Copyright © 1998, American Society for Microbiology. All Rights Reserved.
NOTES
Localization of a Baculovirus-Induced Chitinase in the
Insect Cell Endoplasmic Reticulum
CAROLE J. THOMAS,1,2HELEN L. BROWN,2CHRIS R. HAWES,2BUM YONG LEE,3
MI-KYUNG MIN,3LINDA A. KING,2ANDROBERT D. POSSEE1*
NERC Institute of Virology and Environmental Microbiology, Oxford OX1 3SR,1and School of Biological
and Molecular Sciences, Oxford Brookes University, Oxford OX3 0BP,2United Kingdom, and
MOGAM Biotechnology Research Institute, Yongin-Kun, Kyonggi-Do 449-910, Korea3
Received 17 March 1998/Accepted 10 September 1998
Confocal immunofluorescence microscopy was used to demonstrate that the Autographa californica nucleo-polyhedrovirus (AcMNPV) chitinase was localized within the endoplasmic reticulum (ER) of virus-infected insect cells. This was consistent with removal of the signal peptide from the chitinase and an ER localization motif (KDEL) at the carboxyl end of the protein. Chitinase release from cells, a prerequisite for liquefaction of virus-infected insect larvae, appears to be aided by synthesis of the p10 protein. Deletion of p10 from the AcMNPV genome delayed the appearance of chitinase activity in the medium of virus-infected cells by 24 h and also delayed liquefaction of virus-infected Trichoplusia ni larvae by the same period.
The DNA genome of Autographa californica nucleopolyhe-drovirus (AcMNPV) encodes a chitinase protein similar to chitinase A of Serratia marcescens (57% identity, 65% similar-ity [2, 6]). In virus-infected Spodoptera frugiperda cells, the AcMNPV chitinase gene (chiA) is expressed as a late protein (58 kDa) with endo- and exochitinase activities (6). The AcMNPV chiA gene can be deleted without affecting virus replication in cell culture or insects, but liquefaction or melting of larvae is abolished (7). A similar effect is observed when the cathepsin gene is deleted from the AcMNPV genome (18). We proposed previously that cathepsin removes the protein from chitin in the insect cuticle, thereby facilitating the digestion of exposed chitin by chitinase (7). This mechanism, however, is at variance with a number of observations. At least 90% of the chitinase activity induced in AcMNPV-infected S. frugiperda cells in culture remains intracellular (6). Further, immunoflu-orescence staining with a polyclonal chitinase-specific anti-serum located the protein to the cytoplasm of virus-infected cells. This was inconsistent with the presence of a eukaryotic signal peptide at the N terminus of the chitinase protein (6), which should have served to attach it to the endoplasmic re-ticulum (ER), facilitating entry to the secretory pathway. Clearly, unless AcMNPV chitinase is released from cells in an inactive form, there must be a block in the secretion of this protein. In this study, we have determined the precise location of the chitinase within virus-infected cells.
Localization of AcMNPV chitinase in insect cells.Confocal laser scanning microscopy (CLSM) was used to detect chiti-nase in AcMNPV-infected cells. One million S. frugiperda (Sf9 [19]) cells were seeded in 35-mm dishes which contained 13-mm sterile glass coverslips. The cells were incubated for 1 h at ambient temperature with AcMNPV C6 (17) (10 PFU/cell) or were mock infected with medium. The cells were then in-cubated at 28°C for 48 h. The coverslips were removed from
the dishes, and immunofluorescence staining was performed as described previously (9). The antibodies employed were poly-clonal serum raised against AcMNPV chitinase (6) and fluo-rescein isothiocyanate (FITC)-conjugated anti-guinea pig goat immunoglobulin G (IgG). The preparations were examined un-der a Zeiss LSM410 confocal laser scanning microscope with a 455-nm argon laser and appropriate filter sets (see Fig. 1).
