0022-538X/87/062033-05$02.00/0
Copyright© 1987, American
Society
for MicrobiologyEffects of cDNA Hybridization
on
Translation of
Encephalomyocarditis Virus RNAt
DING S. SHIH,1* IN-WOO PARK,' CHRISTOPHER L. EVANS,' JESSE M. JAYNES,'
ANDANN C. PALMENBERG2
Departmentof Biochemistry andLouisiana Agricultural Experiment Station, Louisiana State UniversityAgricultural Center, Louisiana State University, Baton Rouge, Louisiana 70803,1andBiophysics Laboratory, University ofWisconsin,
Madison, Wisconsin 537062
Received 28 October 1986/Accepted 16 February 1987
Cell-free translation of the RNA of encephalomyocarditis virus was examined after hybridization of chemically synthesized cDNA fragmentsto different sites ofthe 5' noncoding region of the viral RNA. The following resultswereobtained. (i) The bindingof cDNA fragmentstothefirst41 nucleotides,tothepoly(C) tract(between nucleotides 149and263), and tothe sequencebetween nucleotides 309 and 338 didnotaffect translation of the viralRNA; (ii) the binding of cDNAfragmentstothesequencebetween nucleotides 420 and 449causedaslight inhibition;and(iii)the bindingof fragmentstoeight differentsites between nucleotides 450 and theinitiator AUG codon (nucleotide 834)causedhighdegreesofinhibition. The resultssuggestthat thefirst partof the 5' untranslated region, atleasttonucleotide 338, maynotbe requiredfor encephalomyocarditis viral RNA translation; however, the regionnearnucleotide 450 is important for translation of the viral RNA. The possibility that initiationoccursataninternal site is discussed.
Encephalomyocarditis (EMC) virus is a member of the
picornavirus family. Muchis known about the protein
syn-thesispathways ofpicornavirusesand about the products of
thesynthesisreactions. However, an important aspect of the
viral protein synthesis process, the initiation of translation,
isstillunclear at present.
For most eucaryotic mRNAs, the ribosome-scanning model has been accepted to explain the translational
initia-tion process (7, 8). In this model, 40S ribosomal subunits
first bindtothe 5' end of a messenger and then migrate along
theRNAchainuntil they encounter the first favorable AUG
triplet, at whichpoint translation begins. An essential
fea-tureof this scanningmodelisthat the 40S subunitsrecognize
andbindonlytothe 5' endsof themRNAmolecules before
thestartof thescanningprocess.The 5' cap structure,which
is present in most eucaryotic mRNAs, is known to be
involved in the ribosome-binding reaction (forareview, see
reference3).
Unlike most eucaryotic mRNAs, picornaviral RNAs do
nothavea5' nucleotidecap structure. Instead, they havea
covalently linked protein (VPg) at their 5' ends (26). This
genome-linked protein is not required for translation (18).
Therefore, during initiation of picornaviralRNAtranslation,
it is possible that an alternate ribosome recognition and
binding mechanism is involved. Forexample, theribosome
subunits mayrecognizeand initially bindto aninternal site
ofthe viralRNAbeforethe scanningprocessbegins.
We have investigated this possibility by usingEMC virus
as amodel. EMC viralRNAisaparticularlyactive
messen-gerinseveralcell-freeprotein-synthesizing systems(16, 20,
24). In rabbit reticulocyte lysates, the viral RNA is
com-pletely and efficiently translated, and the translation
prod-ucts are accurately processedtoform matureviralproteins
(16, 20, 21,24). Recently,thecomplete nucleotidesequence
of EMC virus was determined (14, 25; A. C.
Palmenberg
*Corresponding author.
tPaper 86-12-0184 of the Louisiana
Agricultural
Experiment
Station, LouisianaState University,BatonRouge.
and G. M. Duke, manuscript in preparation). The RNA is 7,835 nucleotides long, and the translation initiated AUG
codon is located at nucleotide 834 (14). This codon is
followed by an open reading frame of 6,876 nucleotides,
which encodes the viral polyprotein. The initiator AUG is
precededby 10otherAUGtriplets scattered throughout the
5' noncoding region. The present study was designed to
investigatethemechanism by which ribosomes initiate
trans-lationatthe particularAUG triplet in position 834.
