriptioninitiation site is indicatedbyan arrowand markedbyanasterisk(*)in the
sequence.
mapping, perhapsbecause of the low transcription level of thismessage.
3'-endmapping bycDNAsequencingand RACE. Inorder tofurtherverifythat these mRNAswere3'-endcoterminal, two different methods were used: sequencing different batches of cDNA, and RACE. Twenty positive cDNA clones were sequenced at both ends, and the sequence
results indicated that these threegroupsof cDNA(2.85, 1.8, and 1.1kb)usethesamepolyadenylation signal (AATAAA), which is 15 bp upstream of the poly(A) tail. On the other hand, these three cDNAs have differentlengths ofpoly(A) tail. The results of the RACE experiment revealed asingle band of 290 bp, the predicted size. Subsequent sequence
analyses of this RACE-amplified DNA fragment further verified the results (data not shown). Both the cDNA
se-quence and RACE amplification provide supporting data for 3'-end cotermination of the major 2.85-kb and 1.1-kb mRNAs and theminor 1.8-kb mRNA.
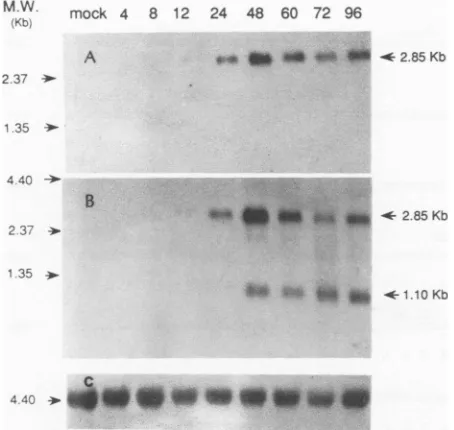
Analysisoftranscriptionpatterns.TotalRNAscollectedat different time points (0, 4, 8, 12, 24, 48, 60, and 72 h p.i.) wereelectrophoresed througha1.2%agarosegelcontaining 6%formaldehyde and analyzed byNorthernhybridization. Thetranscriptionpatternswerederived from theuseoftwo
antisenseoligonucleotide probes,onefrom the 5'end of the 2.85-kb mRNA and another from the 3' end of both
tran-scripts. The5'-end probe detected only the 2.85-kb mRNA (Fig. 4),while the3'-end probedetected both the2.85-kb and 1.1-kbmessages(Fig. 4B). RNAlevelswerequantitatedby densitometerscanningandnormalizedtothat of the internal control, rRNA. Transcription of the 2.85-kb mRNA first appeared at 12hp.i.,reachedamaximumlevelat48 h p.i.,
and thengraduallydecreased. The 1.1-kb mRNAappeared between 24 and 48 h p.i. and remained ata constant level until 72 hp.i. (Fig. 4C). Theincrease intranscriptionat96h p.i.wasprobablyduetovirusreinfection.
Transcription patternin thepresenceofdrugs. To further characterize these two mRNAs, the transcription patterns were studied in the presence of protein and viral DNA synthesisinhibitors. In thepresenceof theprotein synthesis inhibitor cycloheximide (50 ,ug/ml), both the 2.85-kb and 1.1-kbtranscriptsbecameundetectable (Fig. 5). These data suggested that the 2.85-kb and 1.1-kb messages might re-quire HCMV IE gene products for the initiation of their transcription. Therefore, we conclude that these two
mes-sageswerenotIE transcripts.
In the presence of phosphonoacetic acid (100 ,ug/ml), a viral DNAsynthesis inhibitor, the 2.85-kbmRNA accumu-lated to a level comparable to thatin the control (without drug) at 60 h p.i. The transcriptional level of the 1.1-kb
messagein thepresenceofphosphonoaceticacid, however,
decreased to less than one-fifth ofcontrol levels (Fig. 5). These data suggest that the 2.85-kb mRNA is actually an early, or a-type, message ofHCMV, which didnotrequire viralDNAreplicationforexpression. The 1.1-kb mRNAwas expressedat latetimesduring infection(24to48 h p.i.) and requiredviral DNAreplication for maximumexpression; it is clearlya late,or -y-type, message. Theresults show that thesetwotranscriptswereexpressedatdifferent times after infection andwere regulated by differentmechanisms.
Computer analysis of DNA sequence. A 3,158-bp DNA sequence (Fig. 6)was assembled from the sequencing data determined from the analysis of both strands of various
A
5t
C G A G T A T *G A G A T T A T A T C C
3'
., -.
