0022-538X/80/00-0041/09$02.00/0
Revertants of
Adenovirus Type
12-Transformed Hamster
Cell
Line T637 as Tools in the Analysis of Integration
Patterns
DIRK EICK, SILVIA STABEL,ANDWALTER DOERFLER*
Institute of Genetics, University of Cologne, Cologne, Germany
Spontaneously arising morphological revertants of the adenovirus type 12
(Adl2)-transformed hamstercell line T637 had been previously isolated, and it
had been demonstrated that in theserevertants varying amounts of the integrated
Ad12genome wereeliminated fromthe host genome. In this report, the patterns
of persistence of the viral genome in the revertants were analyzed in
detail.
In some of the revertant cell lines, F10, TR3, and TR7, all copies ofAdl2 DNAintegrated inline T637werelost.InlinesTR1, -2, -4 to -6, -8 to -10, and -13 to -16,
only the right-hand portionofoneAd12 genome was preserved; it consisted of theintactrightsegmentofAdl2DNA and was integrated at the same site as in
line T637. In revertantlinesG12, TR1l,and TR12, one Ad12 DNA and varying
parts of a second viral DNA molecule persisted in the host genome. These
patterns of persistenceofAdl2DNAmoleculesin different revertants supported amodel foramode of integration of Adl2DNA inT637 hamstercellsinwhich
multiple (20 to 22) copies of the entire Ad12 DNA were
serially
arranged,separated from each other by stretches of cellular DNA. Theoccurrenceof such
revertantsdemonstrated that foreignDNA sequencescouldnotonly be acquired
but could also be lost from eucaryotic genomes. There was very little, ifany,
expressionof Adl2-specific DNAsequencesintherevertantlines TR7 and TR12. Moreover, Adl2 DNAsequenceswhichwerefoundtobeundermethylated in line T637were completely methylated in therevertant cell lines G12,TR1l, TR12, and TR2. Thesefindingswereconsistent with the absence ofTantigen from the
revertantlines reportedearlier. Henceitwasconceivable that the expression of
integrated viral DNA sequences was somehow dependentontheir positions in
the cellulargenome. In celllineTR637, the earlysegments ofAdl2DNA were
expressed and undermethylated; conversely, in the revertant lines G12, TRll, TR12, and TR2, thesamesegmentsappearedtobeexpressedto alimitedextent
andwerestrongly methylated.
Mammaliancells haveamechanismthat
en-ables them to
incorporate foreign
DNA into their genomes. Theuptake
andintegration
ofviral DNA have sofar been studied most
thor-oughly,
inparticular
in virus-transformed cells (for review,seereference3). Theintegration
of specific genes and ofbacteriophage
DNA intothe genomesof mammalian cellshas also been
reported (9, 19). On the other
hand,
different clonesofmorphological
revertantsof adenovirustype 12
(Adl2)-transformed
hamster cellshavebeen shown tohave losta
portion
or allof theintegratedviral DNA sequences(7,8).
Similarly,
revertants of simian virus 40-transforned (14)
and polyoma-transformed cells (2) have also
been described. Thus, foreign DNA sequences
can also be eliminated from the genomes of
mammalian cells. The
question
arises whetherin theprocessofexcision of
foreign (viral) DNA,
cellular sequences
adjacent
to viral sequencesare removedatthesametime. Since these
cel-lularDNA sequences arepossibly of the repeti-tive type (5, 13), their loss may not affect cell survival.
The T637line ofAdl2-transfonned hamster cells has been studied
extensively
in ourlabo-ratory (5, 10, 13, 16). Approximately 20 to 22
copies of Adl2DNA percellareintegrated into
thegenomeof thesecellsat two orthree
differ-entsites.Most,
perhaps all,
of the Adl2genomes areseparated by cellularDNA sequences. A truetandemarrangementofAdl2 DNA hasnotbeen Qbserved (5, 13).
The fourearly regionsof Adl2 DNA are
ex-pressed asmRNAin T637cells (10).T637cells
synthesize the
Adl2-specific
Tantigens. Spon-taneously arising morphological revertants ofT637cells have been isolated and characterized
(7,8).Therevertantsexhibita
morphology
strik-ingly different from that of T637 cells and areT-antigennegative
according
totheimmunoflu-orescence test (7). Among 18 revertant clones
41
on November 10, 2019 by guest
http://jvi.asm.org/
42 EICK, STABEL, AND DOERFLER
analyzed, 3 seem to have lost most, if not all,
Adl2 DNA sequences. Fourteenclones have lost
considerable amounts of viral DNA. Several of
the 18 clones have not been independent
iso-lates. The revertants are not useful in the
anal-ysis of Adl2 genesessentialforthe maintenance
of the transformed state, because all revertant
clones are aboutas oncogenic as the T637line
orthe BHK-21 parent line from which the T637
cellsoriginated (7).
