Copyright©1976 American Society for Microbiology Printed in U.S.A.
Selective
Decrease in
the Rate
of
Cleavage of
an
Intracellular
Precursor
to
Rauscher Leukemia Virus
p30
by Treatment of
Infected
Cells with Actinomycin
D
G. A. JAMJOOM, R. B. NASO, AND R. B. ARLINGHAUS*
BiologyDepartment, The University of Texas System CancerCenter, M. D.Anderson Hospital and Tumor
Institute, Houston, Texas 77030
Received for publication26January1976
The cleavage ofan intracellular 67,000- to 70,000-dalton precursor, termed
Pr4, toRauscher leukemia virus (RLV) p30 protein proceededata slowerrate
when virus-producingcellsweretreated with actinomycin D (AMD). Treatment
with AMD also caused a slight accumulation of Pr4 in purified early virus
particles produced by a cell line which usually produces virions that contain
little Pr4. The cleavage of other intracellular viral precursorpolypeptides was notaffected bytreatmentwith AMD. Treatment ofinfected cells with cyclohexi-mide,onthe other hand, allowed the cleavage of Pr4toproceedatthe usualrate
for a short period of time before further cleavage was drastically slowed or
prevented. The cleavage of several other viral precursor polypeptides wasalso
inhibited bytreatmentwithcycloheximide. Different lines of evidence suggest
that themechanism of action of AMD isnotduetoapossible indirect effecton
protein synthesis. Thus, the rate ofcleavage of Pr4 was not affected by the
length ofpretreatment with AMDbetween 1 to8h. Inaddition, the combined effectof AMD andcycloheximide, attheir maximal inhibitory concentrations,
wasgreaterthan the effect of either drug alone, indicating the involvement of
two atleast partially different mechanisms in the action of AMD and
cyclohexi-mide. Furthermore, AMD did not affect the pulse labeling ofviral precursor
polypeptides. These results suggest that the interaction with viral RNA, whose production is inhibited by AMD, accelerates the cleavage ofPr4 top30 during virus assembly. A hypothetical model is presented to illustrate the possible advantages ofhavingastepin virusassembly in which genomicRNA interacts
witha precursortocapsid proteins before the cleavage of that precursor.
The cleavage of precursor proteins during the maturation of viral particles is a phenomenon
ofwidespreadoccurrence. Ithas been observed
in many types of viruses, including
bacterio-phages T4 andX, picornaviruses, Sindbisvirus, vaccinia virus, influenza virus, Sendai virus,
and adenoviruses, among others (for reviews see8, 21, 27, 46, 48). Although the purposesof
suchproteolytic cleavage are not definitely de-termined, they evidently include the formation ofthe finalstructural proteins and the facilita-tionand stabilizationof the protein-proteinand
protein-nucleic acid interactions that occur dur-ing assembly.
Differenttypes of proteolytic cleavage can be
distinguished in certain instances. For
exam-ple, inpicornaviruses, nascent chain cleavage, which mayfacilitate the release of segments of thepolyprotein chain from the ribosome (7), is
distinguishedfrom eitherintermediate or
mor-phogenetic-type cleavages. Morphogenetic
cleavage is strongly linked with virus
assem-bly. The mostprominent morphogenetic cleav-age is thatwhichaccompanies thepackagingof
theviralgenome. This type of cleavage in most cases seems torequire an interaction with the
viral nucleicacidsince itusually doesnot occur in emptyparticles.
Proteolytic cleavage also plays a prominent role in the formation of oncornavirus proteins (2, 37, 51; R. B. Arlinghaus, R. B. Naso, G. A.
Jamjoom, L. J. Arcement, andW. L. Karshin,
in D. Baltimore, A. S. Huang, and C. F. Fox,
ed.,Animal Virology, ICN-UCLA Symp. Mol. Cell. Biol., in press). In Rauscher leukemia
virus (RLV) the group-specific core protein,
p30, is made by way of cleavage of several
precursors of higher molecular weight. The most prominent of these are two precursors withmolecularweightsof80,000 and67,000to
70,000, designated Pr3 and Pr4,
respectively.
Less prominent, based on their occurrence in
1054
on November 10, 2019 by guest
http://jvi.asm.org/
CLEAVAGE OF RLV p30 PRECURSOR 1055
small quantities in infected cell extracts, are
larger precursors with a molecular weight of about 200,000,designated Prla+b (2).
The effect of actinomycin D (AMD) on the inhibition of oncornaviral RNA synthesis is well established (4, 10, 33, 50). Treatment of
virus-producing cells with AMD, however,
al-lows for several hours the synthesis of viral proteins and the maturation ofRNA-deficient viral particles that have normal morphology (33). Thus, treatment with AMDmakesit
pos-sibletoexamine the role ofgenomicRNA inthe maturationandprocessing of theviralproteins
duringvirusassembly.
In this paper, we describe the observation
thattreatmentofchronically infected cellswith AMD noticeablyslowed the rateofcleavage of Pr4 and caused its accumulation in infected cells and,to asmallextent, inearlyvirus
parti-cles. Pr4, previously designated p70,is an
inter-mediate intracellular precursor polypeptide, but is present in mature virions produced by severalinfected cell lines (24).
MATERIALS AND METHODS
Cells and virus. RLV-infected NIH Swiss mouse
embryo (JLS-V16) (37) and RLV-infected BALB/c
mouse spleen and thymuscells (JLS-V5) (49) were
usedinthis study. Theculturemedium contained a
modified Eagle amino acid formula and 10% fetal
calf serum, asdescribedpreviously (49). Cellswere
grown in 2-ounce (about 60-ml) prescription glass
bottles or, for virus production, in l-quart (about
0.95-liter) glass bottles. The cultures wereused 3 to
6 daysafter passage and werenearly confluent.
For viruspurification, culturefluid was collected
and clarified at 10,000xg for 10 min.The virus was
pelleted at 78,000 x g for 2 h, suspended in TNE
buffer (0.01 M Tris, pH 7.5, 0.1 M NaCl, 0.001 M
EDTA), and banded by isopycnic centrifugation on a
15 to60% (wt/vol) sucrose gradient in TNE for 16 h
at13,000 rpmin aBeckmanSW27rotor.Torecover
cell-associated viral particles, the cell sheet was
rinsed with rinsing buffer (0.14 M NaCl, 5 mMKCl,
0.3 mM
Na2,HPO47H20,
0.4 mMKH2PO4, 5.5 mMglucose, 4 mM NaHCO3, 100 mg of neomycin per liter) and treated for 5 min with 0.02% trypsin in
rinsing buffer. The resultant cell suspension was
pooled with the culture fluid, and the cells were
pelleted. The supernatant fluids wereprocessed for
virus purification asdescribed above. The sucrose
gradients were fractionated into 1-ml portions, and trichloroacetic acid-insoluble radioactivity and
den-sity were measured. Theradioactivity peak at
dens-ities of 1.13 to 1.16g/cm3waspooled, dilutedinTNE,
andpelletedat78,000 xg for2h.
Labeling of cells and virus.Labeling of cells for
pulse-chase studies was done as follows: the cells
wererinsed with warm Hanks solution and labeled
in Hanks solution containing 100
ACi
of[35S]methionine
per ml (288 Ci/mmol; Amersham/Searle) or in complete growth medium (49) minus
methionine containing 5%dialyzedfetal calfserum
and no tryptose phosphate. The pattern of
radioac-tive proteins was identical in either case. For a
chase, the cells were rinsed with Hanks solution and
incubated in complete growth medium. For virus
labeling, the cells were rinsed with Hankssolution
and incubated for 4 h in growth medium which
contained 45 ,MCi of [35S]methionine per ml, 1/10
Eagle's concentration of unlabeled methionine, 5%
dialyzed fetal calf serum, and no tryptose phos-phate.
Immunoprecipitation and gel electrophoresis. Cell lysis, the preparation of anti-RLV antiserum, and the immunoprecipitation of virus-specific
pro-teins in infected cell lysates were described
previ-ously (37). Sodium dodecyl sulfate (SDS)-polyacryl-amide gel electrophoresis of proteins was done by using the buffer system described by Laemmli (28);
RNA urea-acrylamide-agarose gel electrophoresis
wasdone as described by Floyd et al. (17) except that
SDS was used instead of lithium dodecyl sulfate,
andelectrophoresis was at room temperature. The
gels were processed for fluorography as described by
Bonner andLaskey (6). To obtain a linear response
to radioactivity, the X-ray films were preflashed
(32). To quantitate the amount of radioactivity in
different bands, the films were scanned at 590 nm in
aGilfordspectrophotometer, and the relative areas
ofdifferentpeaksintheresultingcurves were
mea-suredinaDuPont310 curveresolver.
RESULTS
Effect of AMD on viral RNA and protein
synthesis inchronicallyinfected cells. Inhibi-tion ofoncornaviral RNA synthesis by treat-ment with AMD occursrapidly (e.g., reference 4). On the other hand, the synthesis of viral
proteins and the maturation of RNA-deficient particles in murine leukemia virus-infected
cellscontinuesforseveral hours in the presence of AMD (33). These two results are shown in
Fig. 1 for the RLV-infected JLS-V16 cell line used in these studies. In this experiment, the cells were treated with 5,g of AMD per ml for 75 min before they were pulse-labeled for 75
min with mediumcontaining both [H]uridine and
[P5S]methionine.
After a chase for 3 h incomplete medium, the virus waspurified ona
sucrose gradient andthe radioactivity was
de-termined. It can be seen that whereas the
[PH]uridine
radioactivity was drasticallyre-duced ascompared with the
control,
nosignifi-cantdifference had occurred in the
[35Jmethio-nine radioactivity during this period. This re-sult confirms thefinding that viralprotein
syn-thesis andmaturationof viral particles can oc-curintheabsenceof viral RNAsynthesis(33),
indicatingthe stability of the viral mRNA for severalhours (34).