Oligodeoxyribonucleotides were synthesized on an
Ap-plied Biosystems380A DNA synthesizer. After completion
ofthesynthesizing cycles, theoligonucleotide solution from
the synthesizer was incubated with 1 ml of concentrated
NH40H for 4 to 12 h at 50°C to completely remove the
blockinggroups. The solution was thenevaporatedto
dry-nessinarotary evaporator.Thedry residuewasdissolvedin 1 ml of 25 mM triethylammonium bicarbonate (TEAB), pH 7.6,and the solutionwasevaporated. ThisTEABtreatment
stepwasrepeatedtwice. The residuewasdissolvedin 3 ml
of TEAB, and the oligonucleotide fragments were isolated
by using a Sep-pak cartridge (11), whichserved to remove
prematurely terminated sequencesfromthecomplete
oligo-nucleotide chains. The oligonucleotide fractioneluted from
the cartridge was dried, and the 5'-protecting groups were
removedby dissolvingthenucleotidesin1 mlof80% acetic
acidandincubatingthe solutionat roomtemperaturefor25
min. Afterevaporation, the sample was dissolved inwater and dried. Thefully unprotected productwasfurther
purified
by fractionation on a 20%
polyacrylamide gel
underdena-turing conditions. The conditions forelectrophoresisandfor
oligonucleotide
isolation fromthegel
have been describedelsewhere(11).
All hybridizations weredone in reaction mixtures
(8 ,ul)
containing20 mMHEPES
(N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic
acid)
(pH 7.6)-0.1 M NaCl-1 mM EDTA.The RNA concentration was 0.5
mg/ml,
and cDNA was added inafivefold molarexcess.Thereaction mixtureswere heatedat70°C
for10minfollowedby
incubationat45°C
for 2033on November 10, 2019 by guest
http://jvi.asm.org/
Oigo(dG)
z2w
1170011
(Fig. 2A)wasthe result ofhybridizationtotheA10andA12
oligonucleotidefragments.
The effectsontranslation ofhybridizationofseven other
fragments(A4 through A9andAll) areshown(Fig. 2C,2D,
[image:2.612.66.280.69.271.2]and3). The data indicate that fragments A4 through A7 and All caused more than 80% inhibition of translation; frag-ment A8 caused 65% inhibition, and fragment A9 caused 75% inhibition.
Figure 3depictsamino acid incorporation data for A4- and A3-hybridized RNA. As described above, hybridization of A4 caused a strong inhibition of translation. On the other hand, hybridization ofA3, which correspondstotheregion
immediately 5' tothe A4-hybridized region, caused only a low level of inhibition. The translational activity of the A3-hybridized viral RNA was about 70% of that of the control(unhybridized) RNA.
Itwaspossible that the lowdegree of inhibition observed with A3 wasdueto weak orincomplete hybridization with viral RNA. However, this possibility was ruled out by primer extension experiments. Theelectrophoresis patterns FIG. 1. 5'-Untranslatedregion of EMC viral RNA.Symbols:-,
5'-noncoding sequence; _, location of initiator AUG codon; , locations of other AUG triplets; -_ , positions of termination codons;L.J, cDNA fragments. For the indicated fragments, the lengths and theexactnucleotide locations(given inparentheses) of thesites of hybridization were:Al,41(1to41);A2,30(309to338); A3, 30 (420to449); A4,30(450to479);AS,30(480,to509);A6,30 (510to539);A7,30(590to619); A8,33(667to699); A9,34(710to 743); A10, 40 (744to783);All, 30 (784to813); A12, 41(814to854).
3 h.Nonhybridized control samplesweretreatedidentically,
exceptthat cDNA was omitted from the reactionmixtures.