In the present report, we have used the
re-vertants as tools in the analysis ofintegration
patternsof Adl2 DNA in the T637line.
Accord-ing to the patterns of persisting Adl2 DNA
sequences,the18clones fall into five classes. We
haveanalyzedindetail which parts of the viral
genomehave beeneliminated from each of the
revertant lines. The data obtained further
sup-port themodel presented in theaccompanying
paper (13) for the integration of Adl2 DNA in
T637 cells, i.e., multiple viral genomes being integratedpossiblyinto repetitiveDNA. Intact
viral DNA moleculesareseparated bystretches
ofcellular DNA, and multiple cell-virus DNA
combinationsareserially arranged. There is
lit-tle, if any, expression of the remaining Adl2
genomesin therevertantlines, andthepattern
ofmethylationhas becomestrikinglyaltered in therevertantsincomparisonwith the T637line.
MATERIALS AND METHODS
Cells and viruses.Theoriginandthemethods of
propagation oftheT637 cells (6)and ofthe morpho-logical revertants (7) were previously described.
Among the 18 revertant clones, G12 and F10 were
obtained first and independently from lines TRI through TR16.The clones ofthe seriesTRIthrough TR16wereisolated fromasingle culture of T637 cells which had been derivedfrom asingle independently reisolated clone of T637 cells. It was demonstrated that manyofthe TRclones (TRI,-2, -4, -5, -6, -8, -9, -10, -13, -14, -15, and -16) exhibitedidentical
integra-tionpatternsofAdl2DNAand, therefore,were likely torepresent reisolates of the sameoriginalrevertant
clone.
Adl2was propagatedon human KB cellsgrowing insuspensioncultureormonolayerculture.The meth-odsofAdl2propagation, virus purification, and isola-tion ofAdl2DNA weredescribed elsewhere (10).
IsolationoftheEcoRIfragmentsofAdl2DNA.
Isolationproceduresareoutlined in detail elsewhere (5, 13). The same methods were used to purifythe
BamHI orHindIIIfragments ofAdl2 DNA. In later
experiments, EcoRI fragments of Adl2 DNA were usedwhich had been cloned in the bacterialplasmid
pBR325 (S. Vogel, M. Brotz, U. Winterhoff, and W.
Doerfler, manuscriptinpreparation).
Other methods. The methods used in the isolation
ofcellularDNA(16),in nick translation ofAdl2DNA or viral DNA fragments (11), and in the analysisof
viral DNAsequences in cellular DNAby blotting(12), by DNA-DNA hybridization (18), and autoradiogra-phyarealldescribedin detail elsewhere (5, 13,16).
Methylation of integrated Adl2 DNA se-quences. Themethodsofcleavageof DNA fromT637 cellsand from therevertantswiththeMspIandHpaII restrictionendonucleasesaredescribedelsewhere(15). Intact,32P-labeledAdl2DNAwasusedas hybridiza-tionprobe.
RESULTS
Survey of integration patterns of Adl2
DNAinallrevertantlinesand inT637 cells.
The DNAs from all revertant lines (FIO, G12,
and TRi through TR16) and from the parent
line,T637,wereisolated andanalyzed by South-ern blotting upon cleavage with theEcoRI
re-striction endonuclease. The DNA on the blots
was subsequently hybridized with Adl2 DNA
which was32p labeledby nick translation. The
datapresented in Fig. la demonstrate five
dis-tinct patterns of integration in revertant lines
F10, G12, TR12, TR1l, and TR2. Moreover, it wasestablishedthat the revertant linesTRl,-2,
-4, -5, -6,-8, -9, -10, -13, -14, -15,and-16exhibited
identical patterns (Fig. 1). Thus, these latter
isolates probably represented reisolates of the
samerevertantclone.Aspreviously reported (7), inrevertantlines F10, TR3, and TR7, viral DNA
sequencescouldnotbedetected (Fig. la,b), even
thoughamuchmoresensitive methodfor DNA-DNA hybridization (18) was applied in this
studythan in earlier studies (7, 8). In Fig. la,
the five principal integration patterns of Adl2
DNA inrevertantlines FIO, G12, TR12, TRI1,
and TR2 are presented. In the DNA from line
F10,viral DNAsequenceswere notapparent. In
the DNAs from linesG12,TR1l,andTR12, the
FIG. 1. Analysis of the DNA from 18 morphological revertants and from line T637 for the patterns of integration ofAdl2DNA. DNAs from revertant celllines or fromAdl2as indicated werecleavedwith the restriction endonuclease EcoRI, and the fragments were separated by electrophoresis in horizontal 0.5% agarose slabgels. Each gel slot contained 10 LgofDNA. The DNA fragments were then transferred to nitrocellulose filters by the Southern technique and hybridized withAdl2DNA which was32Plabeled bynick
translation. Adl2-specificDNA fragments werevisualized byautoradiography. In the gel slots designated 5 x Adl2, five genome equivalents ofAdl2 DNA mixed with 10 ,ug of B3 DNA used as substrate were
electrophoresed after EcoRI digestion. The calculation of genome equivalents was based on a molecular weight of 20x 1i;forAdl2DNA (4) and a DNA content of 1.3 x 10V jigper T637 cell (6). All methods used areoutlinedindetail elsewhere (5, 13, 16). In (a), theEcoRI map ofAdl2DNA and the sizes of the EcoRI
fragmentsA to Earegivenin kilobasepairs(kbp).
