JOURNAL OF VIROLOGY, Mar. 1977, p. 1019-1030 Copyright© 1977 AmericanSociety for Microbiology
Vol.21,No.3
Printed in U.S.A.
Early Gene Expression of Adenovirus
Type
2:
R-Loop
Mapping of mRNA and Time Course of Viral DNA, mRNA,
and
Protein
Synthesis
PAUL D. NEUWALD,' JURG MEYER,2 JACOB V. MAIZEL, JR., AND HEINER WESTPAHL* Laboratory of Molecular Genetics, National Institute of Child Health andHumanDevelopment, Bethesda,
Maryland20014
Received for publication20July 1976
Adenovirus type 2 DNA was hybridized to early mRNA isolated from the cytoplasm ofinfected cellsprior totheinitiationof viral DNA synthesis.
Result-ing Rloops were visualized in the electronmicroscope, andtheir positions were
oriented with the help of DNA fragments generated by digestion with the
restriction endonuclease Bam HI. Early RNA was found to map (in order of
relative R-loop frequency) with midpoints near positions 0.95, 0.80, 0.03, 0.65,
and 0.09 on the conventional adenovirus map. The time of appearance of
individual viral mRNA's was comparedto the time course of viral protein and
DNAsynthesis. Wepresent a refined map ofadenovirusgenefunctions which is basedonresultsdocumented inthisand the accompanying study by Meyer et al.
(1977), aswell as ondata publishedby otherlaboratories.
Theintricatecontrol systemof initial
adeno-virus type 2 (Ad2) gene expression assigns a
number ofearly viral functions to individual classes of mRNA, transcribed from distinct
re-gionsof the genome (5, 7) which comprise
non-contiguous parts of either viral DNA strand
(32, 38). At least six polypeptides of different
electrophoretic mobilities, specified by these
RNAs, have been observed in extracts of
in-fected cells (4, 16, 19, 36, 37, 44) or incell-free
translations(2, 10, 26, 36, 37). Lewis etal. (26)
allocated these early viral gene functions to specific regions of the Ad2 genomeidentifiedby
restrictionendonuclease cleavage.
Uponentering apermissive cell,the
adenovi-rusneedstobring the host'sregulatorysystem underits owncontrol sothat viraland cellular functions can cooperate in virus gene
expres-sion. The early Ad2 polypeptides observed by
the various laboratories are integrated inthis
processinsome asyetunknownmanner. They
are commonly listed according to molecular weight. The 72,000 molecular weight polypep-tide, seen first by Anderson etal. (1) in
Ad2-infected cells, is the polypeptide subunit (26)
ofa single-stranded DNA-binding protein (42) which is implicated in DNA replication (21,
43). At least five additionalearly virus-specific
'Presentaddress:Biological Markers Program, Freder-ick CancerResearch Center, P.O. BoxB, Frederick, MD 21701.
2Present address: Biozentrum der Universitat Basel, AbteilungMikrobiologie, CH-4056Basel,Switzerland.
polypeptides
(seeTable 2)have been described inthe literature, withmolecular weightsat ornear 50,000, 19,000, 17,000, 14,000, and 11,000.
Each of thesepolypeptides qualifiesas anearly viral geneproduct encodedin one of the early
regions of the Ad2 genome (26). The 19,000
molecular weightcomponent is a
glycopolypep-tide (19). Some of the early polypeptides are
enrichedinnuclear, othersincytoplasmic
frac-tionsoftheinfectedcell (see Table 3).
A subset ofearly viral RNA found in Ad2-transformed cells (15) maycodefor early viral functions involved in the process of virus-in-duced cell transformation. This RNA
repre-sents several early species, ofwhichthose
de-rived from theleft end of the viralgenome (12) aresufficientto elicit cell transformation (14). RNA transcribed from the left end of the ge-nomeduring lytic infection givesrise invitro to twopolypeptides with molecular weights near
50,000 and 14,000 (26). Virus-specific
polypep-tides ofcorresponding molecular
weights
have notyetbeendetected within transformed cells, whereas, in several established lines ofAd2-transformedcells, the 72,000 molecular weight
polypeptide has been unequivocally identified
(22). The functions ofearly virus polypeptides
during lytic infection or in the process of cell transformationremain to beelucidated.
Inthis report, we deal with some
quantita-tiveaspects ofearly Ad2 gene expression.