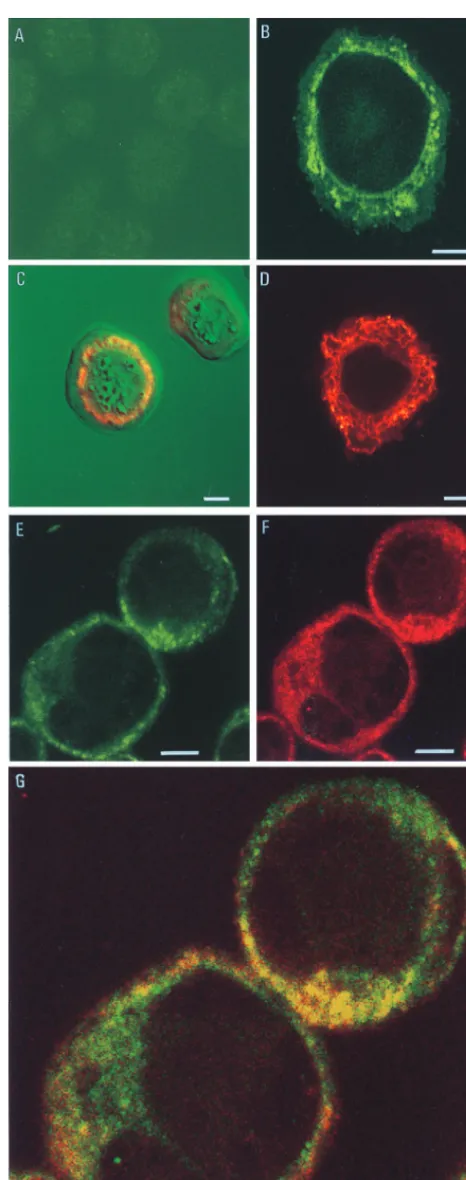
Uninfected Sf9 cells (Fig. 1A) showed no fluorescence when stained with the chitinase-specific antibody. The image pre-sented has been digitally enhanced to reveal the outline of the cells. In an AcMNPV-infected cell, the chitinase was located as a reticulate staining pattern which radiated out from the peri-nuclear region through the cytoplasm (Fig. 1B). The peri-nuclear membrane was also stained, but no fluorescence was observed at the plasma membrane. This distribution of chitinase ap-peared to follow an ER staining pattern. Characteristically, in virus-infected cells, the ER becomes dilated due to the cyto-logical effects of infection and extends from the nucleus as an irregular reticulate structure. Figure 1C is a transmitted light view of an AcMNPV-infected Sf9 cell with a superimposed image of the immunostained chitinase. The polyhedra can be seen within the nucleus of the cell. Surrounding the nucleus and extending from the nuclear membrane is the chitinase, which, in this image, is visualized in red to complement the green background.
Unambiguous evidence for the ER localization of the virus chitinase was provided by double labelling AcMNPV-infected cells with the chitinase-specific antibody and a mouse mono-clonal antibody to an HDEL ER retention motif (12). The HDEL and KDEL motifs, located at the carboxyl ends of proteins, are widely used as markers for the ER in many cell types (14). HDEL is believed to be the principal ER retention sequence in insect cells (9), so the ER can be visualized by using an HDEL-specific antibody. Figure 1D shows uninfected Sf9 cells which have been immunostained with the HDEL-specific antibody and antimouse antibody conjugated with Texas red. The native ER proteins were stained to permit visualization of the characteristic reticulate network staining of this organelle. Figure 1E to G shows AcMNPV-infected Sf9 * Corresponding author. Mailing address: NERC Institute of
Virol-ogy and Environmental MicrobiolVirol-ogy, Mansfield Rd., Oxford OX1 3SR, United Kingdom. Phone: 44 1865 281663. Fax: 44 1865 281696. E-mail: rpossee@worf.molbiol.ox.ac.uk.
10207
on November 9, 2019 by guest
http://jvi.asm.org/
calized. This confirmed that the chitinase was located in the ER of the AcMNPV-infected insect cells. Similar results have been obtained with S. frugiperda Sf21 cells (data not shown). Insect cells infected with an AcMNPV recombinant (AcchiA2) lacking the
complete chiA gene (7) failed to produce a chitinase-specific signal in immunofluorescence studies (data not shown).