We have estimated the rate ofsynthesis of intracellular virus-specific polypeptides by
VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
1,400
1,200
1,000
2
800
u 600 0
cr-400
200
15
14
13
12
II z
E
0-z
5 10 15 20 5 10 15 20
FRACTION NUMBER
FIG. 1. Effect ofAMDon viral RNAandproteinsynthesis.RLV-infectedJLS-16 cells were pretreated for
75 min with 5
lAg
ofAMDper ml in complete medium, pulsed for 75 min with methionine-free mediumcontaining
/P5SImethionine, /PIHluridine,
and 5 Ag of AMD per ml, and chased for 3 h in complete mediumcontaining 5pgof AMD per ml. Virus was collected and purifiedona 15to60% sucrose gradient. Fractions
wereassayed for their density and trichloroacetic acid-insoluble radioactivity. For control virus, the cells were
similarly treated, exceptthat AMDwasomitted.
measuring the amount ofradioactivity after a 15-minpulselabel withradioactiveaminoacids that is immunoprecipitable in cell extracts by antiserapreparedagainst mature virions.Such
measurements have indicated that 5 h of pre-treatment with 5 to 10
gg
of AMD per mlonlyreduced by 15 to 20% the incorporation of I:5S]methionineintointracellularvirus-specific polypeptides, whereas a 7-h pretreatment was
required to reduce incorporationby 50%. These observations suggest that the viral mRNA has
a functional half-life ofapproximately 7 h. A
similar value has been reported by Levin and Rosenak (34).
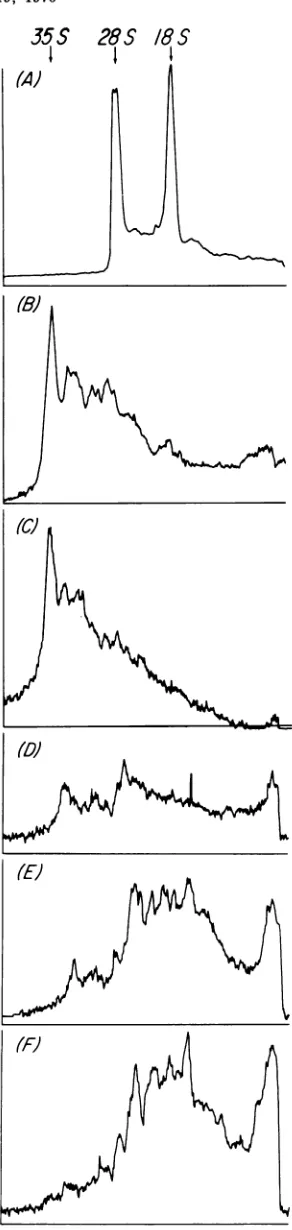
Effect of AMI) on the rate ofcleavage of Pr4 to p30. Figure 2 shows the effect of
pre-treatment of RLV-infected cells with AMD on
the rate ofcleavage of Pr4 and appearance of p30. After a 1-h treatment of infected cells either with AMD in growth medium or with
fresh medium without AMD, cells were pulse labeled for 15 min with
[t5S]methionine
inHankssolution in the presence or absence of5
,ug of AMD per ml. The label was chased in
complete growth medium for different times
while maintaining the presence or absence of
AMD. Cells were then lysed, and the
cytoplas-micvirus-specificpolypeptides wereisolatedby
immunoprecipitation with anti-RLVserum. In
control cultures (Fig. 2B, D, F, and H) Pr4
disappeared during a chase period of3 h at a
ratesimilar tothatatwhich p30appeared dur-ing thisperiod. The fact thatPr4 is aprecursor
top30 has been verified by pulse chase studies
and trypticdigestion (2). In AMD-treated
cul-tures (Fig. 2A, C, E, and G), cleavage of Pr4to
p30 also occurred, but at a much slower rate.
Quantitation of the results of Fig. 2 isshownin
Fig. 4A as the ratio of p30 to Pr4 obtained at
different times ofchase in the presence or
ab-sence ofAMD.
The disappearance of the p30 precursors
Prla +b and Pr3 during the chase was also evident in Fig. 2. However, in contrast tothe disappearance of Pr4, the rates at which Prla+b and Pr3 disappeared were similar in
AMD-treated and control cultures. This shows
that AMD does not affect the cleavage of all
viral precursor polypeptides indiscriminately.
Dose-response analyses of the effect of AMD on the cleavage of Pr4. The effect of pretreatment with different doses of AMD on thecleavageof Pr4 top30 was examined.Table 1 shows the effect of a 1-h pretreatment with differentconcentrations ofAMDonthe p30/Pr4 ratio obtained aftera subsequent 15-min pulse and 1-hchase. It can beseen that the effect of AMDbecame noticeable under the conditions of the experiment at 0.1
gg/ml.
Thisconcentra-tion of AMD is known to inhibit viral RNA
synthesis (10). Theeffect ofAMD washigherat 1
gg/ml,
but nofurther increase was observedwhen theconcentration was raisedto 10 ,ug/ml. This result suggests that the effect ofAMD is
specificand not due togeneral cytotoxic effects, which would beexpected toincrease at higher
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.505.125.408.68.261.2]CLEAVAGE OF RLV p30 PRECURSOR 1057
!I
tJ
"IW.
~~~~~~
- -...FIG. 2. EffectofAMD on the cleavage ofPr4 top30.RLV-infected JLS-V16 cellswerepretreatedwith 5pg
of AMD per ml for1 h incomplete growthmedium.-The culturefluidwasremoved, and the cultureswere
pulselabeledfor 15minwith[35S]methionineinHanksbuffercontaining5pgofAMD per ml.The cell sheet
wasrinsedand incubated incompletegrowthmedium containing 5pgofAMD permlfor15 min(A),30min
(C),1h(E),and 3 h(G).Control cultures: Parallel cultureswereprocessedasabovewithout AMD andchased
for15 min(B),30 min(D),1 h(F),and 3 h(H).Cytoplasmicextracts weretreated with antiserum toRLV
proteins, and the immunoprecipitates wereanalyzed bySDS-polyacrylamide gel electrophoresis (6 to 12%
[image:4.505.58.447.72.388.2]lineargradientgel). Similaramountsofradioactivitywereapplied.
TABLE 1. Dose-response analysis of the effect of
AMD on thecleavageof Pr4a
Concn ofAMD p3O/Pr4
(,ug/ml)
0 1.67
0.1 1.20
1.0 0.54
10.0 0.53
a RLV-infected JLS-V16 cultures were treated
with freshcomplete growth mediumcontaining the
different AMD concentrations for 1 h. They were
then pulsed with [35S]methionine in Hanks buffer
for 15min and chasedin complete growthmedium
for1h, while maintaining thesamedrug
concentra-tions during the pulse and chase. Virus-specific polypeptides were immunoprecipitated from
cyto-plasmic extracts and analyzed by
SDS-polyacryl-amidegel electrophoresis andautoradiography.The
autoradiogram was scanned, and thep30 and Pr4
peakswerequantitated.
concentrations of the drug. The duration of
AMD treatment inthisexperiment (2 hand 15 min)didnotresultinnoticeable changes in cell
morphology. Cells treated with 10 ,ug ofAMD per ml could still make viral proteins and ex-port virusfor several morehours.
Effect of cycloheximide on the rate of
cleavage ofPr4 to
p30.
The clarification of the mechanism by which AMD slows the cleavageofPr4requires an examination of the effect of inhibition of protein synthesis on the cleavage of Pr4 (seebelow). We therefore tested the ef-fect ofcycloheximide onthe rate ofcleavage of Pr4 (Fig. 3). In this experiment, the cells were
pulsedfor 15 minin Hankssolution containing
[35S]methionine
and then chased in thepres-enceorabsenceof100 ,g ofcycloheximideper ml for different periods of time. Virus-specific
polypeptides in the cytoplasmic extracts were
VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.505.54.247.498.562.2]1058 JAMJOOM, NASO, AND ARLINGHAUS
A B C D E F G H
Prla+b- .'Z---
-Pr2a+b
----Pr3 --- _
Pr4 - 4---- awm...2-.
a's0
dMMM
.~
-.
p30 -- - m -;w"n_E_I__
_
p30/Pr4 0 0.14 0.16 0.49 0.49 1.4
mm~~~~--P1B
p----81
SEp12
[image:5.505.65.453.76.344.2]0,68 7' 0.82
FIG. 3. Effect ofcycloheximideonthecleavageofPr4top30.RLV-infectedJLS-V16 cells werepulsedfor
15minwith[35S]methionineinHanks buffer(A) andchased for thefollowingtimes:(B,C)15min;(D, E)30
min;(F,G)60 min;(H, I) 3 h. B,D, F,and Hwerechased incomplete medium;C, E,G,and Iwerechased in
complete medium containing 100
lAg
ofcycloheximideperml.Cytoplasmicextracts wereimmunoprecipitatedwithanti-RLV, and the immunoprecipitatewasanalyzedongelsasinFig.2.
then examined after immunoprecipitation by anti-RLV antiserum.