Thehybridization procedure described abovewas
devel-oped by combining the procedures of Hastie and Held (5)
and Liebhaber et al. (10). The first method (5) involved
heating mRNA and single-stranded cDNA in an aqueous
solutionat65°Cfor30min. The second method(10) involved
heating mRNA anddouble-stranded DNA in a
formamide-containing buffer for10min at70°Cfollowedby incubation
at 45°C for 3 h. Each ofthe methods produces complete
hybridization.
Translation ofthehybridizedRNAsampleswasexamined
by using the mRNA-dependent rabbit reticulocyte system
(17). Reticulocyte lysates were obtained from Green
Hect-are; Oregon, Wis. The conditions fortranslationhave been
describedpreviously (20, 21).
Figure 1depicts schematically the5' region of EMCviral
RNA and the locations of cDNA hybridization sites. The
cDNAfragments, designatedAl through A12, are 30 to 41
nucleotideslong, and their positionsare indicated.
Translation of cDNA-mRNA hybrids. EMC viral RNA was testedforits abilitytodirect translation afterhybridization
to various cDNA fragments. The effects with two of the
fragments, A10 and A12, are shown (Fig. 2A). A12
hybrid-izes to the genomic region encompassing the polyprotein
initiation codon, including 18 upstreamand 20downstream
nucleotides.A10 hybridizes to a region that is 30 nucleotides
5' tothe A12-hybridized region. The data indicate thatA10
andA12caused 93 and77%inhibition of normalEMC viral
polyprotein
translation, respectively.The results of a controlexperiment in which theA12-mRNA complex was
dissoci-atedby heatingat97°Cbefore translation are given (Fig. 2B).
Heating ofthe complexeliminated most of the inhibition. We
therefore concludethatthe decrease in translation observed
0
15 30 45 60 0 IS 30 45 60
2 C D
12 3 4 0 0
0/
9-
6-6-
4-x~~~~
0 153045 60 0 1530 45 60
Time (Min)
FIG. 2. InhibitorycDNAfragments.Theconditions for hybrid-ization and for translationwere asdescribed in thetext.The control RNAsampleswereheated andincubated in thesame manner asthe hybridizedRNAsamples,except that cDNAfragmentswere omit-ted.(A)Effectofhybridization ofA10andA12.Symbols:0,minus RNAcontrol;A, A10;x,A12;0, minuscDNA control. (B) Effect ofdehybridization of theannealedcomplexes ofA12andEMC viral RNA. The hybridization solution containing the annealed com-plexes wasdivided intotwoaliquots. Onewastranslated directly, andtheotherwasheatedat97°C for3min,quickly frozenin adry ice-ethanolbath, thawed,and translated.Symbols: 0,minusRNA control; A, A12-mRNAcomplexes; x,dehybridized mixture; 0, minuscDNAcontrol.(C) Effects of A8, A9, andAll. Symbols:0, minus RNA control; O, All; A, A9; x, A8; 0, minus cDNA control.(D) Effects of AStoA7.Symbols:0,minusRNAcontrol; O, AS; A, A6; X, A7;0,minus cDNA control.
1-0
A2
.A3.
A4.A5
a A A
3m--TB-t5'
,A6,
, 7, ,AA9
.T
1001
1
.. W
I r,
I
t
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.324.539.295.583.2]ofprimer extension products synthesized from RNA tem-plates primed with A3 (nucleotides 420 to 449) or with All (nucleotides 784 to 813) are shown (Fig. 4). The formation of
only one major product was seen in each case. The chain
length was approximately 350 nucleotides for the major
A3-extension product and 750nucleotidesfor themajor All
extensionproduct. Densitometertracings (notshown) of the
gelprofiles confirmed that equal molar amounts of product
DNAweresynthesizedinreactions with the two primers (A3
andAll),indicatingthat the extent ofhybridizationof A3 to RNA wassimilar to that of All. Therefore, although the two
cDNAs differed significantly in the ability to inhibit EMC
viral polyprotein translation, these properties were not
causedby differences in hybridization efficiencies.