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
REVERTANTS OF Adl2-TRANSFORMED CELLS
N_ N
L D- I:- a:
LL 0 F -aLC%
I-X '.-. cr c
a Lue)
-0
..LI N-r(0 <
cc: n: c=c: x
I- -I- fr-I- tO
Kbp 11.8 8.7
A-
B-5.4 C
4.0
D-2.4 E
Eco R I
C D B EF A
C D _ B EEF _A _
q~~~ ~ ~ ~ B
i.-.1
s.
_
h
b
;
x
.._.~~~~~~~~.
--C.2
~~-
- CII 0 (
I~~~~lb -0N
<C,
10
-clubW:.
.b.
_b--r_0
b
I
e
-...*
EcoR I
.:4
--E--A
B
C
--D
E
I
li.
t:..:
.L-EcoRI
s.
VOL. 36,1980 43
--T.
T.
'H& -E
AL
4
Ai. .rl
-A6.
f.
on November 10, 2019 by guest
http://jvi.asm.org/
44 EICK, STABEL, AND DOERFLER
intemal
EcoRIfragments
B,D,
and E of Adl2 DNAcomigrated
with the correspondingfrag-mentsinthe virion marker DNA. The intensities
of these bands in the lines mentioned were,
however,
considerably
reduced in comparison with the T637 parent line. The DNA from all threerevertantlines also containedoff-size
band3of T637 DNA(13). In
addition,
the DNAfrom line TR12carriedaweak off-sizefragment (band 2), and the DNA fromlineTRll containedanAdl2-specific fragment intermediate in size
be-tween the EcoRI fragments A and B of Adl2
DNA (Fig. la). The DNAs from line TR2 and all its subclones contained off-size band3, aband
comigrating
with the EcoRIfragment E,
andaband of
slightly higher
molecular weight than the EcoRIfragment
C of virion marker DNA. The identities ofmostof these bandswere de-terminedby using specific
viral DNAfragmentsas
hybridization
probes.The amountsof Adl2 DNA persisting in the
revertant
lines
were quantitated byphotomet-rically
comparing
intensities of Adl2-specific bands in theautoradiograms
of Fig. 1. This method is described in theaccompanyingreport(13). Theresults of this quantitation indicated that the DNA from therevertantlines TR1,-2,
-4 to-6, -8 to-10, and-13to -16containedabout
one copy ofafragment of Adl2 DNA percell (seebelow). The DNAs from therevertantlines G12,
TR12, TR1l,
and TR2 containedconsid-erably
less than fivecopies
ofAdl2 DNA andmorethanonecopy,probably abouttwo copies,
atleast ofsomepartsofthe Adl2genome.
Analysis
ofintegration
patterns inre-vertants,
using specific
Adl2 DNAfrag-mentsas
hybridization
probes.
Werepeatedthe Southem blotting experiments with the DNAsfrom line T637 and fromthe revertants
F10,
G12,
TR12,TR1l,
andTR2, using 32P-la-beled EcoRIfragment
A, B,C,orDashybridi-zation probe. In some experiments, the
32P-la-beled BamHI
fragments
orsome of the cloned EcoRIorBamHIfragments
of Adl2 DNAwereusedasprobes also. The data presentedin Fig.