>The
cycloheximide
con-centration used was inexcess of that
required
for fastand maximal inhibition of protein
syn-thesis. Figure3B, D, F, and H shows the
pat-tern of viral polypeptides in control cultures after chases for 15 min, 30 min, 1 h, and 3 h, respectively. This is similar to the control chases shown in Fig. 2. When
cycloheximide
wasadded during the chase (Fig. 3C, E, G, and
I), the cleavage ofPr4 continued atthe same rate as the control for about 30 min before it
wasdrastically reduced.Quantitative
measure-mentsof the ratio ofp30 to Pr4 from Fig. 3 is showninFig.4B. Thisresultsuggeststhat the cleavage ofPr4 requiresthe function ofa pro-tein(s) that is initially present in excess
amounts, ifcompared on a functional basisto
the amount ofPr4 present,butthat isquickly
depleted. Candidates for such a protein would
include a cleavage enzyme or a protein that directsastepinviralassemblythatisrequired
for the cleavage ofPr4. However, other more complexmechanisms forthe effectof inhibition ofproteinsynthesis onthecleavage ofPr4 are
alsopossible.
In contrast to AMD,
cycloheximide
alsore-10
0
rK)-C
c-8 6
4
2 A
3 0
TimeofChase(Hrs.)
FIG. 4. Effectof AMD and cycloheximide onthe
cleavage ofPr4 top30inthechase. Graphic
represen-tation of
p30/Pr4
of Fig.2andFig. 3 isshown in(A) and (B),respectively.
duced the cleavage of the precursors
polypep-tides Prla +bandPr2a+b (Fig. 3).
Puromycin,
at 2 x 10-3 M,hadaneffectonthe
cleavage
of viral precursorpolypeptides,
similarto that ofcycloheximide, indicating
thatthe effect is dueto the inhibition ofprotein
synthesis
and notparticularto
cycloheximide
(notshown).
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.505.273.458.403.550.2]CLEAVAGE OF RLV p30 PRECURSOR 1059
Quantitation of the combined effect of
AMD and cycloheximide on the cleavage of
Pr4. The effect ofAMD onthe cleavage of Pr4 could be due either to its direct inhibition of viral RNA synthesis or to the inhibition of a species ofmRNA, with a shorthalf-life, which codes for a protein whose function is required for the cleavage ofPr4. Todistinguish between
these alternatives we have used two ap-proaches, the first of which is to test whether
treatment with AMD can increase the
inhibi-tionof cleavage of Pr4causedbycycloheximide used at levels that are high enough to insure maximal inhibition of proteinsynthesis. Figure 5 shows the amount ofPr4 present in a pulse
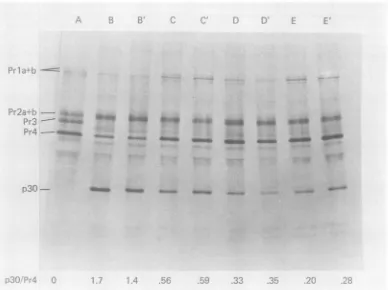
(A), its reduction after a subsequent chase in
duplicate (B, B'), the effect of200
Ag
of cyclo-heximidepermladdedduring
thechase (C,C'), theeffect of10 ,ugofAMD permladded15 minbefore and during the pulse and chase (D, D'), and the effect of the combined AMDand
cyclo-heximide treatment (E, E') on the extent of
cleavage of Pr4 in the chase. Measurements of
theratioofp30 to Pr4 afterthe different treat-ments are indicated beneath the different col-umns.The experimentwasdoneinduplicateto give an idea about the statistical variation in
such quantitative measurements. It can be
seenthat theratioof p30toPr4 waslowerafter the combined treatment with AMD and
cyclo-heximide than aftertreatmentwith eitherdrug alone. This indicates that at least part of the effect ofAMD onthecleavage ofPr4 is notdue
tothe inhibitionof proteinsynthesis.
Although the increase in the inhibition of cleavage ofPr4 after the combined treatment with the two drugs over the separate treat-ments with either drug alone has been
repro-ducible, it seems tobeslight. However, it must
be noted that simple mathematical additivity should notbe
expected
in suchinteracting sys-tems. Moreover, examination of the kinetics of cleavage ofPr4 inthepresenceof thetwodrugs(Fig. 4A, B) points to the difficulty in
demon-FIG. 5. Combinedeffect ofAMD andcycloheximideontheextentof cleavage ofPr4 top3O.RLV-infected
cellswerepulse-labeled for15minwith1:5S]methionine(A)andparallel cultures,induplicate,werechased
for60 min incomplete growth medium (B, B')orchasedfor60 min incomplete growthmediumcontaining
200mgofcycloheximideper ml(C, C').Inthesameexperimentparallelcultureswerepretreatedwith10pgof
AMD permlfor15min incomplete growthmedium and thenpulselabeledfor15minandchasedfor60 min inthe presenceof10pgofAMDper ml(D, D'),orchasedfor60 mininthe presenceof10pgofAMDplus200
pgof cycloheximideperml(E, E'). Cytoplasmicextracts wereprepared andprocessedasinFig.2.
VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.505.59.447.309.599.2]strating large additivity in the inhibition of that cleavage after the combined treatment withthe drugs.
We have also tested the combined effect of AMDand cycloheximide when AMD was added only during the chase, in order to exclude any
possible effectof AMD onthe synthesisof
virus-specific proteins. In this case, the combined effect of the two drugs was similarly greater than the effect of either drug alone (data not
shown).
It must be noted that therateofcleavage of
Pr4 top30in control cultures varies somewhat in different experiments. We have found the
rate ofthe cleavage to be affected by the age and confluency of the cell culture. The varia-tion, however, is not large enough to interfere
withthestatistical significance of the resultsif comparisons are only made among culturesin one experiment and ifthe compared cultures
are similarly prepared and used within a few
hours of each other.
It is to be emphasized, however, that
al-though these results indicate that the
mecha-nisms of action ofAMD andcycloheximideare atleastpartially different, they donotruleout
thepossibility that part ofthe effects ofthese twodrugsonthe cleavage ofPr4maybe dueto acommonmechanism. Thus, itispossible that
part of the effect ofcycloheximideisdue tothe inhibition of the synthesis or migration of viral
RNA. Cycloheximide was found to interfere with total RNA synthesis (14), the synthesis and processing of rRNA in particular (e.g., ref-erences 14, 53) and the nucleocytoplasmic translocation ofRNA (12) in the different sys-tems examined. Similarly, inhibition of RNA synthesis by AMD was found to have an indi-rect inhibitory effect on the initiation of protein synthesis, which is not a result of thedecay of available mRNA (19, 47).
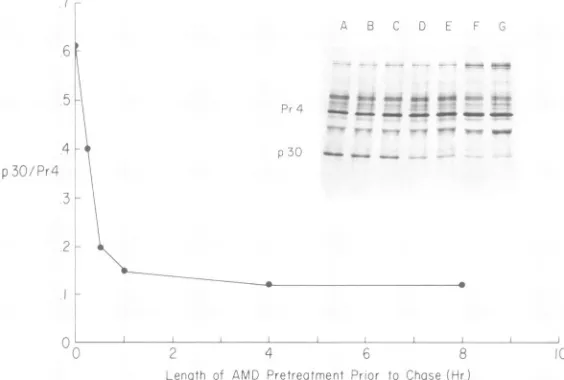
Effect of the length of pretreatment with AMD on the cleavage of Pr4. A better way of testingwhethertheeffectof AMD on the cleav-ageof Pr4 is due to indirect inhibitionofprotein synthesis is to examine the effect of thelength of pretreatment with AMD on the cleavage of Pr4 in a subsequent pulse-chase experiment.
Thus, if AMD causes a depletion of a protein whose function is required for the cleavage of Pr4, then it is expected that the longer the pretreatment with AMD, the slower the rate obtained for thecleavageof Pr4.Figure 6 shows that the extent ofcleavage of Pr4 was
propor-tionaltothelengthof pretreatment with AMD
within30 minof pretreatmentbefore chase. No additional decrease in the rate of cleavage of Pr4, however, was obtained by lengthening the time ofpretreatment up to 8 h. The simplest
interpretation of these resultsisthat the effect
of AMD on thecleavageof Pr4 is not mediated by the depletion of a labile protein with a
short-r
[image:7.505.121.403.411.601.2]0
FIG. 6. Effect ofthelength ofpretreatment with AMD onthecleavage ofPr4 top30.Insert:RLV-infected
cells werepulse-labeled for15 min inHanks solutioncontaining[35S]methionineandchasedfor60 min in
complete growth medium (A). Parallel culturesweretreatedasfollows: (B)AMD(5,pg/ml)addedduringthe
chase; (C)AMDaddedduringthepulse andthechase;(D)AMDadded15min, (E)1h, (F)4h,and(G)8h
before the pulse and chase.Cytoplasmic extracts wereprocessedasinFig.2. The ratiosof p30 toPr4are
plotted versusthelength ofAMDpretreatment.
on November 10, 2019 by guest
http://jvi.asm.org/
CLEAVAGE OF RLV p30 PRECURSOR 1061
lived mRNA, but is more likely due to the
direct inhibition byAMDofgenomic viral RNA
synthesis.
The results also indicate that the mRNA(s) for thecleavage enzyme(s)involved in the mat-uration of p30 has a relatively long half-life that is comparable to the half-life of the viral mRNA.
Ifourinterpretation of the effect of AMD on the cleavage of Pr4, as being a result of the inhibition by the drug of the synthesis of gen-omic RNA, is correct, then the above results indicate that there is a pool ofgenomic viral RNA thatisavailable for catalyzingthe cleav-age of newly made Pr4 and that this pool is
depleted in about 30 min. This period is not
likelytoberequired for AMDto exert its inhi-bition of RNAsynthesis, sinceAMD isknown
toact within afewminutes(e.g., reference 44).