Thehybridization of Al,which corresponds to the first 41
nucleotides,causedno detectable inhibition of translation of
the viral polyprotein (Fig. SA). The same result was also
obtained when the hybridization reaction was done by
heating the solution to a much higher temperature (97°C)
beforetheannealing step (Fig.SB). Similarly, the
hybridiza-tion of oligo(dG) or of A2, which corresponds to the
se-quence from nucleotides 309 to 338, had no effect on
translation (Fig. 5C andD).
The translation data presented above (Fig. 2, 3, and 5)
demonstrated asequential transition ofthe effect of cDNA
hybridization fromstronglyinhibitorytopartiallyinhibitory
to noninhibitory. The area of viralRNA thatis critical for
efficienttranslation seemedto centeraround nucleotide450,
which is the 5'-most nucleotide within a region, to which
cDNA fragmentA4 washybridized, leadingto strong
inhi-bition.
In addition to the hybridization-translation studies
de-scribed above, weexamined translation of EMC viral RNA
after the deletion of specific segments of the 5' noncoding
region byRNase Hdigestionof cDNA-mRNA hybrids. We
found that deletion of the 5' sequence to approximately
nucleotide 338 caused no discernible effect on translation,
but deletion of the sequence extending beyond nucleotide
450diminished the translation activity (datanotshown).
Analysis of translation products. The protein products
synthesized by various hybridized RNA sampleswere
ana-lyzedby sodium dodecyl sulfate-polyacrylamidegel
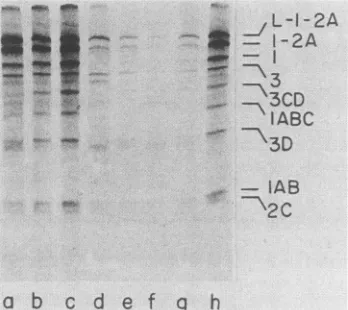
electro-phoresis. Figure 6, lanes a, b, and c, represents 45-min
translation reactions directed by A2-, oligo(dG)-, and
Al- 5-0
4-3~~~~~~~~
0L
o
20 15 30 45 60
Time
(Min)
FIG. 3. Activities ofviralRNAhybridized toA3 orA4. Sym-bols: 0, minus cDNA control; x, A3; A, A4;
0,
minus RNA control.2036-
_..,163
D ;- W:1018
-_3594_
6344
"lWMK:'298
4973
J4277
-3530
-'2027
"1
904
N1
584
N 330
x3
1564
a
b
cFIG. 4. Gel analysis of the primer extension products from A3-and All-hybridized primers. A3 and All were hybridized sepa-rately to the viral RNA. Each hybridized sample (8 ,ug of RNA) was precipitated with ethanol and dried under vacuum. The residue was dissolved in 15 ,ul of water and added to a reverse transcription reaction mixture. The transcription reaction was done by the methodof Ahlquist et al. (1), based on the original procedures of Landet al. (9)and Kacian and Myers (6). The cDNA products were analyzed on2% alkaline agarose gels bytheprocedure of McDon-nellet al.(13).Lanes: a,products from A3-hybridized template;b, products from All-hybridized template; c, radioactively labeled standards. Thepositions of stained molecular weight markers are shownatthe left oflaneaandattheright of lanec.
hybridized RNA, respectively. Also shown are the products from intact virion RNA (Fig. 6, lane h). The protein bands
(Fig. 6, in lanes a, b, c, and h) were indistinguishable in
position, although there was some variation in band
inten-sity. This variation could beattributed tothe fact that the
translation of the different RNA samples was done at dif-ferent times. Variations in the conditions oflysate prepara-tion and in thecomponents of theprotein synthesisreaction mixture couldcertainlyhavecauseddifferencesin the rates of translation andprocessing,which woulddirectlyaffect the product pattern.
Theproducts from RNA hybridized to A8, A10 plus A12, A10, and A12 are shown(Fig. 6, lanes dthrough g, respec-tively). In agreement with the amino acid incorporation data (Fig. 2 and 3), much less of the viral proteinwasproduced in thesesampleswhencomparedwiththe controlsample.