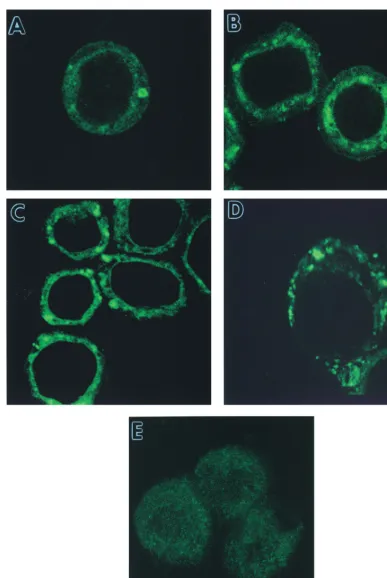
To confirm that the ER localization of chitinase was not confined to very late time points, AcMNPV-infected cells were examined at 12, 24, and 48 h postinfection (p.i.) by CLSM (Fig. 2). Faint ER staining with a chitinase-specific antibody was observed at 12 h p.i. (Fig. 2A), with stronger signals at 24 and 48 h p.i. (Fig. 2B and C, respectively). A strong ER staining pattern with the same antibody was also seen at 24 h p.i. in AcMNPV-infected Trichoplusia ni (Tn368) cells (Fig. 2D). This indicated that the localization of chitinase in the ER was not cell-type specific.
The ER localization of chitinase did not appear to be asso-ciated with proteolytic cleavage of the protein, since a 58-kDa product was evident in virus-infected cells between 12 and 96 h p.i. (Fig. 3). Enzyme assays using a range of substrates (6) showed that the intracellular chitinase was active in each of the samples harvested over the time course (data not shown).
Immunogold labelling of chitinase in AcMNPV-infected cells. Immunogold labelling and electron microscopy were used to confirm the localization of chitinase in AcMNPV-infected cells. The Sf9 cells were AcMNPV-infected with either AcMNPV or AcchiA2or were mock infected with culture medium. The
cells were harvested at 48 h p.i. and fixed, and ultrathin sec-tions were cut as described previously (3, 20). The secsec-tions were incubated successively with antichitinase antiserum (1: 8,000) for 16 h at 4°C and then with anti-guinea pig antiserum (1:20) conjugated to 10-nm colloidal gold for 1 h at ambient temperature. After immunolabelling, sections were post-stained with 2% aqueous uranyl-acetate (5 min) and lead cit-rate (10 min) before examination with a JEOL 1200EXII transmission electron microscope.
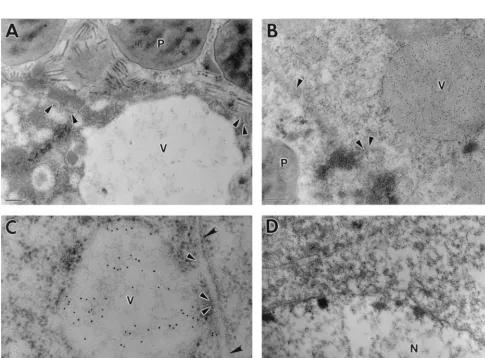
[image:2.612.54.287.66.657.2]In cells infected with wild-type AcMNPV, polyhedra and nonoccluded virus particles were observed within the nuclei (Fig. 4A). Gold particles were observed around the nuclear membrane and also within vacuole-like structures which were apparent throughout the cytoplasm (Fig. 4A and B). In some vesicles, the concentration of gold particles was very high (Fig. 4B). The origin of the vacuoles was uncertain, but they were observed only in AcMNPV-infected cells. They most probably correspond to areas of degenerate or vacuolated ER. In Fig.