Note, however,that sucharesultmeasuresthe pool ofgenomicviral RNA only interms ofits
availability for accelerating the cleavage of newly made Pr4 and not in terms ofits total
amount inthecell.
Theabove results would indicate, moreover,
thattheeffect ofgenomic RNA onthe cleavage ofPr4iscatalyticrather than obligatory, since
cleavage stilloccurred after8hofpretreatment
withAMD, at a rate that was constant between 1 to 8 h, after the drug had attained maximal inhibition of viral RNA synthesis.
Effect ofAMD on the pattern ofsynthesis of viral precursor
polypeptides.
To confirm that AMDdid not haveany effect onthesyn-thesis of the virus-specific
polypeptides,
pulse-labelingand pulse-chaseexperiments were
per-formed in the presence and absence of AMD. Thedrug (5
,g/ml)
wasaddedtoRLV-infected
cells1hbeforeaddition of
[35S]methionine.
Fig-ure 7showsacontrol15-minpulse labeling (A) and a control 15-min pulse-60-min chase
(C).
AMD-treated pulse and chase cultures are
showninFig.7Band D,respectively. No
signif-icant differences were
observed
in the pulse labeling ofAMD-treated (Fig. 7B) and control (Fig. 7A) cultures. In the chase, however, the usual increase inthe amountofuncleaved Pr4 wasseen in AMD-treated cells. AMD thus didnot affect the pattern of synthesis of the viral
proteins, but only the rate of cleavage of Pr4. Occurrenceof uncleaved Pr4 inempty par-ticles. We have previously reported the occur-renceofuncleavedPr4 inparticlesproduced by JLS-V5 and JLS-V9, but to a much lesser ex-tent inparticlesproducedby JLS-V16 cells (24).
The fact that Pr4 occurs in virusparticles
pro-duced naturally by some cell lines indicates thatacertain amount ofuncleaved Pr4 canbe
incorporated into mature virions. Since AMD reduced therate of cleavage ofPr4 ininfected cells producing RLV, we
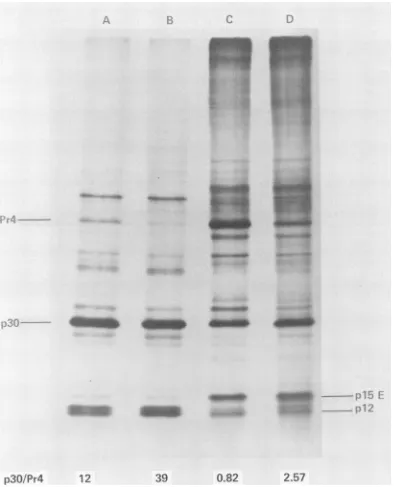
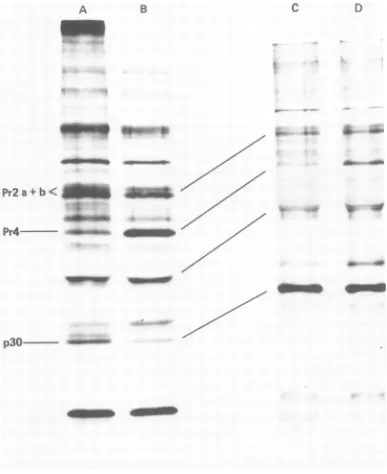
tested
whether it is possible to cause a buildup of Pr4 in RNA-deficient particles produced by RLV-infected JLS-V16 cellsinthepresenceof AMD. Figure 8shows the [35S]methionine-labeled proteins of RLVproduced inJLS-V16 cells inthepresence
of 5 ,g of AMD per ml added 4.5 h before labeling and subsequent virus isolation (Fig.
8A). It is evident that AMD caused a slight
accumulation ofPr4 in virusparticles relative
tocontrolvirus (Fig. 8B). Such a buildup of Pr4 inAMDvirusparticleswasreduced if a period
of chasewasallowedbefore virus isolation (not
shown). Inspection of the virus-specificproteins in the AMD-treated cells at the time ofvirus
isolation (Fig. 8C) showsalargerproportionof Pr4 than was present in virus produced by these cells. This agrees with the idea,
men-tioned above, that the relative amount of
un-cleaved Pr4,inthepresenceof AMD,is
propor-tionaltothe period of chase. Thus, because the
maturation and budding of virus requires a certain time,therelativeamount ofuncleaved
Pr4 should always be higher in cells than in virus.This alsoexplains why it maysometimes be difficult to observe any increase in Pr4 in
AMDvirusparticles (34), especiallyifaperiod ofchase is allowed between labeling
and
virusisolation. Recent observations have
indicated
that incubation of Nonidet
P-40-disrupted
viri-ons at37°C
resultsinapreferential
cleavageofPr4. This cleavage ofPr4 did not occur if the incubationwascarriedout inthe absenceof the
detergent
(datanotshown). These observationscanbeinterpretedtomeanthat the enzymatic activity
required
for the cleavage ofPr4is pres-entinvirionpreparations, butthey alsomeanthataresidual level ofPr4 may notbe accessi-bletothecleavageenzyme(s) intheintactviral
particles.
As mentioned above, treatment with AMD greatly reduces or inhibits the synthesis
and
incorporationofnewRNAinsubsequently
pro-duced viral
particles.
Concerning the preexist-ingpool of viral RNAinthe cell, Levin et al. (33)found that if the cellswereprelabeled
with[3H]uridine
and then treated with AMD for 2 h,then the viralparticlesproduced lacked the60 to
708
viralRNA.Similarly, Paskindetal. (40) found that 4 h of pretreatment with AMDgreatly depletes the pool of preexisting viral RNA in maturing virions. Thus, the particles
whoseproteins areshowninFig. 8A should be mostly, if notcompletely,devoid of viral RNA, sincetheseparticleswerenewly made after4.5
h of AMD pretreatment. Because these
parti-VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
4
-.Av#*j
.'4g_
*- _4-... .
-U., s_,7S
p30/Pr4
0 02.23
1.33
FIG. 7. Effect ofAMD on thepulse labeling ofRLV-specificprecursor polypeptides. Control cultures:
Infected cellswereincubated for 1 h incompletegrowthmedium andthenpulse labeled with[P5S]methionine
for15min(A), andaparallelculture wasthenchasedfor60 min(C). Cytoplasmicextracts werepreparedand
analyzedas in Fig. 2 exceptthat the gels were10%polyacrylamide.AMD treatment:RLV-infectedcellswere
incubated for1 h incomplete growth medium containing5pgofAMD per ml and thenpulselabeledfor15
minwithl'Simethioninein5jAgof AMDperml(B), andaparallelculturewasthenchasedfor60 min in
completegrowth medium containing5 mgofAMDperml(D).
cles contain a level ofp30that issimilarto the
levelof
p30
inuntreated particles,thecleavage of Pr4 to p30 must occur in the formation of empty particles. This confirms the conclusion mentionedabove, that the viral RNAcatalyzesthe cleavage of Pr4 but is not absolutely
re-quiredfor thatcleavagetooccur.
We have considered the possibility that in the absence of genomic viral RNA the viral
capsids are filled with cellular RNA. In this
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.505.68.459.76.538.2]CLEAVAGE OF RLV p30 PRECURSOR 1063
.0
Om
es
-~~_<
*-_ [image:10.505.55.449.76.563.2]p30/Pr4
1
2
39
0.82
2.57
FIG. 8. Effect ofAMDon theamountofPr4 in virusparticles. RLV-infectedcells wereincubated in the presenceandabsenceofAMD(5
pgIml)
for4.5h. The cultureswererinsedtwicewith Hanks balancedsaltsolution and incubated with[35S]methionineinthepresenceand absenceofAMD(5
p.gIml)
for4hingrowthmediumcontaining1120 Eagle methionineconcentrationand5%dialyzedfetal calfserum. Cell-associated
viruswithculturefluidviruswaspurified,and anti-RLVimmunoprecipitates fromthecytoplasmicextracts wereprepared. (A)SDS-polyacrylamidegel electrophoresis ofvirusfromAMD-treatedcells; (B) virusfrom
control cells; (C) anti-RLVimmunoprecipitate from AMD-treatedcells; (D) anti-RLV immunoprecipitate
fr-om
control cells. Similar amountsofradioactiveproteinfrompurified virus(A andB)orfrom immune precipitatesofcytoplasmicextraction(C and D)wereappliedtoa6to12%gradientslabpolyacrylamide gel.VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
case, it is possible that such cellular RNA would substitute for viral RNA in catalyzing thecleavageof Pr4.Toexaminesucha
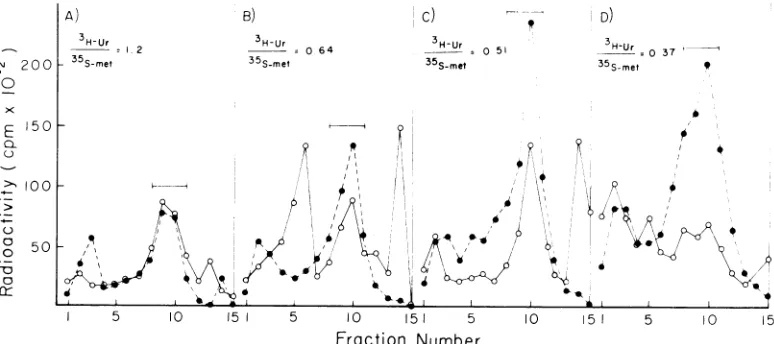
possibil-ity, we have labeled cells for3h withmedium containing both [3H]uridine and
[15S]methio-nine, added AMD for 15 min to stop newviral RNA synthesis, and then collected virusat2-h intervals in the presence ofAMD. The same content of [3S]methionine was maintained throughout the experiment, whereas
13H]uri-dine was removed and replaced with excess cold uridine before virus collection. The col-lected virus was purified on a sucrose
gradi-ent, and the trichloroacetic
acid-insoluble
ra-dioactivity of
[35S]methionine
and [:H]uridinewasmeasured. The
[35S]methionine
radioactiv-ity would be a measure ofthe viral proteins,
according
towhich the[:'Hiuridine
RNAcanbe normalized. Figure 9 shows that the [3H]uri-dine incorporation in viral particles wasre-duced after AMD treatment. Note that the sH/:5S in the virus was not a reflection of the
ratio inthecelldebris orthemedia (Fig. 9and legend). This result indicates that it is not
likely that cellular RNA quantitatively
substi-tutes forviral RNA inviralparticles after the inhibition ofgenomic viral RNA synthesis
by
AMD.