Our data demonstrated thatbinding of cDNA fragmentsto
the beginning portion of EMC viralRNA (5' to nucleotide
338) hadnodetectableeffectonpolyprotein translation, but binding of cDNA fragments to sequences 3' to nucleotide
450 severely inhibited translation. Furthermore, we found
that removal of the first 338 nucleotides by RNase H
digestionhad nodiscernible effectonsubsequenttranslation
activity of the remaining RNA fragment, while removal of
nucleotide sequencesextending beyond nucleotide 450
vir-tuallyabolished the messengeractivity.
Our resultsare incomplete agreementwith experimental data from other studies.The Lfragment generated byRNase Hdigestion ofoligo(dG)-hybridizedfoot-and-mouthdisease virus RNA serves as very active mRNA in vitro and pro-duces the same
products
asfull-length
virion RNA (5).Deletion and recombination experiments with cloned
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.373.504.75.269.2] [image:3.612.92.266.532.696.2]9o
0S=
20<
0X 2- =
0 15 30 45 60 0 15 30 45 60
FIG5. DoihbtrcDAfamns0Afeto 1y
Th Cyrdzto 0ecinmxuewshaet9° nta fa
'4
3-
2-0 15 30 4560 01530 45 60
Time
(min)
FIG. 5. NoninhibitorycDNA fragments. (A) Effect ofAl. Hy-bridization was doneunder standard conditions. (B) Effect ofAl. Thehybridizationreaction mixturewasheatedat970C instead ofat thenormal 70°C. (C) Effect of oligo(dG). The oligonucleotide was added at afivefold molarexcess of the entire poly(C) region (115 nucleotides). (D) effect ofA2. Symbols: 0,minus RNAcontrol; x, hybridized samples; 0,minuscDNAcontrol.
cDNAs have shown that the translational properties of transcribed EMC viral mRNAs reside exclusively in the 5'
noncoding portion ofthe viral genome, specifically in the
segmentextending 3'from(approximately)nucleotide 460 to
.L-1-2A
-_I-2A
_ - I
_
73CD
ITABC
,
\3D
I-AB
\2C
[image:4.612.93.267.500.655.2]a
b
cd
ef
gh
FIG. 6. Sodium dodecyl sulfate-polyacrylamide gel electropho-resispatternsof translationproducts from hybridizedRNA samples. Allsamples werewithdrawnafter 45 min of translation. Lanes: a
throughg,productssynthesized from viralRNAhybridizedtoA2, oligo(dG),Al, A8, A10 plus A12, A10, and A12, respectively; h, products from a control RNA sample. The viral proteins are
indicatedattherightof lane h.
theAUG codon thatbeginsthepolyprotein. The 5'-most 460 nucleotides, the intactpolyprotein reading frame,and the3' noncoding regions are not necessary for efficient mRNA
translation (15; Palmenbergetal., inpreparation).
Thus itis reasonabletoconclude thattheinitialsequence (about 400 bases) of theEMC viralRNAisnotnecessaryfor
translation; however, the region near nucleotide 450 is
important. On thebasis ofthisconclusion,wepropose that
for the initiation of EMC viral RNA translation the 40S ribosomal subunits may first bind at asite near orincluding
nucleotide 450 and that from this site the subunits migrate
toward the initiator AUG siteand beginprotein synthesis.