A
.;A.4.;
on November 9, 2019 by guest
http://jvi.asm.org/
[image:6.612.147.484.79.322.2]STRUCflURALLY TRICISTRONIC GENE OF HCMV 2049
MW.(Kb;f mock 4 8 12 24 48 60 72 96
A *
4o
_ 0 *2.85 Kb2 37 *
M W (Kb) 9.49 -_
7.46 -_
Mock CH PAA LATE
4.40
-135 4
2.37
--4.40 *
1.35-_
B
2.85Kb
237 *
1.35 *
60
*I*6*
1.10KbC
[image:7.612.341.525.76.221.2]440 4
FIG. 4. Transcriptionpatternanalysis ofthe 2.85-kb and 1.1-kb mRNAsby Northern blothybridization. TotalRNAwasprepared
frommock- and HCMV-infected HEL cellsatvarious times(0, 4, 8, 12, 24, 48, 60,72,and96hp.i.)asindicated aboveeach lane. Eight
microgramsoftotalRNAwassubjectedtoelectrophoresis through
a1.2%agarosegel containing 6% formaldehydeandthentransferred
tonitrocellulosepaper. (A) The resulting filterwasprobed withan
antisenseoligonucleotide primerlocatedatthe 5' endof the2.85-kb mRNA(seeFig.1D, primer 520). (B)The filterwasreprobed withan
antisenseoligonucleotide primer (primer 136)locatedatthe 3'end of both mRNAs. (C) The blot was probed with yeast rRNA as an
internal control. The arrows on the right indicate the messages
detected, and the arrows onthe left indicate thepositions of size
markers(in kilobases).
restricted genomic or cDNA fragments with an M13mpl8 subcloning system and primer extension approach. This DNAsequence includes a 2,290-bp cDNA fragment and a 1,008-bp genomic regionupstreamof the5' end of the 2.3-kb cDNA. ThesetwoDNAfragmentsoverlaptailtohead,with 140bpatthe3' end of the1,008-bpupstreamgenomic region and at the 5' end of the 2,290-bp cDNA sequence. ORF analysesof thesequencesshown inFig.1FandFig.6were
donewith the DNA Inspector II program. Three
indepen-dentnonoverlappingORFs of Towne strainHCMV, equiv-alent to
Us18, Us19,
andUs20
of AD169 strain HCMV, werederived. Atypical polyadenylation signal (AATAAA, bp3138to3143)waslocated15bpupstream of thecommonpoly(A) tail. All of these ORFs have a typical consensus
Kozaksequence(12)around the translation initiation codon. Sequence comparison with AD169. The sequences of the Towne and AD169 strains of HCMV revealed 99.3%
homol-ogy.The Towne strainisanHCMVstrainisolated from the urine ofacongenitallyinfectedbaby. The AD169 strain isa
prototypeofHCMV isolated from adrenalglandsalmost 25
years agothat has been subculturedinvitro formore than 280passages(17).Thesequencecomparisonbetween these twostrains revealed 20 base mismatches(Fig. 6). No inser-tions or deletions were found. Nine of the 20 mismatched bases are locatedwithin noncoding regions. Ten of the 11
othersingle-basemismatches do not resultin anyaminoacid
sequencealteration, andtheothermismatchchangesamino acid residue 238 of
Us20
from alanine in AD169 to valineinFIG. 5. Transcription inthepresenceofprotein and viralDNA synthesisinhibitors.Eight microgramsof total RNAfrommock- and HCMV-infectedcellstreated withcycloheximide (CH;50 p.g/ml)for 10horphosphonoacetic acid (PAA;100 ±g/ml)for 60 horuntreated
cellsat60 hp.i.(LATE)wasappliedtoeach laneforNorthernblot RNAanalysis.Theresultingfilterwasprobed with the 2.3-kb cDNA
probe.Thearrows ontherightindicatemessagesdetected, and the
arrows onthe leftindicate the migration of sizemarkers.
the Towne strain. The results of this experiment show that the products of this tricistronic gene are very well
con-served.