The RNA present in viral particles after
AMD treatment wasexamined under
denatur-X
E 150
Q0
U
> 100
.)
0 50
-o
0
cr
A) ~ 3H-Ur I.2
N 200_35S-met
B)
3H-Ur
= 0 64 35S-met
ing
conditionsby electrophoresis
inurea-acryl-amide-agarose gels (Fig.
10). The resultsindi-cate that whereas virus collected
during
thefirst 2 h after AMD treatment contained 35S
subunit viral
RNA,
virus collected atsubse-quent
periods
wasdevoidofthis RNA. A simi-larfinding
hasbeenpreviously reported (33).It isinteresting,however, that particles collectedafter several hoursofpretreatmentwith
AMD.
containedabroad
peak
of RNA in theregion 15to 28S. An increase in the 4S RNAcontent of AMD virus has alsobeenreported (33). Part of thiscellular RNAcouldbe present dueto
con-tamination ofthe viral preparation
by
cellulardebris,
whereas another part could be due totheentrapmentofcellularmaterial (e.g.,
ribo-somes)inside the virusenvelope. However,the
possibility exists that some cellular RNA is
encapsidated in place of viral RNA. Further
work isrequiredtoinvestigate this
possibility.
Effect of AMD on the cleavage of Pr4 in
JLS-V5 cells. The effect ofAMDonthe rate of
cleavage
of Pr4top30wasevenmorenoticeable inJLS-V5 than in JLS-V16cells. JLS-V5 cellsproduced virus particles that contained a
higheramountofuncleavedPr4 than ispresent
in virus produced by JLS-V16 cells (24).
Also,
the rate ofcleavageof Pr4 top30wasnoticeably
slower in JLS-V5 than in JLS-V16 cells (not
shown).
Treatment ofJLS-V5 cells with AMDc) D)
3H-Ur 51 3HHUr =
35S-met 35S-met 9
5 10 15 5 10 151 5 10 15 5 10 15
Fraction Number
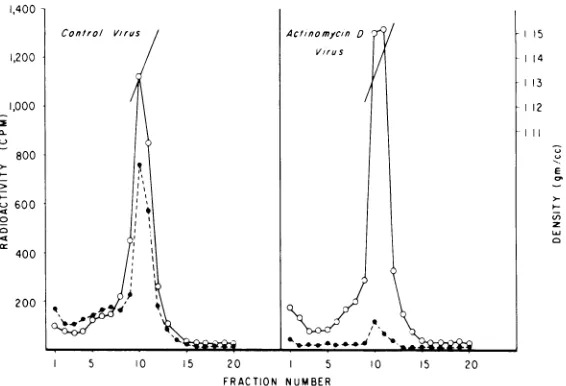
FIG. 9. Quantitative measurement of RNA content in AMD viral particles. RLV-infectedJLS-V16cells
were labeledfor3 h with [3H]uridine and [15Slmethionine in medium containing 1/10 Eagle methionine
contentand 5%dializedcalf serum. AMD (5/IgIml)and 6
M.M
unlabeled uridine were then added for 15min. Themedium wasthen replaced with a portion of the same medium lacking[:'H1uridine.
Medium changes of the same volume weremadeat2 h (A),4h(B),6h(C), and 8 h (D). Virus was collected from the different mediumchangesandpurifiedon a15 to 60%sucrose gradient. Fractions of the gradient were assayed for theirdensitiesandtrichloroacetic acid-insolubleradioactivity.Thebarsindicate the viral peakat adensity of1.13
to1.15glcm3. The ratio of:PHIuridine(3H-Ur) and[35Slmethionine(35S-met) ofthe viruspeakis indicated
withinthebars. The 3H-UrI35S-"'e'ratioof the celldebrismaterial thatpelletedthroughthe sucrose gradient was(A)2.1, (B)1.9,(C)2.6, and(D)2.1, whereas in the supernatantgrowthmedia, aftervirusisolation,it
was(A) 0.81, (B)0.51, (C)0.48, and (D) 0.43.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:11.505.70.457.389.561.2]CLEAVAGE OF RLV p30 PRECURSOR 1065
(Fig. liB) slowed the cleavage rate of Pr4 in
these cellsto agreaterextentthan insimilarly treated JLS-V16 cells (Fig. liD). These results
suggestthat theincreased accumulation ofPr4 in JLS-V5 virus is due to the slower rate of
cleavageofthisprotein intracellularly.A
num-ber of factorscouldcausesuchaneffect,among
them a limitingamount of genomic RNA syn-thesis relativetoviralproteinsynthesis.
However, examination of virusparticles pro-duced by JLS-V5 aftertreatmentwith AMDdid notrevealadrastic accumulation of Pr4,as was seenin the cells(notshown). Thissuggests that
although it ispossibletoincorporate some
un-cleaved Pr4 intomature virions,cleavage ofPr4 top30to alarge extentisprobably an essential
step inassembly.
Effectofcordycepin andcytochalasin B on
therateofcleavage of Pr4. Itwasalso possible
to cause a slowdown of the cleavage ofPr4by
treatingcells with 10 ,ugofcytochalasin B per
ml (Fig. 12D) or200 ,ug ofcordycepin (3'-deoxy-adenosine) perml (Fig. 12C). The reason that wetested thesetwodrugswasthat, duetosome
oftheir known effects, theymayinterfere with the availability of viral RNAinthecytoplasm. Thus, cytochalasin B causes cell enucleation
(42), whichwould interfere with the migration
of nuclear RNA to the cytoplasm. Cordycepin inhibits poly(A) additiontoheterogeneous
nu-clear RNAand,as aresult, intereferes with the
processing and migration of that RNA to the cytoplasm (1). This could apply to viral RNA, whichisknownto contain poly(A) (20, 31, 43).
Cordycepin has been showntoinhibit induction ofmurineleukemia virus production by 5-iodo-2'-deoxyuridine (54), to depress virus produc-tion invirus-producing cell (54), andtoinhibit transformationby murinesarcomaviruses(35, 54).
However, we have not determined that the effects of cytochalasin B
and
cordycepin
onthecleavage
of Pr4 areactually
duetotheir effectsonviral RNAsynthesis, anditispossible that the effects ofthese drugsmay be dueto other cytotoxic effects that these
drugs
may have. Thus,cordycepin
wasfoundtohaveinhibitoryFIG. 10. RNA in viral particles isolated after
treatment with AMD. Viralparticles werepelleted
from the peakfractions of the gradients describedin
Fig. 9, suspended in TNE, made 1% inSDS, and
extracted with one-fourth the volume ofphenol and
one-fourth the volume ofchloroform. The RNAinthe
aqueouslayerwasprecipitatedwith twice thevolume
of ethanol, boiled for1 min, andelectrophoresedon
2%polyacrylamide-0.5%agarose ureagels(17)for6
hat75V.(A)Marker rRNA; (B) marker RLV RNA;
(C-F)RNAextractedfromthe viralpreparations
A-DofFig. 9,respectively.Arrow indicatespositionof
35S mengovirusRAVA.
VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
[image:12.505.75.221.54.669.2]1066 JAMJOOM, NASO, AND ARLINGHAUS
C
a.
Pr2
a +b <
Pr4
_
..
.4o A4
a.-4
Ja
o- .a*
p3-FIG. 11. Cleavage ofPr4top30 inRLV-infectedJLS-V5cells and JLS-V16 cellstreated withAMD.
RLV-infected JLS-V5cells (B) and JLS-V16 cells (D) were incubated for90min incomplete growthmedium,
containing 5 ,ug ofAMDperml, thenlabeledfor75mininAMDwith[35S]methionineinmediumcontaining
1/10 Eagle's methionine concentration, and finally chasedfor 3 h inAMDcompletemedium.Controlcultures
ofJLS-V5 (A) andJLS-V16(C) wereprocessedinthesamefashionexceptthatAMDtreatmentwasomitted.
Anti-RLVimmunoprecipitatesfromRLV-infectedJLS-V5cells (Aand B)wereanalyzedon6to12%gradient gels;immunoprecipitatesfromRLV-infectedJLS-V16 cells (Cand D) wereanalyzedon10%gels.
effectsontotal RNAsynthesis (35)and protein
synthesis (52). Further studies are needed to
determine the mechanismby which these two drugsinterfere with the cleavage of Pr4.
In our studies, treatment with cordycepin
also resulted in the appearance of a distinct
high-molecular-weight
polypeptide that isim-munoprecipitable
withanti-RLV antisera(Fig.A
B
D
flw..
*,""I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:13.505.67.454.81.555.2]CLEAVAGE OF RLV p30 PRECURSOR 1067
A
BC
D
91]I a ji) <
_~ *w
_
1.