If our internalinitiation hypothesisis correct, an obvious questioniswhich factors are responsible for ribosome
rec-ognition and binding? The Shine-Dalgarno sequence is
thought to act as a ribosome-binding site in procaryotic
mRNAs (22, 23). This conserved, purine-rich sequence is
complementary to the 3' end of bacterial 16SrRNA. Since
the 3' ends of eucaryotic 18S rRNAs also contain a
con-served sequence (2, 4) which is very similar to the 3' sequence ofthe 16S rRNA, itisreasonable tosuggest that the3' endof18S rRNA maybe involvedin theinitiation of
translation ofsome eucaryotic mRNAs. In fact, the
hypo-thetical involvement of 18S rRNA in translation initiation
has already been proposed (2, 4, 19). For picornavirus
RNAs, the possibility of mRNA-rRNA interaction is
particularly relevant because of the lack of the 5' cap
structure ofthe viral RNA. Examination ofthe EMC viral RNA sequence reveals a partial complementarity
(GAAGCUUCUUGAAG [underscores indicate a
potential
basepairing])existingbetween EMC viralnucleotides483to
496 andthe 3' endof rabbit reticulocyte 18S rRNA (2,
4).
This limited
complementarity
may not be sufficient foreffective ribosome
binding.
However,thepossibility
of othersequence
complementation
between the 5'noncoding
re-gions of EMC virus and different parts of the 18S rRNA
cannotbe ruled out. Inthis
regard,
itisinteresting
to note that arecentstudyshowed thatpoliovirusRNA canhybrid-ize(understringentconditions)to aspecific region ofthe18S
rRNA as well as to several regions of28S rRNAof
higher
eucaryotes (12).
The studies presented here provide new insight into the
mechanisms of translation of EMC viral RNA and suggest
avenues for future experimentation. In light of the high
translational efficiency of this particular mRNA, we hope
that clarification of the translational mechanism may serve
as amodel forotherexperimentalsystems.
We thank Simon Chang for assistance in the synthesis of the cDNAfragments.
This workwassupported in part by the Agency forInternational Development, the U.S. Department of Agriculture Competitive Research Grants program, and theLouisianaAgricultural Experi-mentStation.
LITERATURE CITED
1. Ahlquist, P.,R. Dasgupta, and P. Kaesberg. 1984. Nucleotide sequenceof thebrome mosaic virus genome and its implications for viralreplication.J. Mol. Biol. 172:369-383.
2. Azad,A. A., and N.J. Deacon. 1980. The 3'-terminalprimary structureof fiveeucaryotic 18SrRNAsdetermined by the direct chemical method of sequencing. The highly conserved se-quences include an invariantregion complementary to eukary-otic5S rRNA. Nucleic AcidsRes. 8:4365-4376.
3. Banerjee, A. K. 1980. 5'-Terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 44:175-202. 4. Hagenbachle, O.,M.Santer,J. A.Steitz, and R. J. Mans. 1978.
on November 10, 2019 by guest
http://jvi.asm.org/
Conservation oftheprimarystructure atthe 3' endof18SrRNA from eukaryotic cells. Cell 13:551-563.
5. Hastie, N. D., and W. A. Held. 1978. Analysis of mRNA population by cDNA. mRNA hybrid-mediated inhibition of cell-free protein synthesis. Proc. Natl. Acad. Sci. USA 75: 1217-1221.
6. Kacian, D., and J. Myers. 1976. Synthesis of extensive, possibly complete,DNA copiesofpoliovirus RNA in high yields at high specific activities. Proc. Natl.Acad. Sci. USA73:2191-2195. 7. Kozak, M. 1978. How do eukaryoticribosomesselectinitiation
regions inmessenger RNA?Cell 15:1109-1123.
8. Kozak, M. 1981.Possibleroleofflankingnucleotidesin recog-nition ofthe AUG initiation codon by eukaryotic ribosomes. Nucleic Acids Res. 9:5233-5252.
9. Land,H.,M.Grez, H. Hauser, W.Lindenmaier,andG. Schutz. 1981;5'-Terminalsequencesof eukaryoticmRNA canbe cloned in highefficiency. Nucleic Acids Res. 9:2251-2266.
10. Liebhaber, S. A., F. E. Cash, and S. H. Shakin.1984. Transla-tionally associated helix-destabilizing activitiy in rabbit reticu-locytelysate. J. Biol. Chem.259:15597-15602.