2 canbe
summarized
asfollows.(i) Off-size band 3 (13; Fig.3) in revertant
lines
G12, TR12,
TR1l,
and TR2contained sequencesthat were homologoustoEcoRI fragmentA of
Adl2 DNA. Homology to fragment A was also
found in the DNA from line G12 in a band
comigrating with EcoRI fragment B, and in
DNAfrom line TRll in the
Adl2-specific
bandelectrophoresing
in a position between EcoRIfragments
Aand B (Fig. 2a). In the DNA fromlineTR12, off-size band 2hybridized to EcoRI
fragments
AandC.(ii) In the DNA from lines G12, TR12, and
J. VIROL.
TR1l, the
Adl2-specific
bandcomigrating with the authentic EcoRIfragment Brepresented
in factfragment B. In the DNA from lineTR2, theAdl2-specific
bandelectrophoresing
in a posi-tion slightly above EcoRI fragment Crepre-sented
part of the EcoRIfragment
B ofAdl2 DNA (Fig. 2b). This fragment showed weak homology to fragment A also, suggesting that the truncated Bfragment
might
still be linkedtothesamecellular DNA site viaashort
frag-ment from the
original
left terminus of Adl2 DNA(Fig. 2a).
The weakhomology
to EcoRI fragment B in off-size band 3(Fig.
2b and3)
might have been due to a contaminant in the probe.(iii)EcoRI fragment Cwasmissing altogether fromthe DNA of line TR2.
Homology
toEcoRI fragmentC of Adl2 DNAwasfound in the DNA from line TR12 in off-size band 2 and in the DNA from line TRll in theposition
betweenE&oRI
fragments A and B(Fig. 2c).
In the DNA fromlineG12,
abandcomigrating
withEcoRI fragment B alsodisplayed
homology
toEcoRIfragment
C. The weak apparenthomology
of off-size band 3 in the revertant cell lines toEcoRI
fragment
C couldpossibly
beexplained
by hybridization of the Cfragment
due to theinverted
terminal repeat.(iv) The DNA from lines
G12,
TR12, and TRll contained the authenticDfragment.
This fragmentwasabsent from theDNAofline TR2 (Fig.2d).(v) None of the EcoRIfragmentswas detect-able in theDNAof lineF10.
All of the data presented are summarized schematically in Fig. 3. All DNA bands with
homologies
toEcoRIfragments
A,B,C,
andD wereidentifiedby hybridization tothe isolated EcoRIfragments
of Adl2 DNA as indicated (Fig. 3). The identities of the EcoRIfragments E and F were deduced from the fact thatthey comigrated with the authentic virion markerDNAfragments.
Right-hand
end of Adl2 DNA ispre-served intact in revertant cell lines. The data presented in Fig. 4 demonstrate that the right terminal 2.9 to 3 kilobase pairs (kbp) of
Adl2
DNA were persisting intact in the T637cell line andinthe revertantcell lineTR2. The
rightterminalHindIII fragment E ofAdl2DNA
(2.9 to 3kbp)was
32p
labeled bynicktranslationand was used inhybridization experimentswith
the DNAs from cell line T637 and from the
revertantlineTR2. TheDNAs of these
cell
lineswere cleavedwith the restriction endonuclease
MboI or PstI; the fragments were electropho-resed, blottedonto
nitrocellulose
filters,andhy-bridized to theprobe (Fig. 4).The 1.97- and
on November 10, 2019 by guest
http://jvi.asm.org/
REVERTANTS OF Adl2-TRANSFORMED CELLS
fc f-4 "l
x -o
LO H
N-LL 9 H- - H
a
A- .. r
A
'.k
xU} - CS a:
Ln . LL 0
::_ ~ ~ 49
"lIL"l
N
- N
3 Cl - CN
4 CO N -
-x - - cr cc
-U LLOH
B44_
...
.w~~~~t
N
X N
n
c
N
o N _ N
LL CD
d
-a
a *
EcoRI EcoRI
FIG. 2. Analysisofintegrationpatternsof fiverevertantlines and ofT637cells,usingEcoRIfragmentA, B, C,orDofAdl2DNAasthe hybridizationprobe. Experimentalconditionsweresimilartothose described
inthelegendtoFig. 1.The DNAs ofrevertantlinesFIO, G12, TR12, TR11, and TR2wereanalyzed, using EcoRIfragment A(a),B(b),C (c),orD (d)ashybridizationprobe. Thesefragmentswere32plabeled bynick
translation.