_
pr'I- t <
t)
tia-4
oi3G0
-p30/Pr4
0
-_3-_~~~~~~~l
-_
0'w
4
0.82
__v_s|p_ -5
F-0.75
FIG. 12. Effect of cordycepin and cytochalasin B on the cleavage ofPr4 top30. RLV-infected cells were
pulse labeled for 15 min with[35S]methionine(A)and chased for 60 min (B) .Cordycepin treatment: A culture
waspretreated for1 h with 200
Mg
ofcordycepin per ml and pulse labeled and chased in the presence ofcordycepin (C). CytochalsinB treatment:Aculturewaspretreatedfor1 h with 10 pg ofcytochalasinB per
ml; the cells werecentrifuged for2 min at2,000 xg, resuspended,and thenpulselabeledfor15minand
chased for60min (D). The cytoplasmic extracts were processed as in Fig. 2.
12C, arrow),which is not usually observed. The
identity ofthispolypeptide is under investiga-tion.
DISCUSSION
We have shown thatAMD significantly
re-duced the rate ofcleavage ofintracellular Pr4 to p30 while not affecting the processing of
other p30 precursors, such as Pr3 or Prla +b.
The results are consistent with the conclusion
that AMD exerts its effect on the cleavage of Pr4by preventing the synthesis of viral geno-mic RNA, thereby preventing interaction of viral RNA with viral precursor proteins during virus assembly. This interpretation makes it possible toclassify thecleavage(s)of Pr4 top30 as amorphogenetic cleavage (21) and onethat
is acceleratedbyinteraction with viralRNA.
Pr4, previously called p70, has been detected inmature RLVparticles in variable amounts,
VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
[image:14.505.56.449.78.500.2]dependingon thecell type producing thevirus
and the conditions of cell growth (24). It has beenpossibletoslightlyincrease the amountof
Pr4 in virus preparations produced by RLV-infected JLS-V16 cells by pretreatment of
cul-tureswith AMD. However, evenlong pretreat-ments (up to 4h)with AMDdidnotpreventthe almostcomplete cleavage ofPr4 to p30insuch
viruspreparations. Theseresultsindicate that theinteraction with viral genomicRNA is not
absolutely required for the cleavage of Pr4 to
p30 and supportsthe idea that the viral RNA only accelerates the rate ofcleavage ofPr4 to
p30.
Three findings indicate that the effect of AMD on the cleavageofPr4 is not dueto the indirect inhibition ofprotein synthesis. First, thecombined effect of AMD and
cycloheximide
was greaterthanthe effect of eitherdrug alone. Second, the length of the
period
ofpretreatmentwith AMD between1 and8hdidnotaffect the subsequentrateofcleavage ofPr4. Third, AMD did not affect the pattern of viral protein
syn-thesis during pulse-labelingexperiments.
Our results agree with the finding reported by other investigators (34, 40) on the presence
oftwo non-equilibrating pools ofvirus-specific
RNAininfectedcells: (i)mRNA thatfunctions
in translation; and (ii) genomic RNA that is
packagedintovirions. BecauseAMDexerts its maximal effect on Pr4 cleavage after treatment
for1h,wededuce that the pool ofviral mRNA,
which has a functional half-life of 7 h, is not
structurally involved in the interaction which accelerates Pr4 cleavage. Thismeansthatit is
only
the genomic RNA which functionsinthe enhancement ofPr4 cleavage.Anotherpoint inthisstudyconcernsthepool sizeof genomic viral RNA thatisavailable for
accelerating the cleavage of newly made Pr4.
Our results indicate that suchapoolisrapidly depleted. Thus, whereas less inhibition of
cleavage of Pr4 to
p30
was observed in cells pretreated withAMD for 15 to 30 minthanfor 60min, longerpretreatment with AMD (up to 8h) hadnofurther inhibitory effect thandida 1-h treatment on the rate ofcleavage of Pr4 to
p30.
However, these results do not measure the rate of depletionof total genomic viral RNA in the cell, which is known to require severalhours(about 4) to bedepleted (40). We suggest
that this latter pool of genomic viral RNA is
already associatedwithviralproteinsandthus cannotaffect thecleavage of newly made Pr4.
Our results suggest a long half-life of more
than 8 h either for the enzyme(s) involved in theprocessing of Pr4 or for the mRNA(s) coding
for this enzyme(s). Thedecreaseinthe rate of
cleavageof Pr4 after treatment with inhibitors
ofprotein synthesis may indicate that the en-zymes themselves donot havea long half-life.
It is possible, however, that inhibitors of pro-teinsynthesis interferewiththecleavage ofPr4
by a more complex mechanism that oversha-dows the direct inhibition of the synthesis of such processing enzymes.
Thedependence of the cleavageofprecursors to capsid proteins on the interaction with the viral genome is a commonpattern inviral
as-sembly (e.g., reference 22, 25, 46). Such a de-pendencemay stemfrom the role thatcleavage playsin the packaging of the genome. For in-stance, in T4the
packaging
of theviral DNAisaccompanied
by the cleavage of the core pro-teins p22and IPIII (30). Moreover, thecleavage ofthe major head proteinp23leads to astruc-turaltransformation that resultsin an expan-sion of the empty
head,
which isprobably
re-quired
for thepackaging
ofthe DNA(29).How-ever, a
relationship
betweencleavage
andpackaging
of thegenomeisnotclearinseveral other systems. For example, inpoliovirus
the cleavage of theprecursorproteinvp0tovp2andvp4 seems to require the interaction with the viral RNA, since it does not occur in empty
capsids. However, this cleavageis not
required
to bring about the initial association between the viral RNA and the
capsid
proteins, sinceRNA-containing provirion particles have been foundtocontainvp0(15). InRLV,asmentioned
above,
the cleavage of Pr4 to p30,although
catalyzed by
the genomicRNA,
still occursintheformation ofempty cores, thereby suggest-ingthatthe role of this cleavage isnot in pack-aging of thegenome. However, examination of the cleavageproducts of Pr4, together with the
nature of the influence ofgenomicRNAonthe cleavage ofPr4 that was discussed above, has leadus to aconsideration ofamodel thatpoints to several advantages of having adependence of therateofcleavage ofPr4 onthe interaction withthe genomicRNA. Wediscuss this model
inthe sectionbelow.
Theresults presented above have suggested
tousthat the interaction between thegenomic
RNA and Pr4 is a prominent step in RLV
as-sembly. Suchinteraction islikelytoresultfrom a direct affinity between Pr4 and the viral RNA, althoughanindirectinteraction is
possi-ble. Although directinteraction ofpurifiedPr4 topurified viral RNA hasnotyetbeen demon-strated, several recentfindings havepointedto the likelihoodthat such interaction does occur.
Thus, Pr4 has been shown to contain two pro-teinsthat have affinityfor the RNA:
plO,
whichisfoundinassociationwith theviral RNA (16)
andwhich is richinarginine andlysine, anda
protein that migrates onguanidine
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 19, 1976
ride agarosecolumns in the p12 region (L. Kar-shin, inpreparation). (The p12 region in
SDS-polyacrylamide
gels contains another compo-nent, designated pl2E, which migrates onGuHCl agarose columns in the void region. This latter protein contains a
methionine-con-taining tryptic peptide present in pl5E and
Pr2a+b, the precursor of the viralgp69/71
[38].)
The othercomponents ofPr4arep30 andp15 (2;
Arlinghaus et al., in press). Sen et al. have demonstratedastrain-specific binding ofp12 to
genomic RNA (45). Davisetal. have
described
abasic proteininRLVwithamolecularweight of 9,800 that binds to viral RNA and to other heterologous single-stranded RNA and DNA
(9).Thisprotein isthoughtobethesame asthe
plO thatis a component ofPr4. Inaddition, a
70,000-dalton
precursorproteinthatsharesan-tigenic specificities with RLV p30 has been foundinmouse L929cells, whichproducealow level ofa C-type virus. This protein exhibits DNA-binding properties and hasbeenisolated by affinitychromatography on
single-stranded
DNA columns (39). RLV p30 does not bind to
single-stranded
DNA under the same condi-tions (39). Recently, this precursor protein has also been found to share antigenicdetermi-nantswith RLVp15, p12, andplO(C.W. Long,
personal
communication)and is thusvery simi-lar to our Pr4. Theseresults suggestthat Pr4 probably interactsdirectly with the RNA. Our results provide evidence for the occurrence ofaninteraction betweenthe viral RNAandPr4 incells. Whatarethepossible advantages ofan
arrangement in viral assembly in which the cleavage ofa precursor
polypeptide
isaffected
by the interaction of that precursor with the genomic RNA? To answer this question, we
propose the model
depicted
in Fig. 13 for theCLEAVAGE OF RLV
p30
PRECURSOR 1069assembly
of the RLV core. In thismodel,
a"procapsid" structure, consisting of subunits
composed
ofuncleaved
Pr4, isformed before
interactionwith the viral RNA. The formation ofthisstructureisdetermined only by the
spec-ificity of the interactions of the protein sub-units. ThePr4components thatprobably inter-act with the RNA, e.g., plO and p12, are de-pictedontheinside,and the other components,
e.g.,
p30,
are on the outside. Entrance of the RNA into this structure results inneutraliza-tion of the positive charges of the basic region plO of the molecules in the center and allows the proteinsubunitstocome in acloser
proxim-ity and in a more favorable orientation. This facilitates the cleavage ofPr4. Cleavage ofPr4 removes the electrostatic (and
possibly
thesteric)hindranceand allows the
p30
subunitstoforma morestable association. Cleavage ofPr4 occurs both inthe presence ofviral
RNA,
and,at a slower rate, in its absence. Specificity is
determined by theinteractionof theRNA
bind-ing components (e.g.,
p12
andplO)
with the viral RNA. In the absence of the viral RNA, charge neutralization isachieved
bycounter-ions orbyspeciesof host cell RNA.