Two-fdimensional protein structure analysis. Three ORFs
(US18, Us19,
andUs20)
were determined from DNAse-quenceanalysis. Two-dimensional structure analysis of the putative
Us18
polypeptide reveals seven hydrophobicseg-ments. Each ismorethan20 aminoacidslongandmayspan
the cell membrane. One strong antigenic site was found withinoneof thetwopotential glycosylation sites, where the hydrophilic moiety is also obvious. The N-terminal end of this peptide is hydrophilic, whereas the C-terminal end is morehydrophobic.ThisORF has40leucineresiduesamong
274 aminoacids. Theputative
Us19
polypeptidehasonly six hydrophobicsegments,but oneof these isalongstretchof 50hydrophobicamino acids. Nopotentialglycosylationsites werefound in theUS19
polypeptide,buttwohighlyantigenic clusterswith hydrophilicmoieties were found between the first and the second and between the second and third hydrophobic domains from the N-terminal side of thispep-tide. Both the N- and C-terminal ends of this peptide are hydrophobic. The putative peptide contains 52 leucines
among240 amino acid residues. Two-dimensional structure analysisof
Us20
revealedsevenhydrophobicsegments,two potential glycosylation sites, and twohighlyhydrophilicas wellashighly antigenicmoieties located around amino acid residues 50 and 250. TheputativeUs20
peptidecontains 47 leucinesamong342 amino acid residues. Thehydrophobicity profilesof thesepeptidesshow characteristics of membrane-associated proteins: long stretches of hydrophobic amino acids connected by alength of random coil or hydrophilic amino acids.Western blot analysis and indirect immunofluorescence tests.Antibodiestothebacteriallyexpressedfusionproteins were able to reactwithvirus-specific products in Western blotanalyses.The results obtained with HCMV-infectedcell
lysates showed that
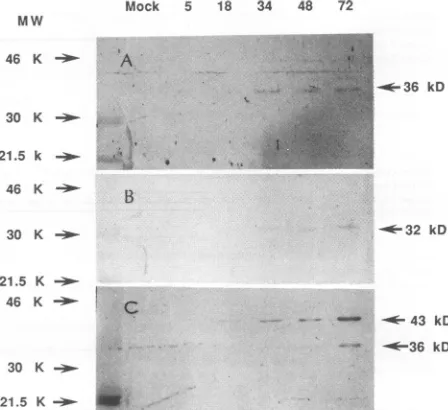
anti-US18-GST
antibodies detected a 36-kDavirus-specificband(Fig. 7A). Thisprotein appeared at 34hp.i. andcoincided with the latetranscriptionof the1.1-kbmRNA,a-y-class transcript.The
Us19-GST
antibody detecteda32-kDavirus-specific peptidewhichappearedata_l _ - 2.85 Kb
_1.1 Kb
VOL.67,1993
on November 9, 2019 by guest
http://jvi.asm.org/
[image:7.612.64.291.78.293.2]2050 GUO AND HUANG
NcoI IR1
CA*IUCCA TA A GIN1II=O=CTACGTA7TrGCC=
i2 IR1
G C C
IR-3_1 DR1,_ r
CGCCG 1T rVCT=CGAGGCGGTA,AMAATGCP GTGAATC1 AGOCGGIGCG7CACGIGCATOTG7GGTACGCXGACAClT%CPGACATGA
IR3
CAP2.85 K0rtA
SnsI IR4_ -6IR4
MAGAGTTTAl.Q:C=TCGGCGACA7GACClt T3TlACCACITG7GTACCTACA ATGCOCI
R V I S R A R S A C. T W T S C T S L S P C S T S C P
IR5 IR6 IR6
DR2 ,_DR2,_
G A a s T G T~~~~~~GA37Ga3GCGrACCA: ACGTGCa:XCACTI= P Q Q R R R P S S S P N R R V R G V T T S P C P T R
r minor DR3 e_ DR4 DR3 __ Xho
TrrGCGAGACCGTCGTCACCALAGGCGkGCTACGCOs iG=aCATG'A GGCT9og
r*2ndtransl. init.
L C R DR RHIM Q A Q E A N A L L L S R M E A L E W
WATCTAGCIT.GClAGCTIGGCOTIOGGAAGCG CI±CA AAAC OAACl CG I F Q L A F S P G L G S V F W L G F P Q N R N F C V TCTGCATGC ATCACG7ACACGTIGGGCAACGAACACCCTAGrAACGrCACGGroCC1I7ATCM¶O¶IGTI7GCCAAM
C M F I T Y T LN.A.TG N E H P S V L F I Y L L A N
r-__cDNA2.3 kb
.=CGArJCCCGrAGCGOCCAC
P S P A A P T
TATABOX
.~O~ AA G
S L V Y K R R ,TCAAAAAGTTCACCGrATG
F K K F T V W 3GAGAACTACPCCICTI
E N Y F F L .AGCCrGACGGCGGCCATCTI S L T A A I F
H
M~
IR5
ACI CTCArACGA0GGTCGC
L L R R R S L
GGrAGGT1XGCCGCAGGGC
V G A P Q R L GCTGCGCGIGTACXCATCT L R V Y A I F
CACCGlrxCGTGOZCATCG
T V L V P I V CCAGA7G7GCTCTAAAGCC
A
Q M C S E S R
GCGTACrAGTAG= AI G T C TGIACC C I G
, C G T ~~~~~~~~~~~GACCAC
A V L V G S Y V M T L A L F I S F T G L A F L G G R D R R R W K C I S C V Y V V M
PstI
TGTGC=GIrI CGTrCTCTGCAGCCPGCCAICT CTElIAAGATAATAGT
L L S F L T L A L L S D A D W L Q K I V V T L C A F S I S F F L G I L A Y D S L
TCATGGA7TCII'TC7lCCAOCTAACCAATGATCCTGCCGICTGCACCIGACGCA C CACCGTGATA
M V I F F C P P N Q C I R H A V C L Y L D S M A I F L T L L L M L S G P R W I S
TATA BOX
GTCTTGGCGCGAr,C711CGkCAACOSGAC'TlrAArGCCACGr CACGACGGA AGTCCTARGATCCGCCGr=CAGCA0CGTaCA=CMCAGC
C T
L S D G V P L D L T A A S T T G K S * M L A