kbp (not visible) MboI fragmentsaswellasthe
1.52- and 0.83-kbp PstI fragments comigrated
with the corresponding Adl2 virion marker
DNAfragments, whereas the terminal 1.16-kbp
MboIandtheterminal1.15-kbpPstIfragments
were displaced to off-size positions due to the
linkage of viral to cellular DNA (Fig. 4). We
conclude that the right terminal 2.9- to 3-kbp
45
N
Hc
b
id
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.510.114.406.71.563.2]46 EICK, STABEL, AND DOERFLER
Ad12 T637 TR1 2 3 4 5 6 7 8 9 10 11 12 13 14 i-AC
2-AC
3-
--AC
-A - - -A -
--AC
-
-B(A)--A
15 16 G12 F10 T637 Adl2
1-AC 2-AC
---A 3-A
-AC
-B I E IF A I
FIG. 3. Schematic representation ofAdl2-specific DNA bands in the DNAs of line T637 and of 18
morphologicalrevertants. Theidentitiesof all bandswereindicatedby the letters Athrough F, which refer
tothe EcoRIfragments ofAdl2 DNA. TheEcoRImapof Adl2 DNAisalsodepicted.
segment ofAdl2 DNA waspreserved intact in the DNAs of the T637 line and of the revertant cellline TR2. Therewas averylimited number ofoff-size bands in line T637 and only one off-size band in line TR2. The latter off-off-size band wasidentical to oneoff-size band inline T637,
suggesting that the sites ofintegration in line
T637 and in the revertantwerethesame.
Expressionand methylation oftheearly
regions in the revertant cell lines. It has
beenpreviously reportedthatthe revertantcells
do not express the Adl2-specific T antigen as determinedby immunofluorescence (7, 8).
Pre-liminarydata(S.Schirm and W.Doerfler,
man-uscript in preparation) indicated that in the
revertantlines TR7 andTR12,verylittle,ifany,
Adl2-specificRNA couldbedetectedby
hybrid-ization of32P-labeled Adl2 DNA or ofthe left
terminal BamHI fragment A (see Fig. lb in
reference 13) to total cytoplasmic RNA
ex-tracted fromrevertantcelllines TR7andTR12
andcovalently linked to diazobenzyloxymethyl
paperbythemethod of Alwineetal.(1).InT637
cells,onthe otherhand,Adl2-specificsequences
werereadilyfoundbythesamemethods. Itwas
concludedthat Adl2 DNAdidnotappeartobe
expressed extensively as mRNA in revertant
lines TR7andTR12. Forline TR7,thisfinding
was expected since Adl2 DNA sequences did
notpersistinthis line(Fig. lb).
Wehavereportedearlierthat incellline T637
theearly regionsof Adl2DNA correspondingto
EcoRI fragments A and Careundermethylated,
whereas the late regions in EcoRI fragment B
are strongly methylated (15). Similar results
havebeen obtained with Ad2 DNAsequencesin Ad2-transformed hamster cells (17). It was,
therefore, of interesttoinvestigatetheextentof
methylationof theAdl2 DNAsequencesinthe
revertantcell linesG12, TR12, TR1l,andTR2. TheDNAsofcelllinesT637, G12,TR12, TR11, and TR2 were cleaved with the restriction
en-donuclease MspI or HpaII, and the fragments
were electrophoresed and transferred to
nitro-cellulose filtersasdescribed (15, 17). The DNA
on the filters was then hybridized with 32P-la-beled Adl2 DNA. The results of these experi-ments (Fig. 5) demonstrate that theAdl2 DNA sequences were almost completely methylated in the revertants and undermethylated in the T637 line aspreviously found (15). The restric-tion endonuclease MspI cleaved at all of the
CCGG sites in Adl2 DNA in the revertants, whereas the enzyme HpaII did not (Fig. 5).
Thus,the Adl2 DNAsequencespersistinginthe
revertants mighthave come under completely
different control as a consequence of their
re-positioning inthe reversionprocess. Itwasalso
conceivable, though unlikely,thatunmethylated
sequences ofAdl2 DNA might have been lost preferentially.
DISCUSSION
The analysisofthepatterns ofintegration of
Kbp
11.8 A 7
BI-54C
40
DI--B(A) - - - -B(A)
24 E
0.6F
-A
IECO
RD
I
C
I D I
J. VIROL.
-B(A)
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.510.66.458.62.305.2]REVERTANTS OF Adl2-TRANSFORMED CELLS
nB'o ft b!~~~ci af a :
CY-9.F
:I iI
41mb
FIG. 4. Presenceoftherightterminalsegmentof
Ad12 DNA in celllines T637 andTR2. TheDNAs
from Adl2 orfrom cell lines T637 and TR2were
cleaved with restrictionendonuclease MboIorPstI
asindicated. Thefragmentswereseparated by
elec-trophoresison2.0%agaroseslabgelsandtransferred to nitrocellulose filters bythe Southernprocedure.