FIG. 1. Immunofluorescent staining of S. frugiperda cells. Sf9 cells were in-fected with AcMNPV (10 PFU/cell) or were mock inin-fected with medium. After 48 h, the cells were immunostained as described in the text and examined with a confocal laser scanning microscope. (A) Mock-infected cells stained sequentially with guinea pig chitinase-specific antiserum (1:10,000) and FITC conjugated with anti-guinea pig IgG polyclonal antiserum (1:40). (B) AcMNPV-infected insect cells stained as described for panel A. (C) Transmitted light and immunofluo-rescence image of an AcMNPV-infected insect cell immunostained as described
for panel A. The pattern of immunostaining is depicted in red. (D) Mock-infected insect cell stained with mouse anti-HDEL monoclonal antibody (1:10) and FITC-conjugated goat anti-mouse IgG (1:40). (E to G) Colocalization of chitinase and ER proteins detected by double labelling of AcMNPV-infected Sf9 cells. Antichitinase antiserum and anti-HDEL monoclonal antibody were used as primary antibodies, followed by detection with, respectively, the following FITC-conjugated or Texas red-FITC-conjugated secondary antibodies: chitinase (E), HDEL (F), and dual staining (G). Bar55mm.
on November 9, 2019 by guest
http://jvi.asm.org/
4C, note the outer nuclear membrane, which appears to be continuous with the membrane encompassing the vacuole con-taining gold particles. Virtually no gold particles were detected in the region of the plasma membrane in AcMNPV-infected
cells (data not shown). In cells infected with AcchiA2, no
immunogold labelling was detected in either the nucleus or cytoplasm, and the perinuclear region was completely devoid of gold particles (Fig. 4D). The vacuole-like structures
ob-FIG. 2. Temporal production of chitinase in AcMNPV-infected insect cells. Sf9 cells were infected with virus; stained with a primary guinea pig chitinase-specific antiserum; FITC conjugated with anti-guinea pig IgG at 12 (A), 24 (B), and 48 (C) h p.i.; and examined by CSLM as described in the text. T. ni (Tn368) cells were similarly infected and stained at 48 h p.i. (D). Mock-infected Sf9 cells were stained with the same antibodies (E).
on November 9, 2019 by guest
http://jvi.asm.org/
[image:3.612.107.494.65.643.2]served in AcMNPV-infected cells were absent. There was no labelling in mock-infected cells or in AcMNPV-infected cells in which the primary antibody treatment was omitted (data not shown).
escens chitinase A (Fig. 5). Figure 5 also shows putative signal
peptides for four other baculovirus chitinases: Bombyx mori NPV (BmNPV) (10), Orgyia pseudosugata MNPV (OpMNPV) (1), Heliothis zea SNPV (HzSNPV) (8), and Choristoneura
fumiferana MNPV (CfMNPV). The signal peptides for BmNPV,
OpMNPV, and CfMNPV are similar to the AcMNPV se-quence, but the HzSNPV sequence is very different. It is prob-able that these sequences would also be cleaved from the nascent protein in virus-infected cells.
Identification of ER retention signals in baculovirus chiti-nases.The association of the AcMNPV chitinase with the ER
[image:4.612.61.279.67.162.2]PFU/cell). Virus-infected cells were harvested between 12 and 96 h p.i., and the pellets from 1-ml culture volumes were fractionated in a 12% polyacrylamide gel before immunoblotting with antichitinase antiserum (1/10,000) and alkaline phosphatase-conjugated goat anti-guinea pig IgG polyclonal antiserum (1/1,000). The position of the 58-kDa chitinase band is indicated with an arrow.