CATG=YxTCCCGCTAGAATGGAC0GTCGAGGAGGT=CCCrCACCTAGAAGA'GGGTATGGCSAGOSCTGC3CCCGTCC7K)TCCCCA(TTACOGCCACOCGTCGCAGC
H V V P L E W T V E E V V P Y L E R L A V W L R A S V L V A F Q L T A T V A L S
GTr.CTCAGCC.ITG~TGCCOCACCGr.TG..C3GCTGArCGCATGGGGGGACGGGA0ACACCCCGCGC ACCACCAOTGATTGCCA GGCIrCCTSTGC V L S W W L M P P P V A E L C E R G R D D D P P P L S H L S L V V P V G C L F L
CT,GCTACGCGGsICCGTCGkTCGAOC GTTGlUCCCGGmAACTCOCTCITGTIGCrACAGCT(rCGCA0GCGCTAGCCITCAC IGCrC-7G'AGCCA'MCG:AGGCC
L L R G P S I D R C P R K L P L L L A Y C L P H A L A F L T L L M C Q P S P Q A
DR5,._ DR5 ,_
IIIIIrGGCGC(33CloGC(rlA3TGAlCTAGCICGGCCllTlCGC' lleTrCqYGACC.,-GCGTCICGCCG(CIr-GCTACCGTC(GrICYVCTG
F V G A A L L A L A V D L S C L G A S L L G C D P G A S L R R L W L P S V L S L Spl/IR7 DR6 ,_ DR6 ,
CTCTGCGCTACGGG=GCGCTCGGSCCGC G:CCCCTCGTrIr=GTTACA a3CGACGkCGACGIrACMr.AC(TGT<TAATwACGA7CG7ATC
Spl/IR7
L C A T A L G L W L L R A A A P F F L G L H A T T L L T V T L M L I H D L S L I
120
240
360
480
600
720
840
960
1080
1200
1320
1440
1560
1680
1800
1920
Towne Genomic
AD169Genomic
TowneGenomic AD169 Genomic TownePeptide
Towne Genomic AD1 69 Genomic TownePeptide
Towne Genomic
AD169 Genomic TownePeptide
Towne Genomic AD169Genomic TownePeptide
AD169Peptide
Towne Genomic AD1 69 Genomic TownePeptide
AD169Peptide
Towne Genomic AD169 Genomic TownePeptide
AD169Peptide
TowneGenomic Towne cDNA AD169 Genomic TownePeptide
AD169Peptide
TowneGenomic Towne cDNA AD169 Genomic TownePeptide
AD169Peptide
Towne cDNA AD169 Genomic TownePeptide
AD169Peptide
TownecDNA
AD169 Genomic
TownePeptide
AD169Peptide
TownecDNA AD1 69 Genomic TownePeptide
AD1 69Peptide
TownecDNA AD169Genomic TownePeptide
AD169Peptide
Towne cDNA AD169 Genomic
TownePeptide AD169Peptide
Towne cDNA
AD1 69Genomic
TownePeptide
AD169Peptide
Towne cDNA AD169 Genomic
TownePeptide
AD169Peptide
OG ;c
SG :G
IY LQ
on November 9, 2019 by guest
http://jvi.asm.org/
STRUCTURALLY TRICISTRONIC GENE OF HCMV 2051
DR7 ,_ DR7 ,_
2040
T C Q S S F P E S F Q P S L R L Y V E N V A L F I G M Y H L L R L W L W S P *
F CAP1.1 KbufA
CAATBOX TATA BOX
2160
r-11CDNA 1. 1 kb
M G D T A S V S E H H E S P T V T I V P L H R S H A L V A EQ Q L F Q W
w T CZI~ZZ~CGGGACAGTGCG
T
L K R F K L L M E V Y H G L V W Q L A C T L T V C L L A W L A F P DV Q G Q C A
A A T
N G I V P A L S S I V P V S T L A M L R G F A E F R P H T T N F A H L T V A C L
~~ ~~CN3CGGTIlrllACC¶ACIQG
L I N T G I T V C T G F C G E R R V I G L S F A L V M V F F V L C S G L T Y L A
CCGGCACAAZ0ACAACTGTCATCACA3CGGAC
C C
G N N_L_P R W K V I GI G Y G W S V I V F Y L L L Y F S P V L W V S K I Y S G L
Y V L V V T A A S A V L I Y E T L D L I Y Q R G T L S K N S V C V S V V L Y T I
7 ATCGGI'rC ATAMACA CTCPr ATAAA
A G
V M S L L N M S V A I F S G H V W V Q Q Y A E K H G G R I D G V S L L S L L *
AAGAT1 GGAITGC IC,CCCI=GAAAGGTC
C G A T T
POLY ASIGNAL
CGTT1ITmATI'CAAC6T _ _ _CGTkAskssskhshksskkhksAkkkk_kkhs
2280
2400
2520
2640
2760
2880
3000
3120
3209
FIG. 6. Nucleotidesequenceof thestructurally tricistronicgeneencodingthe2.85-kbmessageand theputative polypeptidesUs18,Us19,
andUs20.The2,290-bp cDNA(2.3-kb cDNA)and1,008 bpofgenomicsequencelocatedupstreamofthiscDNAwerecombined andare
shown inthisfigurewithanoverlap of 140 bp. Sequence mismatches between the Towne andAD169 strainsarenoted intheAD169genomic
sequence.Thepolypeptidesequencesareindicated belowthe DNAsequence.Thetranscription initiationsitesidentifiedby primerextension andS1nucleasemappingareindicatedbysolid bentarrows.The minorcapsite forthe 2.85-kb mRNA is also indicated. Thebeginningsof
thetwocDNAs used in thisstudyarealso marked withopenbentarrows. Upstreamsequenceanalysisof the2.85-kb and 1.1-kb mRNAs revealedseveral indirectrepeats(IR)and directrepeats(DR),whicharenumberedsequentially.Solidhorizontalarrowsindicate the IR and
openhorizontalarrowsindicate the DRupstreamofUS18andUs20.Thepolyadenylation signalsequenceandpotential glycosylationsites
areunderlined. Thepossiblesecond translation initiation codon forUs20isindicatedbyabentarrow.