The DNA was then hybridized to the 32P-labeled HindIII fragmentEofAdl2 DNA whichcontained
the rightterminal 2.9to 3kbp ofAdl2DNA. The
numbers on theleft margin ofeach autoradiogram designate the sizes in kilobasepairs ofindividual
DNAfragments. Themapson the bottoms ofeach
graph indicate the relative positions of the right
terminal MboI and PstIfragmentsandtheir sizes in kilobasepairs. Mapsweretakenfromreference5.
Adl2 DNAinmorphologicalrevertants of T637
cellshas revealed five different modesof
persist-ing Adl2 genomes (Fig. 3 and 6). In lines F10,
TR3, and TR7, apparently all viral DNA
se-quences have been lost. In line TR2 and its
identicalsubclonesTR1, -4 to-6, -8 to-10,and
-13to-16,allviralgenomeshave been lostexcept
aportionofoneviralDNA molecule. This
inter-pretation is consistent with and based upon
model (a) in the accompanyingpaper (13), i.e.,
a serial arrangement ofmultiple copies of the
entire Adl2 genome integrated into cellular
DNAseparated fromoneanotherby repetitive
cellular DNA sequences. An episomal location
of the integrated Adl2 DNA copies on avery
FIG. 5. Extentof methylation ofAdl2DNAin line
T637and in revertant lines G12, TR12, TR11, and TR2. TheDNAsofAdl2orof thecelllinesas
indi-cated were cleaved with restriction endonuclease MspIorHpaII. Fragmentswereseparated by
electro-phoresison1.3%agaroseslabgels.AU other
condi-tions were described elsewhere (5, 13, 15, 16);
32p-labeled,intact Adl2 DNAwasusedashybridization
probe.
large extrachromosomal element would also
ex-plain the occurrence ofrevertants. At present,
there isnoevidenceforsuch episomalstructures.
Inthe process of deletion of viral and cellular DNA, the entire block of DNA comprising all
the viral and cellular DNAsequencesis excised,
leaving only one viral DNA equivalent which
hasbeen truncated intheprocessinsideEcoRI
fragment B (Fig. 3 and 6). This model is in
agreementwiththepersistence of off-size band
3 homologous to EcoRI fragment A, of
frag-mentsE andF, and ofatruncated Bfragment
(Fig. 3).It hasalso beendemonstrated that the
right terminalsegmentofAdl2 DNAcomprising
2.9 to 3kbp persists intact incelllines T637 and
MN..
47
., ..
.1
A..-am
.7.---ft
.r,.,2
0
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.510.58.234.74.354.2] [image:7.510.278.427.78.420.2]48 EICK, STABEL, AND DOERFLER
S////S//SXX//X//SSSH%A /C/ D%//g
I~~~~~~~~~
I
B|E |F |
EFll
A
//XX/
I~
~~~~~~~~~
;
lll
E-QL/////////W/WSA
C|D
B|E|F|
Ali/i,//
A C D B EF A
S/SE///S/,/m/H
A
g
C | D I
B
|I
E|F|
777//;
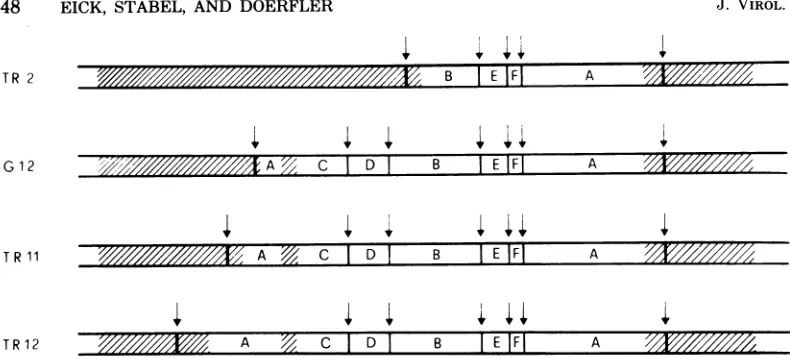
FIG. 6. Proposed models for the integrcltion of Adl2DNA infourrevertantlines. The letters AthroughF refertothe EcoRI fragments of Adl2 DNA. The shadedareasrepresentcellularDNAsequences;thevertical
arrowsindicatethe known viralortheunmappedcellular EcoRI sites.TR2, G12, TR11, andTR12referto individualrevertantlines(see text).
TR2 (Fig. 4). The sites of linkage of Adl2 to
cellular DNA in lines T637 and TR2seemtobe identical.