Incorpora-tion of hostcell RNA is not quantitative (i.e.,
doesnotresultinanRNA/proteinratiosimilar
totheratioof the normal viralparticle), but the
extentofincorporation and the species of RNA
incorporated
are not yet clear. Alarge portion of Pr4 must be cleaved in order to allow themajorcapsid protein subunitstoassume aclose proximity to permit the formation ofa stable
particle,
although a number ofuncleaved
Pr4moleculescanbe incorporated.
Inadditiontotheevidenceontheinfluence of RNA on the cleavage of Pr4, further evidence forthismodelcanbe derived from the present
Pr4 (Uncleaved)
"Procopsid"Structure
Mature Proteins p30, p15, plO, p/2
Assembled Core
[image:16.505.116.396.480.596.2]VRNA
FIG. 13. Hypotheticalmodelof RLVcoreassembly.Thismodelisbasedonthepresumptive evidencethat
the viralgenomicRNA interactswiththeuncleaved precursorPr4ratherthan with thecleavedmatureviral
proteins. Protein subunits ofPr4forma loose andopen arrangement which ispenetrated by thegenomic
RNA. The RNA binding proteins (e.g.,p12 and plO)aredepictedontheinside. Thepositivecharge(ofp1O)
prevents theuncleavedPr4 subunitsfrom formingacompact structure.Charge neutralization (e.g., by the
RNA)facilitatesthecleavageofPr4. Cleavageisrequiredtoallowadjacent subunitstoget incloseproximity
so as toformastablecompactstructure.
on November 10, 2019 by guest
http://jvi.asm.org/
study. Thus, the proposal that the protein-pro-tein interaction is the only major factor that
determines the core structure is based on the very close similarity in composition between
particles containing or lacking the viral RNA
(Fig. 8; 34). Capsid assembly in several viral
systems has been shown to be determined
mainly by protein-protein interaction (22, 25). The importance of cleavage for the formationof
amatureparticle is evidenced bythedifficulty
ofbuildingahigh ratio ofPr4 top30in mature
virus particles, despite several hours of
pre-treatment with AMD, even in virus produced
by cells(e.g.,JLS-V5) that exhibitahigh
inter-cellular Pr4/p30 ratio. No evidence exists yet regarding the proposed procapsid structure,
which may ormay notbe stable enoughto be
isolated.
This. model offers several advantages for
viral assembly. First, the covalent linkage of the majorcoreprotein, p30,toproteins havinga
high affinity for the RNA, e.g., the basic
pro-tein plO, would facilitate encapsidation. This
would beparticularly essential if p30 hadalow
affinityfortheviral RNA. Thearrangementof a basic region in the capsid or core protein
seemstohave been conservedduringevolution.
Thus, inRNA bacteriophages there isa
cluster-ing ofbasic and acidic amino acids (26). Second, the arrangement of the positively charged proteins on the inside results in the
presenceofacenterwithastrongattraction for
theRNA.This arrangement, atthesametime,
ensuresthat theprotein subunits donotforma
compact structure. The existence of an open
structure is ofimportance for thepackaging of
RNA viruses because of the secondary struc-ture of theRNAgenome (26). The assemblyof
DNA viruses,ontheother hand, takes place by the formation of an empty capsid which
ex-pands in size as packaging of the genome
pro-ceeds (23, 29). A similar arrangement of the basic amino acids on the inside of the capsid
structure has beenproposed by Matthewsand
Cole (36) for the capsid off2 bacteriophage. Third, since RNA catalyzes the cleavage of Pr4 and since cleavage is, to a large extent, visualized as required for assembly, the model
predicts that the formation of RNA-containing cores proceeds faster than the formation of
empty cores. This would be of obvious
advan-tage for infectious core formation. Since our
results (Fig. 1) and earlier results by Levinet al. (33)donotindicateasignificant reductionin
the rate of maturation of virions after
treat-mentwithAMD, itmaybepostulated thatcore
formation is not the limitingstep in virus
as-sembly. However, recent resultsby Levin and
Rosenak show areduction, after treatment of
cells with AMD, in the amount of produced
virus, as more accurately determined by radio-immune assay of p30 and the level of activity of the reverse transcriptase (34).
The fourthadvantage of the model is that, if there is low affinity between the major core protein and the ribonucleoprotein complex
in-side it, the genome can then be free to move
inside thecapsid. Such movement may be im-portant for assuming a structurally favorable arrangement. Structural rearrangement of ge-nomic RNA has been described to occur in a murine sarcoma-leukemia virus after matura-tion (11). Moreover, the lowaffinityofthe
ma-joI core protein to the RNA complex would
facilitate the uncoating and release of the ge-nome in the next cycle of infection.
Insummary, the advantages in this model of
havinggenomic RNA intereact with a precur-sor to coreproteins, rather thantothe mature
cleaved proteins themselves, are to ensure the existence ofan open procapsid structure that
can be easily penetratedby the genomic RNA, to make full core formation faster than the
formation ofempty cores, and to allow
encapsi-dation of the RNAin acoat structure for which itmighthave low affinity, thereby facilitating its structural rearrangement and subsequent
release. Further work is required to test the
differentaspects of the model.
Finally, our present results suggest that it may be useful to consider a possible role of genomic RNA in the cleavage ofviral precur-sors in other situations. For example, Eisen-man et al. (13) reported the presence of a stable
precursor-like polypeptidein Rous sarcoma
vi-rus-transformed hamster cells and suggested that the lack of cleavage ofsuch a precursor
mightbe due to theabsence of the appropriate
proteolyticenzymes in thehamster cells.
Simi-larly, the cleavage of precursor polypeptides proceeds slowlyin some cell lines, e.g., mouse L929cells, whichproduce low levels of C-type
particles (39), or, as we noticed, in C243 cells
(5), which produce noninfectious C-type parti-clesthat are devoid of 60to70S viral RNA (41)
(unpublished observation). If genomic viral RNA plays a role in the cleavage of oncorna-viral proteins, then it would be useful to con-sider the absence of viral genomic RNA, as
opposed to viral mRNA, as a possible mecha-nism that plays a role in the cleavage of the
precursor polypeptides.
ACKNOWLEDGMENTS
Thiswork wassupportedinpartby PublicHealth Serv-ice contractCP-61017 and grantCA-15495, bothfrom the National Cancer Institute, andagrant from The Robert A. WelchFoundation (G-429). One of us(G.A.J.) isa
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 19, 1976
toral fellow supported by the Riyadh University, Saudi Arabia.
Wethank J. Levin and C. Long forcommunicating re-sultsbeforepublication, J. Davis for useful discussions, and JamesSyrewicz for excellent technical assistance.
LITERATURE CITED
1. Adesnik, M., W.Salditt, W. Thomas, and J. E. Darnell. 1972. Evidence that all messenger RNA molecules (except histone messenger RNA) contain poly(A) se-quencesandthat the poly(A) has a nuclear function. J.Mol. Biol. 71:21-30.
2. Arcement, L. J., W. L. Karshin, R. B. Naso, G. A. Jamjoom, and R. B. Arlinghaus. 1976. Biosynthesis ofRauscher leukemia viral proteins: presence ofp30 and envelope p15 sequences in precursor polypro-teins.Virology 69:763-774.
3. Bader, J. P. 1970. Synthesis of the RNA of RNA-con-taining tumor viruses. I. The interval between syn-thesisandenvelopment. Virology 40:494-504. 4. Bases, R. E., and A. S. King. 1967. Inhibition of
Rauscher murineleukemia virus growth in vitro by Actinomycin D.Virology 32:175-183.
5. Bassin, R. H., L. A. Phillips, M. J. Kramer, D. K. Haapala, P. T. Peebles, S. Nomura, and P. J. Fis-chinger. 1971.Transformation of mouse 3T3 cells by murine sarcoma virus: release ofvirus-like particles intheabsence of replicating murine leukemia helper virus. Proc. Natl.Acad. Sci. U.S.A. 68:1520-1524. 6. Bonner, W.M., and R. A. Laskey. 1974. A film
detec-tionmethod fortritium-labelledproteins and nucleic acids inpolyacrylamidegels. Eur. J. Biochem. 46:83-88.
7. Butterworth, B.E., L.Hall, C. M. Stoltzfus,and R. R. Rueckert. 1971. Virus-specific proteinssynthesizedin
encephalomyocarditisvirusinfected HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 68:3083-3087.
8. Casjens, S.,and J.King. 1975. Virusassembly. Annu. Rev.Biochem.44:555-611.
9. Davis, J., M. Scherer, W. P. Tsai, and C. Long. 1976. Low-molecular-weight Rauscher leukemia virus pro-tein with preferential binding for single-stranded
RNA and DNA. J. Virol. 18:709-718.
10. Duesberg, P. H., and W. S. Robinson. 1967. Inhibition of mouseleukemia virus (MLV) replicationby Acti-nomycin D.Virology 31:742-746.
11. East, J. L., P. T. Allen, J. E. Knesek, J. C.Chan, J. M. Bowen, and L. Dmochowski. 1973. Structural rear-rangement and subunit composition of RNA from released Soehner-Dmochowskimurinesarcoma viri-ons.J. Virol. 11:709-720.
12. Eckert, W. A., W. W. Franke, and U. Scheer. 1975. Nucleocytoplasmic translocation of RNA in Tetrahy-menapyriformisand its inhibition by ActinomycinD andcyclohemixide. Exp.Cell Res. 94:31-46. 13. Eisenman, R., V. M. Vogt, and H. Diggelman. 1974.