verylow levelat18 hp.i.andwasclearlydemonstratedat34 h p.i. (Fig. 7B). The
anti-Us20-GST
antibody detected a major 43-kDavirus-specific product at 18 hp.i. (Fig. 7C). This translation result also matched the 2.85-kb mRNA transcription data. A minor bandof36kDawas foundatalate time(34h)in virus-infectedlysates.This 36-kDaprotein mightresult from theuseof the second translation initiation codonatlater stages of virus infection.
Antibodies generated in a rabbit against the bacterially expressed fusion proteins
(Us18-, Us19-,
and US20-GST)gave positive immunofluorescence staining in HCMV-in-fected cells. This reactionwas not found in mock-infected cells orat 12 h after HCMVinfection. The anti-Us18-GST
antibody detected positive immunofluorescence only at 48 and 72 h p.i. (Fig. 8). This result suggests that this
virus-specific productwastranslatedat48h,whichcoincided with thetranscriptiondata for the 1.1-kb mRNA. The
Us19-GST
antibodiesdetectedpositiveimmunofluoresenceat24, 48,and 72 hp.i.,with patterns similartothat with the
anti-Us20-GST
antibody (datanotshown).Thisresult indicates that theUs19
virus-specific products appear earlier than the
Us18
virus-specific products.TheUS20-GST
antibodiesdetectedpositive immunofluorescent reactionsat24, 48,and72 hp.i.,liketheUs19-GST
antibodies, but the reactionwasstronger than itwas with the
Us19-GST
antibodies at 24 h p.i. (data not shown). This result indicates that the Us20 virus-specific products appeared earlier than the Us18 and US19 virus-specific products. The early translation of the Us20 virus-specific products also matches the 2.85-kbmRNA transcrip-tion data.VOL. 67, 1993
Towne cDNA AD169 Genomic TownePeptide AD169Peptide
Towne cDNA
Towne cDNA AD169Genomic TownePeptide
AD169Peptide
Towne cDNA AD169Genomic TownePeptide
AD169Peptide
Towne cDNA AD1 69 Genomic TownePeptide AD169Peptide
Towne cDNA AD169 Genomic TownePeptide
AD169Peptide
Towne cDNA AD169 Genomic TownePeptide
AD169Peptide
TownecDNA AD169Genomic TownePeptide
AD169Peptide
Towne cDNA AD169 Genomic TownePeptide
AD169Peptide
Towne cDNA
TownecDNA
ACC'=AGAGTrC7q'rO::.CAG'rrrICAGCC
on November 9, 2019 by guest
http://jvi.asm.org/
[image:9.612.69.555.69.474.2]DISCUSSION
MW
Mock 5 18 34 48 72
Genomic repetition is a common characteristic of the humanherpesvirusgroup, but gene clusters inthe
Us
region are a unique characteristic in HCMV and herpes simplex virus (3, 24). Before the HCMV genome had been com-pletelysequenced, weoriginally speculatedthat the Towne strain EcoRI D fragment located within theUs
regionencoded theHCMV counterparts ofherpessimplexvirusgD andgC. Inourfirst attemptto screenforgD-relatedcDNA, a2.3-kb cDNAwasisolated fromaXgtllcDNA
library.
This cDNA has been used as a tool to characterize thecorre-spondinggene. Tooursurprise, insteadofisolating gCorgD homologs, we identified the
US18, US19,
andUS20
tricis-tronic gene.In the study of the transcription products related to the 2.3-kb cDNA, twomajormessages (a2.85-kb mRNA and a
1.1-kb mRNA) and a minor 1.8-kb mRNAwere identified.
Detection of the minor message by the 2.3-kb cDNA probe wasnotconsistent,varyingfromonepreparationtoanother. FromDNAsequencingand ORFanalysis,theminor 1.8-kb productmight bederivedfrom the secondORF,which does nothaveastrongclassicalpromoter-enhancer sequence and mightrequire special cellularorviral products for maximal expression. Theexpression of thisgroupof messagesmight
be under very tight control. The presence of the 1.8-kb cDNA reflects the existence of this weak message.