In lines G12, TR1l, and TR12, one entire Adl2genomehasbeen conservedintheprocess ofexcision. In addition, the adjacent viral ge-nome,which isseparated bycellularsequences, has been truncated at different sites inside the
right terminal EcoRI fragment A. Truncation
might also have occurred inside the cellular
DNA sequences separating the two persisting viral DNA molecules. The schemespresentedin
Fig. 6 (G12, TR11, and TR12) indicate the
hy-pothetical sites of truncationwithin theEcoRI
fragmentAof the secondpersistingAdl2 DNA
molecule. The nucleotide block consisting ofa
truncatedfragment A,ofcellularsequences,and of the left terminalfragmentC of theviral DNA molecule persisting intact varies in length in each of the threelines,dependingonwhere the break insidefragmentAhasoccurred(Fig.3 and
6).InthehypotheticalschemespresentedinFig.
6, the exactlocations of theEcoRI sites in the
cellular DNA sequences andthelengthsof the cellular DNA separating viral DNA sequences arenotknown.
The patterns ofpersistenceof theviral DNA sequencesremaining in the genomes of the
re-vertantsanalyzedsupportthegeneralmodel(a)
proposed for the mode of integration of Adl2
DNA in T637 cells (13). The mechanism of excision of DNA sequences from transformed cellsisnotknown. Ifthe modelselaborated here are correct, viral and cellular sequences have been eliminatedjointly. Thefact thatasmany asabout 20 viralgenomescanbeexcisedjointly
strengthens the notion that many Adl2 DNA
copies areintegratedinto cellularsequences en
blocasdepictedinmodel(a) inthe
accompany-ingreport(13).We havenotyetdeterminedhow and at whatsite(s)theremaining truncatedviral genomes arereconnectedtocellular DNAupon
completion ofthe excision event. It cannot be
ruledoutatthepresenttime that theremaining
viral sequences in revertants have been
com-pletely excised and reintegrated at a different
site. However, we consider this possibility
un-likely,because the sizesof off-sizeband 3 in the
T637 line and the corresponding band in the revertantlinesareapparently identical (Fig. 3).
Moreover, the sites ofintegration at the right
andperhapsthe leftends ofAdl2 DNA inlines
T637 andTR2arethesameor verysimilar (Fig.
4).
Itcannot be stated how frequently DNA
se-quences areexcised fromvirus-transformed cells orfrom mammalian cells in general. Investiga-tions on this fascinating subject may be
facili-tatedbytheuseofvirus-transformed cells. It is quite interestingto notethat the
revert-antsanalyzed donotseemtocontain the
Adl2-specificTantigen that line T637 synthesizes (7,
8). By DNA-RNA hybridization techniques
us-ingaverysensitivemethod(1),wehave detected
minute amounts, ifany, of Adl2-specific RNA
in the cytoplasm of the revertant line TR12.
Apparently, the Adl2 genome isnot expressed
in these revertants. In line T637, on the other
hand, all four early regions of Adl2 DNA are
expressedasmRNA (10),and these transcripts
canreadilybedetected by themethods used in
the presentstudy. Wehave alsoshown that the
extent ofDNA methylation ofAdl2 DNA
se-quencesinalloftherevertantcell lineshas been
TR 2
G12
TR11
TR12
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.510.66.461.58.237.2]markedly enhanced incomparison with that in the parent line T637 (Fig. 5). Thisfinding would alsoimply that the Adl2 genome copies persist-ing in the revertants have become genetically inactivated, perhaps as a consequence of their repositioning in the cellular genome. We have reported previously (15, 17) on an inverse cor-relation between thelevels of DNA methylation and the extents ofexpression of the same seg-ments of viral genomes in adenovirus-trans-formed cells. It is rather lesslikely that some of the Adl2sequences in line T637 had been un-dernethylated to begin with and that these Adl2 molecules had been lostpreferentially.
The findings presented on the altered levels ofexpression and methylation of Adl2 genes in revertantssuggest thatexpression of viral genes intransformedcells may be somehow dependent ontheir
position
in thecellular genome.ACKNOWVLEDGMEN1TS
We thankJiirgen Groneberg for a gift ofcellular DNA preparations, Hanna Mansi-Wothke for the preparation of media,BirgitKierspelfortypingthemanuscript,andSabine SchirmforagiftofpurifiedBamHIfragmentsofAdl2 DNA. Thisresearchwassupported bythe Deutsche Forschungs-gemeinschaftthroughgrantSFB74.
ADDENDUM INPROOF
(i) Theresults ofrecentexperiments have shown that inthe revertantlinesG12 and TRll sequencesat
the right terminal 2,900 to 3,000 base pairs ofthe persistingAdl2 DNA have beenrearranged andthus
are nolongercolinear with thevirionDNA sequences,
ashas beenfound for the parent line T637.
(ii) In the cytoplasmicRNAsof therevertantlines, very few, ifany, sequenceshavebeendetectablethat hybridized withAdl2 DNA.