11. Lo,K.-M., S. S. Jones, N. R. Hackett, and G. Khorana. 1984. Specific amino acid substitutions in bacteriorhodopsin: replace-mentofarestriction fragmentinthe structuralgenebysynthetic DNAfragments containing altered codons. Proc. Natl. Acad. Sci. USA 81:2285-2289.
12. McClure, M. A., and J. Perrault.1985.Poliovirusgenome RNA hybridizes specifically to higher eukaryotic rRNAs. Nucleic AcidsRes. 13:6797-6816.
13. McDonnell, M. W., M. N. Simon, and F. W. Studier. 1977. Analysis of restriction fragmentsofT7 DNA anddetermination of molecular weights by electrophoresisinneutral andalkaline gels. J. Mol. Biol.110:119-146.
14. Palmenberg, A. C., E. M. Kirby, M. R. Janda, N. L. Drake, G. M.Duke,K. F.Potratz, andM.S.Collett. 1984. The nucle-otideanddeducedamino acidsequencesofthe encephalomyo-carditis viralpolyprotein coding region. NucleicAcidsRes. 12: 2969-2985.
15. Parks, G. D., G. M. Duke, and A. C. Palmenberg. 1986. En-cephalomyocarditis virus 3Cprotease:efficient cell-free expres-sionfrom clones which link viral5'noncodingsequences to the P3 region. J. Virol.60:376-384.
16. Pelham, H. R. B. 1978. Translation of encephalomyocarditis
virus RNA in vitro yields an active proteolytic processing enzyme.Eur. J. Biochem. 85:457-482.
17. Pelham,H. R.B.,andR.J. Jackson. 1976. Anefficient mRNA-dependent translation system from reticulocyte lysate. Eur. J. Biochem. 67:247-256.
18. Sangar, D. V.,D. N.Black,D.J. Rowlands,T.J.R.Harris,and F. Brown. 1980. Location of the initiation site for protein synthesis on foot-and-mouth disease virus RNA by in vitro translation of defined fragments of the RNA. J. Virol. 33:59-68.
19. Sargan, ,D. R., S. P.Gregory, andP. H. W. Butterworth. 1982. A possible novel interaction between the 3'-end of 18S ribo-somal RNA and the 5'-leader sequence of many eukaryotic messengerRNAs. FEBS Lett. 147:133-136.
20. Shih,D.S.,C. T.Shih,0. Kew, M. Pallansch, R. R.Rueckert, and P.Kaesberg.1978. Cell-free synthesis and processing of the proteins of poliovirus. Proc. Natl. Acad. Sci. USA 75:5807-5811.
21. Shih, D. S., C. T. Shih, D. Zimmern, R. R. Rueckert, andP. Kaesberg. 1979. Translation of encephalomyocarditis virus RNA inreticulocyte lysates: kinetic analysis of the formation of virion proteinsand a protein requiredforprocessing.J. Virol. 30:472-480.
22. Shine, J., and L. Dalgarno. 1974.The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA:complementarityto non-sensetripletsandribosomebindingsites.Proc.Natl.Acad. Sci. USA 71:1342-1346.
23. Steitz,J.A., and K. Jakes. 1975.Howribosomes select regions in mRNAbasepair formation between the3' terminus of 16S rRNA and themRNA during initiation of protein synthesis in Escherichiacoli.Proc. Natl. Acad. Sci. USA 72:4734-4738. 24. Svitkin,Y. V., and V. I. Agol. 1978. Complete translation of
encephalomyocarditisvirusRNA and faithfulcleavage of virus-specific proteins incell-free systemfrom Krebs-2 cells. FEBS Lett. 87:7-11.
25. Vartapetian, A. B., A. S. Mankin, E.A, Skripkin, K. M. Chumakov, V. D. Smirnov, and A. A. Bogdanov. 1983. The primaryandsecondarystructure of the5'-endregionof enceph-alomyocarditis virus RNA. A novel approach to sequencing long RNA molecules. Gene 26:189-195.
26. Wimmer, E. 1982. Genome-linked proteins of viruses. Cell 28:199-201.