Us-ing the R-looptechnique ofWhiteandHogness
(personal communication) we visualized
indi-1019
on November 10, 2019 by guest
http://jvi.asm.org/
1020 NEUWALD ET AL.
vidualclassesofearly mRNA
hybridized
with Ad2DNA,established
their mappositions,
andcompared their relative
frequencies
atvarioustimes after infection. At the same
time,
weexaminedthe timecourseof invivosynthesisof earlyandlate Ad2
polypeptides.
MATERIALS AND METHODS
Cells and virus stocks. The propagation and infec-tion ofKBcells with purified virus is describedin
theaccompanyingarticle (28).
Purification and electron microscopy of early
Ad2 mRNA.Poly(A)-containingRNA wasextracted atvarious times after infection from the cytoplasm of cells incubated inthe presence ofcycloheximide
(10 ,ug/ml) aspreviouslyreported (9, 11, 28).
Thehybridization ofearly Ad2 mRNA with Ad2
DNA and thevisualizationof Rloops in theelectron
microscopearedescribedinthe accompanying
arti-cle (28). In all of the hybridizationsofthis report,
theDNAconcentration was10 ,ug/ml and the RNA
concentration was 100 ,tg/ml. The use of higher
RNAconcentrationswasimpracticalsinceextensive
RNAbackgroundinterfered with the visualization
of Rloops.
In vivo labeling of proteins and preparation of cell extracts. ThepolypeptidesofAd2-infected cells were labeled with [35S]methionine as follows. At
varioustimes afterinfection, aliquotsof6x 106cells
wereremoved from both infected and mockcultures. The cells were sedimented by centrifugation,
washed once with 10 ml ofmethionine-freemedium,
and resuspended in 10 ml of methionine-free medium to which was added 0.3 mCi of L
[35S]methionine (233 Ci/mmol, New England
Nu-clear). The cells werelabeled for1 hat37°C,
sedi-mented bycentrifugation,washed once with 10 ml of
phosphate-bufferedsaline(PBS), andsuspendedin1 ml of 0. 15MKCl,5mMMgCl2,3mMCaCl2,0.1mM EDTA, 0.01 MTris-hydrochloride(pH 7.5). Thecells werelysed andfractionatedasdescribedpreviously
(9), andthe nuclei wereresuspendedin 1mlofPBS.
Thesupernatantcytoplasm was clarified by a second
centrifugation at 12,000 x g for 5 min. Both the
nuclear andcytoplasmic fractionswerediluted with 1 volume of2 x electrophoresis sample buffer (1x samplebuffer= 1% [wt/vol]sodiumdodecylsulfate
[SDS], 1% [vol/vol]
f8-mercaptoethanol,
10% [vol/vol]glycerol,0.01% [wt/vol]bromophenolblue, 0.05
MTris-hydrochloride[pH6.8]),boiled for2min, and
appliedto 13% SDS-polyacrylamidegels for electro-phoresis (9, 11). Quantitationofindividualproteins was obtained bycutting the bands outofthe dried gel andcountingthe gelslicesin 10mlofFilter-Solv
liquid scintillation fluid (Beckman). Radioactivity
inthecorrespondingregion of themock-infectedcell
sample wassubtracted from the counts in each pro-teinband cut out of theinfected cell sample. The use
J. VIROL.
of
long
(26cm) slabgels
allowedsuperior separationoftheindividualproteinbands and thusfacilitated
quantitation
by
thismethod.TimecourseofDNAsynthesis.Theappearanceof
Ad2-specific
DNA was determinedby
DNA:DNAhybridization
on nitrocellulose membranes. KBcellswereinfectedasdescribed (28),and10-ml ali-quots
containing
3 x 106cells wereremoved every hour from 0to9hafterinfection,
andagain
at25h afterinfection,
andlabeled with 50 ,uCi of[methyl-3H]thymidine
(12Ci/mmol,
Schwarz-Mann) for 30 min at370C.
Thelabeled cellswere sedimentedby
centrifugation,
suspended in 1 ml of 0.3 N NaOH,and
placed
inboiling
waterfor 10 min. Thelysate
wascooledto00C
andneutralized with1ml of0.3 NHCl,
0.01MTris-hydrochloride
(pH
7.5). DNA wasextractedtwice withan
equal
volume ofphenol (pH
7.5),precipitated
with 2.5 volumes of ethanol at-20'C,
suspended
in200 1.d of1.5 mMNaCl,
0.15 mMsodium citrate(pH
6.8), heated for 15min at1000C,
rapidly
cooledto0C,
andhybridized
with3 ,I.g ofpurified
Ad2 DNA bound to nitrocellulosefilters (17).
Radioactivity
inhybrids,
measured atgiven
timepoints,
wasnormalizedtothe total DNA in eachsample
to correctfor minorvariations in theamountofDNA addedtoeachfilter.