Synthesis of avian RNA tumor virusstructural pro-teins. Cold Spring Harbor Symp. Quant. Biol. 39:1067-1075.
14. Fakan, S. 1971. Inhibition of nucleolar RNP synthesis by cycloheximide asstudiedby high resolution ra-dioautography. J. Ultrastruct. Res. 34:586-596. 15. Fernandez-Thomas, C. B., and D. Baltimore. 1973.
Morphogenesis of poliovirus. II. Demonstration of a new intermediate, theprovirion. J. Virol. 12:1122-1130.
16. Fleissner, E., and E. Tress.1973. Isolation of a ribonu-cleoprotein structure fromoncornaviruses. J. Virol. 12:1612-1615.
17. Floyd, R. W., M. P. Stone, and W. K. Joklik. 1974.
Separation ofsingle-stranded ribonucleic acids by
acrylamide-agarose-urea gel electrophoresis. Anal. Biochem.59:599-609.
CLEAVAGE OF RLV p30 PRECURSOR 1071
18. Gillespie, D., S. Marshall, and R. C. Gallo. 1972. RNA of RNA tumor viruses contains poly(A). Nature
(Lon-don) NewBiol. 236:227-231.
19. Goldstein, E. S., and S. Penman. 1973. Regulation of protein synthesis in mammalian cells. V. Further studies on the effect of Actinomycin D on translation control inHeLa cells. J. Mol. Biol.80:213-254. 20. Green, M., and M. Cartas. 1972. The genome of RNA
tumorviruses containspolyadenylic acidsequences. Proc. Natl. Acad. Sci. U.S.A. 69:.791-794.
21. Hershko, A., and M. Fry. 1975. Post-translational
cleavage of polypeptide chains: role in assembly. Annu.Rev. Biochem. 44:775-797.
22. Hohn, T., and B. Hohn. 1970. Structure and assembly of simple RNA bacteriophages. Adv. Virus Res. 16:43-98.
23. Hohn, B., M. Wurtz, B. Klein, A. Lustig, and T. Hohn. 1974. PhagelambdaDNA packaging in vitro. J. Su-pramol. Struct. 2:302-317.
24. Jamjoom, G. A., W. L. Karshin, R. B. Naso, L. J. Arcement, and R. B. Arlinghaus. 1975. Proteinsof Rauscher murine leukemia virus: resolution of a 70,000dalton, non-glycosylatedpolypeptide contain-ingp30 peptidesequences. Virology68:134-145. 25. Klug, A. 1972. The polymorphism of tobacco mosaic
virusproteinanditssignificance for theassemblyof the virus, p. 207-215. InCiba FoundationSymp. 7: Polymerization inbiological systems, p. 207-215. El-sevier,Amsterdam.
26. Knolle,P., and T. Hohn. 1975. Morphogenesis ofRNA
phages, p. 147-201.In X. Zinder(ed.),RNAphages.
ColdSpringHarborMonogr.Ser.ColdSpringHarbor Laboratory, ColdSpringHarbor,N.Y.
27. Korant, B. D. 1975.Regulationofanimalvirus
replica-tionby protein cleavage, p. 621-644. In E. Reich(ed.),
Cold Spring Harbor symposium on control of cell proliferation, vol.2:Proteasesandbiologicalcontrol. ColdSpringHarborLaboratory, ColdSpringHarbor,
N.Y.
28. Laemmli, U. K. 1970. Cleavage ofstructural proteins during the assembly of the head ofbacteriophageT4. Nature(London)227:680-685.
29. Laemmli, U. K., L. A. Amos, and A. Klug. 1976. Corre-lation betweenstructural transformation and cleav-age of the major head protein of T4bacteriophage.
Cell 7:191-203.
30. Laemmli, U.K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. The DNA packaging
events.J. Mol. Biol. 80:575-599.
31. Lai, M. M. C.,and P. H. Duesberg. 1972.Adenylic
acid-rich sequences inRNAsof Rous sarcomavirusand
Rauscher mouse leukemia virus. Nature (London) 235:383-386.
32. Laskey, R.A.,andA.D.Mills.1975.Quantitative film detection of3H and 14C in polyacrylamide gels by
fluorography. Eur. J. Biochem. 56:335-341. 33. Levin, J.G.,P. M.Grimley,J. M.Ramseur,and I. K.
Berezesky.1974. Deficiency of60 to70SRNAin mu-rine leukemia virus particles assembled in cells
treatedwithactinomycinD.J. Virol. 14:152-161. 34. Levin, J. G., and M. J. Rosenak. 1976. Synthesis of
murineleukemia virus proteinsassociatedwith viri-ons assembled in Actinomycin D-treated cells: evi-dence forthe persistence ofviral messenger RNA. Proc.Natl. Acad. Sci. U.S.A. 73:1154-1158. 35. Lovinger, G. G., R. A. Klein, R. V. Gilden, and M.
Hatanaka. 1973. The effect of cordycepin on cell transformation by RNA tumor viruses. Virology
55:524-526.
36. Matthews, K. S., andR. D.Cole. 1972.Shellformation bycapsidprotein off2bacteriophage. J. Mol. Biol. 65:1-15.
37. Naso, R. B., L. J.Arcement, and R. B. Arlinghaus.
on November 10, 2019 by guest
http://jvi.asm.org/
1975. Biosynthesis of Rauscher leukemia viral
pro-teins.Cell4:31-36.
38. Naso, R. B., L.J. Arcement, W. L. Karshin, G. A. Jamjoom,and R. B.Arlinghaus. 1976. A fucose-defi-cient glycoprotein precursor to Rauscher leu' mia
virusgp69/71.Proc.Natl. Acad. Sci. U.S.A. 73:2326-2330.
39. Oroszlan, S., C. W. Long, and R. V. Gilden. 1976. Isolationofamurine type C virusp30precursor
pro-tein by DNA-cellulose chromatography. Virology
72:523-526.
40. Paskind, M. P., R. A. Weinberg, and D. Baltimore. 1975. DependenceofMoloneymurineleukemia virus productiononcell growth. Virology67:242-248.
41. Phillips,L. A., V.W. Hollis, Jr., R. H. Bassin, and P. J. Fischinger. 1973. Characterization ofRNAfrom non-infectiousvirionsproduced bysarcoma positive-leukemianegativetransformed 3T3 cells.Proc. Natl. Acad. Sci. U.S.A. 70:3002-3006.
42. Prescott, D. M., D. Myerson, and J. Wallace. 1972. Enucleationofmammalian cellswith cytochalasin B. Exp. CellRes. 71:480-485.
43. Ross, J., S. R. Tronick, and E. M. Scolnick. 1972. Polyadenylate-rich RNAinthe 70S RNA of murine leukemia-sarcomavirus.Virology49:230-235. 44. Scherrer, K., H. Latham, and J.E.Darnell.1963.
Dem-onstrationofanunstable RNA andofa precursorto
ribosomal RNAinHeLacells. Proc.Natl. Acad. Sci. U.S.A.49:240-248.
45. Sen, A., C. J. Sherr, and G. J. Todaro. 1976. Specific binding of the type C viral core protein p12 with purified viral RNA.Cell 7:21-32.
46. Showe, M. H.,and E. Kellenberg.1975.Control mecha-nisms in virusassembly,p.407-438.InControl
proc-esses in virusmultiplication, 25th Symp. Soc. Gen. Microbiol. p. 407. Cambridge University Press, Cam-bridge, England.
47. Singer, R.,and S. Penman. 1972.Biological stabilityof HeLa cell mRNA inActinomycin. Nature(London) 240:100-102.
48. Smith, A. E. 1975. Control of translation of animal virusmessenger RNA, p. 183-223. In Control proc-essesinvirusmultiplication, 25th Symp. Soc. Gen. Microbiol. Cambridge University Press, Cambridge, England.
49. Syrewicz, J. J., R. B. Naso, C. S. Wang, and R. B. Arlinghaus. 1972. Purification oflarge amounts of murineribonucleicacid tumor viruses in roller bottle cultures. Appl. Microbiol. 24:488-498.
50. Temin, H. M. 1963. The effects of Actinomycin D on growth of Rous sarcoma virus in vitro. Virology 20:577-582.
51. Vogt, V. M., and R. Eisenman. 1973. Identificationofa large polypeptide precursor of avian oncornavirus proteins. Proc. Natl.Acad. Sci. U.S.A.70:1934-1938. 52. Weiss, S. R., andM.A.Bratt. 1975.Effectofcordycepin (3'-deoxyadenosine) on virus-specific RNA species
synthesized in New Castle disease virus-infected cells.J.Virol. 16:1575-1583.
53. Willems, M., M. Penman, andS. Penman. 1969. The regulation ofRNA synthesis and processing in the nucleolus during inhibition of protein synthesis. J. Cell Biol. 41:177.
54. Wu, A. M., R.C. Ting, M. Paran, andR.C.Gallo.1972. Cordycepin inhibits induction of murine leukovirus production by 5-iodo-2'-deoxyuridine. Proc. Natl. Acad. Sci. U.S.A. 69:3820-3824.

![FIG. 3.15completemin;with min Effect of cycloheximide on the cleavage ofPr4 to p30. RLV-infected JLS-V16 cells were pulsed for with [35S]methionine in Hanks buffer (A) and chased for the following times: (B, C) 15 min; (D, E) 30 (F, G) 60 min; (H, I) 3 h](https://thumb-us.123doks.com/thumbv2/123dok_us/1554747.108071/5.505.273.458.403.550/completemin-effect-cycloheximide-cleavage-infected-methionine-hanks-following.webp)