LrERATURE CIMD
1.Alwine, J.C., D. J. Kemp,and G. R. Stark. 1977. Method for detection ofspecificRNAs in agarosegels bytransfer todiazobenzyloxymethyl-paperand hybrid-ization with DNAprobes.Proc.Natl.Acad. Sci. U.S.A. 74:5350-5354.
2.Basilico, C., S. Gattoni, D. Zouzias, and G. Delia Valle.1979.Loss ofintegratedviral DNA sequences in polyoma transformedcells is associated withanactive viralAfunction.Cell 17:645-659.
3. Doerfler, W. 1977. Animal virus-host genome interac-tions.Compr. Virol.10:279-399.
4. Doerfler, W.,W.Hellmann,and A. K.Kleinschmidt. 1972.The DNA ofadenovirus type12and its denatur-ationpattern. Virology47:507-512.
5. Doerfler, W., S. Stabel, H.Ibelgaufts,D. Sutter,RI Neumann, J.Groneberg, K. IL Scheidtmann, R. Deuring, andU.Winterhoff. 1979.Selectivityin in-tegration sites ofadenoviral DNA. ColdSpringHarbor Symp.Quant. Biol. 44:551-564.
6.Fanning, E.,and W.Doerfler.1976.Intracellularforms ofadenovirusDNA. V.Viral DNAsequencesin hamster cells abortivelyinfected andtransformedwith human adenovirus type 12. J.Virol. 20:373-383.
7. Groneberg, J., andW.Doerfler. 1979.Revertants of adenovirustype12-transformedhamstercells have lost partof the viral genomes. Int. J. Cancer 24:67-74. 8.Groneberg, J., D.Sutter,H.Soboll, andW.Doerfler.
1978.Morphologicalrevertantsof-adenovirus type 12-transformed hamster cells. J. Gen.Virology40:635-645. 9. Mantei, N., W. Boll, andC.Weissmann.1979. Rabbit ,8-globin mRNA production in mouse L cells trans-formed with cloned rabbit,-globin chromosomalDNA. Nature(London)281:40-46.
10. Ortin, J.,K. H.Scheidtnunn,RIGreenberg,ML West-phal, and W.Doerfier. 1976. Transcription of the genome of adenovirus type 12. HI.Maps of stable RNA fromproductivelyinfectedhumancelLsand abortively infected and transformed hamster cells. J. Virol. 20: 355-372.
11.Rigby,P. W.J.,ML Dieckmann,C.Rhodes,and P. Berg. 1977. Labeling deoxyribonucleic acid to high specific activityin vitrobynicktranslationwithDNA polymeraseI. J.Mol. Biol. 113:237-251.
12. Southern, E. M. 1975. Detection ofspecificsequences among DNAfragments separated by gel electrophore-sis. J. Mol.Biol.98:503-517.
13. Stabel, S.,W.Doerfler,and R.RIFriis.1980. Integra-tion sites ofadenovirustype12 DNA in transformed hamstercellsand hamster tumor cells. J. Virol. 36:22-40.
14. Steinberg, B.,R.Pollack,W.Topp,andM.Botchan. 1978.Isolationandcharacterizationof T antigen-nega-tive revertants from a line of bansformed rat cells containingonecopyofthe SV40genome.Cell 13:19-32.
15. Sutter,D., andW.Doerfler.1980.Methylationof inte-grated adenovirus type 12 DNA sequences in trans-formed cells is inversely correlated with viral gene expression.Proc.Natl.Acad.Sci.U.S.A. 77:253-256. 16. Sutter,D.,M.Westphal,andW. Doerfier. 1978.
Pat-terns ofintegration ofviral DNA sequences in the genomes ofadenovirus type 12-transformed hamster cells. Cell 14:569-585.
17.Vardimon,L,ILNeumann,L.Kuhmann,D.Sutter,
andW. Doerfler.1980. DNAmethylation and viral gene expression in adenovirus-transformed and -in-fectedcelis.NucleicAcidsRes. 8:2461-2473. 18.Wahl,G. M.,M.Stern,andG.R.Stark.1979.Efficient
transfer oflargeDNAfragmentsfromagarosegelsto
diazobenzyloxymethyl-paperandrapidhybridizationby using dextran sulfate. Proc.Natl.Acad.Sci. U.S.A.76: 3683-3687.
19. Wigler,M.,R.Sweet, G.K.Sinn,B.Wold,A.Pellicer, E.Lacy,T.Maniatis, S.Silverstein,and R.Axel. 1979. Transformation ofmammalian cells with genes fromprocaryotes.Cell 16:777-785.
49