Mean-while, the detection of 2.85-kb mRNA suggested that the 2.3-kb cDNAisnot afull-length cDNA,since
approximately
628bp of the 5' upstream sequencewasmissing. Also, the 1.1-kb mRNAcould represent the 1.1-kb cDNA.
As determined by primer
extension,
the 2.85-kb mRNA has twotranscription initiationsites;oneisrightat the first translation initiation codon of theUS20
ORF, and the other is 243bp downstream from the first site. There isa typicalTATA sequence (TATAAGA) 21 bp upstream of the first transcription initiationsite, but no typical CAAT sequence wasobserved. Therewere twoGC boxes andadirect repeat located about 40to80bpupstreamof thistypicalTATAbox. On the otherhand, no typical TATA
[TATA(A/T)A(AJT)]
box or CAAT
[GG(C/T)CAATCT]
box upstream of thesecond cap site was found. It is unclear why the major transcription initiationsite for the 2.85-kb mRNA islocated right at the first translation initiation codon of
Us20.
Ingeneral, leader sequences are20to 100nucleotideslongon most vertebrate mRNAs except for mRNAs derived from proto-oncogenes, which have very long leader sequences
(12).Inaddition,the ATG codon isnotwellrecognized when it occurs close to the cap (12). Therefore, the translation regulation of the
Us20
ORF needs further investigationbefore anyconclusionscan bemade.
Thetranscription initiation site of the 1.1-kb mRNAwas 33bp upstream of the translation initiation site.Intheregion upstream ofthistranscript, there is an atypical TATA box sequence(GATAAGA)andanatypical CAAT box sequence
(CGTCAATCA)
within 22to28bp and 50to58bp upstreamofthe cap site, respectively. Promoter activity upstream of the 2.85-kb mRNA was detected during HCMV infection
(data not shown). The promoter activity of the region upstream of the 1.1-kb mRNAis still under investigation. Withregard to the
Us19
ORF, there are typical TATA andGCboxes upstream of the translation initiation site and a minor 1.8-kb mRNA which could correspond to a transcript
initiatingnear these potentialtranscription start sites. How-ever, this message was not clearly demonstrated by
Si
mapping analysis.
46 K
A--*-36 kD
30 K
--.. 1,
21.5 k -.3m- "M,¶I:
46 K-- B
30 K
-1-21.5 K
--46 K -i C
--32 kD
-- - -. 43 kD
-K-36 kD
[image:10.612.329.553.79.284.2]30 K -E
FIG. 7. Western blot analysis of HCMV-infected HEL cellswith
anti-Us18-GST, anti-Us19-GST, and anti-Us20-GST antibodies. DNAfragmentscorresponding to ORFsUs18, Us19,andUs20were isolatedby restriction digestion and then subcloned into the pGEX-GSTgene fusion system forexpression in E. coli(21). Thefusion proteins obtained were used to generate antibodies in rabbits for immunological analysis of these peptides. HCMV-infected HEL cells(2x 105cells) harvestedatthetimes indicatedabove the lanes (hoursp.i.)wereused for Westernblotanalysis.Arrows ontheright indicate virus-specific bands, and arrows on the left show the positionsofsize markers (inkilodaltons). Antibodiesderivedfrom the (A) Us18, (B) Us19, and (C) Us20 fusion proteins detected virus-specific bands of 36, 32, and 43 kDa,respectively.
Sense-strand oligonucleotides targeting either the 5' end orthe 3' end of the 2.85-kb mRNAprobes didnotdetect any antisense messages transcribed from this genomic map re-gion (data notshown). An antisenseoligonucleotide (primer 136) targeting the 3' end of the 2.3-kb cDNA could detect boththe 2.85-kb and 1.1-kbmessages. This result suggested that these two messages were transcribed in the same direction. Thetranscription pattern of this structurally tri-cistronic gene is very similar to that of HCMV tegument proteins pp65 and pp7l, in which two messages are tran-scribed from the corresponding genomic region. A large 4.0-kb mRNA and a small unspliced 1.9-kb mRNA are transcribed witha3'-coterminal end.The pp65 matrix tegu-ment protein is translated from a 4.0-kb mRNA, and the pp7l high-matrix tegument is made from a 1.9-kb mRNA (18).
Transcriptionkineticswerestudied with RNAs harvested at designated time points and after appropriate drug treat-ment. These studies revealed that the 2.85-kb and 1.1-kb mRNAs belong to two different classes of transcript. The 2.85-kb messageisanearly or ,-type mRNA which does not require virus DNA replication for maximal expression. The 1.1-kbmRNA,expressed late(48 h p.i.) after virus infection and requiring viral DNA replication for maximum expres-sion, isalateor y-type message.This differential expression occurs even thoughthese transcripts are derived from the samegenomic map region and overlap at the 3' end. This is common in the transcriptional regulation of early and late HCMVmRNAs,inwhichearly and late mRNAs are trans-lated afterimmediate-early mRNAs (2, 5, 22, 23).