FIG. 4. Immunogold staining of virus-infected insect cells with chitinase-specific antiserum. Sf9 cells were infected (10 PFU/cell) with AcMNPV (A to C) or AcchiA2(D). Scale bars5200 nm. (A) Polyhedra (P) can be seen within the nuclear membrane (arrowheads). Gold particles are visible around the nuclear membrane
and within a vacuole-like structure (V) in the cytoplasm. (B) Dense gold staining was observed in the cytoplasmic vacuole and also in a perinuclear distribution following the nuclear membrane (arrowheads). (C) The vacuoles frequently appeared to be continuous with the outer nuclear membrane (black arrowheads), with the inner nuclear membrane remaining intact (white-bordered arrowheads). (D) The nucleus (N) is bounded by a normal membrane lacking attached vacuolar structures.
on November 9, 2019 by guest
http://jvi.asm.org/
[image:4.612.58.544.323.681.2]of virus-infected cells and cleavage of the signal peptide sug-gested that secretion of chitinase was inhibited. This indicated that a specific signal may be preventing transport of the chiti-nase out of the ER. Analysis of the amino acid sequence of the AcMNPV chitinase revealed a KDEL motif at the C terminus of the protein (Fig. 5). This tetrapeptide sequence motif is a known ER retention-retrieval signal (11).
The OpMNPV and CfMNPV chitinase proteins also possess KDEL sequences at their C termini, but the BmNPV chitinase has an RDEL sequence motif (Fig. 5). This is consistent with published observations that the tetrapeptide sequence is not strictly observed and that certain changes to the motif still enable retention of proteins in the ER (15). Therefore, it appears that the KDEL motif (or a closely related sequence) is present in several baculovirus chitinases. The exception to this observation is the chitinase of HzSNPV, which has HNEL at the C terminus. This partial conservation may be sufficient to serve the same role as the KDEL motif. The HzSNPV chiti-nase also has a 13-residue extension at this end of the protein, relative to the AcMNPV chitinase. The significance of this is unknown. S. marcescens chitinase A does not possess a KDEL motif at its C terminus (Fig. 5).
Release of chitinase from virus-infected cells.Our model for AcMNPV-induced liquefaction of virus-infected insect larvae requires chitinase to be released from cells to attack the cutic-ular chitin (7). It was reported that Sf21 cells infected with AcMNPV mutants lacking an intact p10 gene did not progress to cell lysis (23). We examined the release of active chitinase from Sf9 cells in a suspension culture infected with AcMNPV (10 PFU/cell) or a mutant (AcUW1.p102) lacking an intact
p10 gene (22).
Figure 6 shows that at 24 h p.i., there was approximately sixfold more chitinase in the medium supporting the growth of AcMNPV-infected cells than in that of AcUW1.p102-infected
cells. Only background levels of chitinase activity were re-corded in the latter samples. Between 48 and 96 h p.i., how-ever, similar levels of exo- and endochitinase were in the media of AcMNPV- and AcUW1.p102-infected cell cultures. The
viabilities of both virus-infected cell cultures declined from nearly 100% at 24 h p.i. to 0% at 96 h p.i. However, differences between the AcMNPV- and AcUW1.p102-infected cell
cul-tures were observed when the total numbers of cells at each time point were assessed. For AcMNPV, an initial concentra-tion of 1.13106cells/ml remained static at 24 h p.i., declined
slightly by 72 h p.i. (106 cells/ml), and then was reduced to
about 23105cells/ml by 96 h p.i. In contrast, the
concentra-tion of cells in cultures infected with AcUW1.p102 slightly
increased to 1.53106cells/ml by 48 h p.i. and declined only to
83105cells/ml at 96 h p.i.
Infection of second-instar T. ni larvae with AcMNPV poly-hedra resulted in host liquefaction at 5 days p.i. This process, however, was delayed by 1 day in insects infected with AcUW1.p102.
Generally, proteins that are located in the organelles of the secretory pathway encode signals for their retention at the correct location. Proteins resident in the lumen of the ER usually carry a signal motif (KDEL or a closely related se-quence) at their carboxy terminus. The sequence KDEL is predominantly found in mammalian cells (11), HDEL is found in the yeast Saccharomyces cerevisiae (13), and (H/K/R)DEL is found in plants (21). The precise sequence of the motif varies, although generally conservative changes are seen (R for K or D for E) (15). The demonstration of similar protein retention signals in widely divergent species suggests that it is a universal feature of eukaryotes (15). The signal serves to retain proteins in the ER lumen of mammalian (11), yeast (4), and plant (5) cells. Proteins escaping the ER interact with a KDEL receptor in the cis-Golgi and are returned via a retrograde vesicle-mediated pathway to the ER. If the sequence is deleted, or if it is extended by the addition of other amino acids, the protein is secreted from the cell instead of remaining in the lumen of the ER. Conversely, if the tetrapeptide sequence is added to the C terminus of non-ER resident proteins such as lysozyme, a known secretory protein, the enzyme is retained in the ER lumen instead of being secreted (11).