VIROL.
on November 9, 2019 by guest
http://jvi.asm.org/
STRUCTURALLY TRICISTRONIC GENE OF HCMV 2053
FIG. 8. Indirectimmunofluorescencestudy with antibodies againstHCMVfusion proteins. The rabbitantibodies used in this study were preabsorbedwith HEL cellular lysates. (Inset a)Mock-infectedHEL cells.(Ato D)HCMV-infected HEL cells at 12, 24, 48, and 72 hp.i., respectively,werereacted first with the antibody against the fusion protein and then withbiotin-conjugated goat anti-rabbit immunoglobulin Gantibody andstainedwithfluorescein-labeled avidin. Arrows indicate positive staining. Only results forUs18areshown. Panels a, A, and B weredeliberately overexposedtoshow cellmorphology.
The tricistronic 2.85-kb mRNA was transcribed after 12 h
p.i.andreachedthe maximumtranscriptionlevel at 24 h p.i. The first5'-end translationalproduct of the 2.85-kb mRNA, the
Us2O
polypeptide,wasdetectedat18 h p.i., and its levelincreased slightly over the infection period. The
Us19
polypeptide couldbedetectedat averylowlevelat18 hp.i.,
buthigherlevels of
Us19
weredetectedbetween 24and 36 hp.i. by immunofluorescence and Western blot analyses.
Transcription of the 2.85-kb mRNA decreased after it reached its maximum level. In contrast, translation of this message increased continuously over an extended period.
This result probably reflects the stability of the translation
products or an increase in the translation efficiency of the 2.85-kb mRNAatlate stagesof infection.
Aspointedoutabove,averyminor 1.8-kb mRNAspecies
wasinconsistently detectedwhenthe2.3-kb cDNAwasused as a probe in Northern hybridization. Several attempts to map transcription initiation by S1 mapping andprimer ex-tension failed. These results suggest that anextremelylow level oftranscription,if any, of the 1.8-kbmRNAmayoccur atthe later stages ofinfection. Thisspeculationis alsobased upon the isolation of a 1.8-kb cDNA, derived from this
region, from the cDNAlibrary ofHCMV-infected cells. At thistime,wedonothavedefinitive datatoconclude whether the 32-kDa
US19
peptideis derived fromthe 2.85-kborthe 1.8-kbtranscript. The32-kDaUs19
peptidecouldbarelybe detected at18hp.i.butwasdetected atalevelcomparable
to that of the 43-kDa
Us20
peptide. If the stability and thelevelsoftranslation efficiencyarecomparablebetween these transcripts, then the level of mRNA for
Us19
should becomparabletothatfor
Us20.
Immunofluorescentstaining of theUs19
peptide indicated that it appeared as early as theUs20
peptide,at24 hp.i.,butwaslessabundantthanUs20.
From these data, we believe that the
Us19
ORF may betranslatedfromthe 2.85-kb mRNA.Expressionof this ORF mayrequire avirus-induced factor(s) for maximal activity.
The monocistronic 1.1-kb mRNAwas translated at a later timep.i.,andits translationsubsequentlyremainedconstant
throughout theinfection. This correlatedwellwith the late translation of the
US18
virus-specific polypeptide.Antibodiesprepared againstthe threepeptides,expressed
as fusion proteins, detected virus-specific proteins in
HCMV-infected cell lysates. The differences between the sizes determined by Western blot analysis and the sizes
predictedfrom sequence analysis may be dueto posttrans-lational modification. These antibodies gavepositive
immu-nofluorescencewhen testedduringthe late stages of HCMV infection in HEL cells, but the virus neutralization assay resulted in lowneutralization activities. The low neutraliza-tionactivity of these antibodies could be dueto
inappropri-ate folding of the fusion proteins or the lack of posttrans-lational modification of thesevirus-specific products
expressedin E. coli.It isalso
possible
thattheseproteins
arenotessentialforinfectivity.
Expression
of these ORFswith VOL. 67, 1993on November 9, 2019 by guest
http://jvi.asm.org/
[image:11.612.77.543.76.389.2]HUANG
abaculovirusvectorin insect cells is underwayfor further functional and antigenic studies.
ACKNOWLEDGMENTS
WethankTimothyKowalik,BretWing,andDavidY.Huang for useful discussions andShu-mei Huongfortechnical assistance.
Thisinvestigationwassupported inpartbyPublicHealth Service grants AI-12717 and CA-21773 from the National Institutes of Health.
REFERENCES
1. Berk, A. J., and P. A. Sharp. 1977.Sizing and mapping ofearly
adenovirus mRNAsby gelelectrophoresis of S1 endonuclease-digested hybrids. Cell 12:721-732.
2. Chang,C. P., D. H. Vesole, J. Nelson, M. B.A. Oldstone, and M.F. Stinski. 1989. Identification and expression ofahuman cytomegalovirus earlyglycoprotein. J. Virol. 63:3330-3337. 3. Chee,M.S.,A. T.Bankier, S.Beck, R.Bohni, C.M.Brown,R.
Cerny,T.Horsnell, C.A. HutchisonIII, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P.Tomlinson,K. M. Weston,and B.G.Barrell.1990.Analysisof theprotein-coding content of thesequence of HCMV strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169.
4. Chou,P. Y., and G.D. Fasman. 1974. Prediction ofproteins conformation. Biochemistry 13:222-245.
5. Davis, M.G., andE.-S.Huang.1985.Nucleotidesequenceofa
humancytomegalovirus DNAfragment encoding a 67-kilodal-tonphosphorylated viralprotein. J. Virol. 56:7-11.
6. Gretch,D.R., B. Kari, R. Gehrz,and M. F. Stinski. 1988. A multigene family encodes the human cytomegalovirus glyco-protein complex gCII (gp47-52 complex). J. Virol. 62:1956-1962.
7. Harlow, E., and D. Lane.1988. Antibodies-alaboratory
man-ual, p. 90-91. Cold Spring Harbor Laboratory, Cold Spring Harbor,N.Y.
8. He, Y. S., L.Xu, and E.-S.Huang. 1992. Characterization of human cytomegalovirus UL84 earlygene andidentification of itsputative proteinproduct.J.Virol. 66:1098-1108.
9. Kilpatrick, B.A.,andE.-S.Huang. 1977. Human cytomegalo-virus genome: partial denaturation map and organization of genome sequences. J.Virol. 24:261-276.
10. Kingston, R. E. 1987. Guanidinium method for total RNA preparation,p.4.2.1.-4.5.3. InF. M.Ausubel,R. Brent,R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl(ed.),Currentprotocolsinmolecularbiology. John Wiley &Sons,New York.
11. Kingston, R. E. 1987.Primer extension,p.4.8.1.-4.8.3. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J.A.Smith,and K.Struhl(ed.),Current protocols in molecularbiology.JohnWiley & Sons, New York.
12. Kozak, M. 1987. Ananalysis of 5'-noncoding sequences from 699 vertebratemessenger RNAs.Nucleic AcidsRes. 15:8125-8132.
13. LaFemina, R., and G. S.Hayward. 1980. Structuralorganization of theDNAmolecules from humancytomegalovirus,p.39-55. In R. Jaenisch, B. Fields, and C. F. Fox(ed.), Animal virus genetics. Academic Press,NewYork.
14. LaFemina,R.,andG. S.Hayward. 1983.Replicative forms of humancytomegalovirusDNAwithjoined terminiarefound in permissively infected human cells but not in non-permissive Balb/c-3T3mousecells. J. Gen. Virol. 64:373-389.
15. Mocarski, E. S.,L. Pereira,and L.McCormick.1988. Human cytomegalovirus ICP22, the product of the HWLF1 reading frame, isanearly nuclear protein that is released from cells.J. Gen. Virol. 69:2613-2621.
16. Nathans, J., and D. S. Hogness. 1983. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovinerhodopsin. Cell 34:807-814.
17. Rowe, W. P., J. W. Hartley, S.W.Waterman, H.C.Turner, and R.J.Huebner. 1956. Cytopathogenicagentresembling human salivary gland virus recovered from tissue culture of human adenoids.Proc.Soc.Exp. Biol. Med. 92:418-424.
18. Ruger,B., S.Kiages,B.Walla, J.Albrecht,B.Fleckenstein,P. Tomlinson, and B. Barrell. 1987. Primarystructure and tran-scription of thegenescoding for thetwovirionphosphoproteins pp65andpp7l of human cytomegalovirus.J.Virol. 61:446-453. 19. Sanger,F., S.Nicklen,andA. R. Coulson. 1977. DNA sequenc-ing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 75:5463-5467.
20. Seldon,R. F.1987.Analysisof DNA sequencesbyblottingand hybridization,p.2.9.1-2.9.9. In F. M.Ausubel,R.Brent,R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl(ed.),Currentprotocols in molecularbiology. John Wiley & Sons, New York.
21. Smith,D.B., and K. S. Johnson.1988.Single-steppurification of polypeptides expressedinEscherichia coliasfusionwith gluta-thioneS-transferase. Gene 67:31-40.
22. Stinski, M. F. 1978. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-in-ducedpolypeptides. J.Virol. 26:686-701.
23. Wathen, M. W., and M. F. Stinski. 1982.Temporalpatternsof humancytomegalovirus transcription: mapping the viralRNAs synthesized at immediate-early, early, and late times after infection. J. Virol.41:462-477.
24. Weston, K. 1988. An enhancer element in the short unique region of human cytomegalovirus regulatesthe production of a group of abundant immediate early transcripts. Virology162: 406-416.
25. Weston, K., and B. G. Barrell. 1986. Sequence of the short unique region, short repeats and part of the long repeats of human cytomegalovirus. J. Mol. Biol. 192:177-208.