This study has described the identification of a KDEL motif at the C terminus of the chitinase protein of AcMNPV. This sequence motif is not observed in the S. marcescens chitinase, but has been identified as KDEL in the chitinase proteins of OpMNPV and CfMNPV and as RDEL in BmNPV. Our data do not prove that the KDEL motif in chitinase is solely respon-sible for retaining the protein in the ER, and we will have to perform site-directed mutagenesis on the chitinase gene to alter this sequence and determine the effect on protein location.
The retention of chitinase within virus-infected cells is sur-prising given that in the insect larva, our model for the mech-anism of liquefaction requires that the enzyme should be ex-tracellular to attack cuticular chitin (7). We have not examined virus-infected cells from larvae to determine if the chitinase is also retained within the ER. It may be advantageous for the virus to delay liquefaction of the host until the maximum yield of polyhedra has been attained. If chitinase were secreted from virus-infected cells as soon as it was synthesized (7 to 10 h p.i. [6]), the insect might disintegrate too rapidly and reduce
poly-FIG. 5. Alignment of the signal peptides and carboxyl termini of the predicted amino acid sequences of the chitinases from S. marcescens (16), AcMNPV (6), OpMNPV (1), BmNPV (10), CfMNPV (U72030), and HzSNPV (8). The prokaryotic signal peptide for S. marcescens comprises the first 23 residues. The eukaryotic signal peptide of AcMNPV chitinase is underlined, and the cleavage site of the signal peptidase is indicated by the vertical arrow. The corresponding sequences in the BmNPV, OpMNPV, CfMNPV, and HzSNPV chitinases are shown. The KDEL (RDEL for BmNPV and HNEL for HzSNPV) putative ER retention signals are underlined and shown in boldface type. The length of each protein is indicated at the end of each line.
on November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.88.472.69.185.2]hedron production. The eventual release of chitinase probably occurs after cell lysis, which, at least in part, is mediated by production of the p10 protein. Cells infected with a virus un-able to synthesize p10 released active chitinase 24 h later than did AcMNPV-infected controls and remained intact for longer. The same p10 deletion mutant virus also induced liquefaction 1 day later in insects. Although we need to examine cell lysis in virus-infected insects, these preliminary observations suggest that chitinase release is also associated with p10 production in
REFERENCES
1. Ahrens, C. H., R. L. Q. Russell, C. J. Funk, J. T. Evans, S. H. Harwood, and G. F. Rohrmann.1997. The sequence of the Orgyia pseudotsugata multinucleo-capsid nuclear polyhedrosis virus genome. Virology 229:381–399. 2. Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee.
1994. The complete DNA sequence of Autographa californica nuclear poly-hedrosis virus. Virology 202:586–605.
3. Cole, L., F. M. Dewey, and C. R. Hawes. 1996. Infection mechanisms of Botrytis species: pre-penetration and pre-infection processes of dry and wet conidia. Mycol. Res. 100:277–286.
4. Dean, N., and H. R. B. Pelham. 1990. Recycling of proteins from the Golgi compartment to the ER in yeast. J. Cell Biol. 111:369–377.
5. Denecke, J., R. De Rycke, and J. Botterman. 1992. Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 11:2345–2355.
6. Hawtin, R. E., K. Arnold, M. D. Ayres, P. M. D. A. Zanotto, S. C. Howard, G. W. Gooday, L. H. Chappell, P. A. Kitts, L. A. King, and R. D. Possee. 1995. Identification and preliminary characterisation of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology 212: 673–685.
7. Hawtin, R. E., T. Zarkowska, K. Arnold, C. J. Thomas, G. W. Gooday, L. A. King, and R. D. Possee.1997. Liquefaction of Autographa californica nucleo-polyhedrovirus-infected insects is dependent on the integrity of virus-en-coded chitinase and cathepsin genes. Virology 238:243–253.
8. Hoa Le, T., T. Wu, A. Robertson, D. Bulach, P. Cowan, K. Goodge, and D. Tribe.1997. Genetically variable triplet repeats in a RING-finger of Heli-coverpa species baculovirus. Virus Res. 49:67–77.
9. Macdonald, H., J. Henderson, R. M. Napier, M. A. Venis, C. Hawes, and C. M. Lazarus.1994. Authentic processing and targeting of active maize auxin-binding protein in the baculovirus expression system. Plant Physiol. 105:1049–1057.
10. Maeda, S. 1994. GenBank accession no. L33180.
11. Munro, S., and H. R. B. Pelham. 1987. A C-terminal signal prevents secre-tion of luminal ER proteins. Cell 48:899–907.
12. Napier, R. M., L. C. Fowke, C. R. Hawes, M. Lewis, and H. R. B. Pelham. 1992. Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J. Cell Sci. 102:261–271. 13. Pelham, H. R. B. 1988. Evidence that luminal ER proteins are sorted from
secreted proteins in a post-ER compartment. EMBO J. 7:913–918. 14. Pelham, H. R. B. 1989. Control of protein exit from the endoplasmic
retic-ulum. Annu. Rev. Cell Biol. 5:1–23.
15. Pelham, H. R. B. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15:483–487.
16. Perrakis, A., I. Tews, Z. Dauter, A. B. Oppenheimer, I. Chet, K. S. Wilson, and C. E. Vorgias.1994. Crystal structure of a bacterial chitinase at 2.3Å resolution. Structure 2:1169–1180.
17. Possee, R. D. 1986. Cell-surface expression of influenza virus haemagglutinin in insect cells using a baculovirus vector. Virus Res. 5:43–47.
18. Slack, J. M., J. Kuzio, and P. Faulkner. 1995. Characterisation of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa califor-nica multiple nuclear polyhedrosis virus. J. Gen. Virol. 76:1091–1098. 19. Summers, M. D., and G. E. Smith. 1987. A manual of methods for
baculo-virus vectors and insect cell culture procedures. Texas Agricultural Experi-ment Station bulletin no. 1555. Texas Agricultural ExperiExperi-ment Station, Col-lege Station, Tex.
20. Van den Bosch, K. 1991. Immunogold labelling, p. 181–218. In J. L. Hall and C. Hawes (ed.), Electron microscopy of plant cells. Academic Press, London, United Kingdom.
21. Vitale, A., A. Sturm, and R. Bollini. 1987. Regulation of a plant glycoprotein in the Golgi complex: a comparative study using Xenopus oocytes. Planta 169:108–116.
22. Weyer, U., S. Knight, and R. D. Possee. 1990. Analysis of very late gene expression by Autographa californica nuclear polyhedrosis virus and the fur-ther development of multiple expression vectors. J. Gen. Virol. 71:1525– 1534.
23. Williams, G. V., D. Z. Rohel, J. Kuzio, and P. Faulkner. 1989. A cytopatho-logical investigation of Autographa californica nuclear polyhedrosis virus p10 gene function using insertion/deletion mutants. J. Gen. Virol. 70:187–202. FIG. 6. Chitinase activities in the media of virus-infected cells and total cell
numbers. Suspension cultures of Sf9 cells (106cells/ml) were mock infected or
inoculated with AcMNPV (A) or AcUW1.p102(B) (10 PFU/cell). Medium from each culture was harvested at the times indicated and assessed for exo- and endochitinase activities (6). The cell concentration at each time point was also determined (C).