RESULTS
Visualization
andmapping
ofRloops
gen-erated
by early
mRNAinAd2DNAmolecules.In thefirst sectionofthis
report
wedescribed
the
visualization
ofearly
Ad2 mRNA inviral DNA:RNAhybrids. Figure
1depicts
full-sized
Ad2 DNA moleculescarrying
severalloops
whichweregenerated
by
Ad2 mRNAannealing
to
antiparallel
regions
ofthedouble-stranded
DNA andthereby displacing
thehomologous
DNA sequences. One branch of each Rloop
thusformed containeddouble-stranded
nucleic acid(thick
contour);
the otherbranch containedsingle-stranded
nucleic acid(thin contour).
RNA which didnotparticipate
inthe hydridi-zation reactiondisplayed
ahighly
condensedsecondary
structureseenespecially
intheback-ground
oftheupperelectronmicrograph.
TheRNA used in this series of
experiments
wassynthesized
in human cellslyrically
in-fectedwitha
high
dose of Ad2. We foundthat,
underourconditionsof
infection,
theindividual
stages
ofthelytic cycle
werebothsynchronized
and
condensed,
as evidencedby
the fact thatDNA
synthesis
(seeFig.
8, top
panel,
andTable4) occurred earlier and at a more
rapid
ratethanwas
reported previously
(40).
Thisallowedfor a more accurate assessment of the time
FIG. 1. WholeAd2 DNAcontainingRloops formed byhybridizationto RNAisolatedfromAd2-infected cells 8 h after infection. DNA (0.1 pg) andRNA (1.0 Pg) were incubated in 10
pi
ofR-loop buffer at 52°C for 4 days, and examined in the electron microscope. The bars indicate 1jim.
Abbreviations:ds, double-strandedsideofloop; ss,single-strandedsideof loop.
on November 10, 2019 by guest
http://jvi.asm.org/
* 'h1 ', @ '
S J * * 7.. J t *
.7 .4 *
t'It * - I*I*,,g
I
A.4-H i'- *.-. l j ;. . *
- ._'t'1 -,P ~ $5
f
J.A
- D1*f-i.'
*'
'r
;%~
\)-
Is** . , *s!
. .*4 .
,
..4
.
!
l
4'~~~~~~~
''S
'X~
I,f
II|,
*' ' *''X l1.,
e/
''"
3*'L
r
.'
.
4?
>
;
'1I~~~12
on November 10, 2019 by guest
http://jvi.asm.org/
1022 NEUWALD ET AL.
course of Ad2 genetic expression. Since early
and late mRNA synthesis followed each other
within a narrow time span in cells infected in
this manner, wehadtotake properprecautions
to obtain early RNA preparations that were
free ofcontaminating late RNA. We chose cy-cloheximide as an inhibitor of late viral RNA synthesis(18)becausenoquantitativeor
quali-tativeinfluenceof thisdrugonearly viralRNA
synthesis wasnoticed inprevious experiments (6, 16, 26).
RNA was extracted from the cytoplasm at
different times after infection, and purifiedon oligo(dT)-cellulose (3)to selectfor poly(A)-con-taining Ad2 mRNA (33). R loops were gener-ated under conditions defined previously (28,
46) and examined in the electron microscope.
Figure2summarizes observations ofAd2 DNA
molecules carrying early Ad2 mRNA extracted
at2, 4, 6,or8hafter infection. The histograms
show the average size, position, andfrequency
of Rloops. Moleculeswithmultiple loops facili-tated theorientationof thevarious Rloops with
respect to each other. Molecules containing a
single loop were aligned such that the R loop wascoincidentwith one of the positions onthe multilooped molecules. For example, the
left-most R loop (midpoint near position 0.03) was
closertotheend ofthe DNAmoleculethanwas
the right-most loop (midpoint near position
0.95). The orientation with respect to the
con-ventionalAd2 DNA map wasbased upon
exper-iments which will be described below, and
which confirmed the results obtainedinFig. 2.
Equal amounts ofcytoplasmic RNA were used
for each of the four hybridizations. Thus, the histograms show the relative amounts of each species of early Ad2 mRNA contained in the
cell at the indicated times after infection. As
expected, the R loops occurredin theearly
re-gions (32, 38)of the Ad2 DNA, and mapped, in
orderof relativefrequency, nearpositions 0.95,
0.80, 0.03, 0.65, and 0.09. These positions refer
to the midpoints ofeach histogram peak dis-playedinFig. 2. At veryearly times (2 h after infection), R loops were infrequently observed
between positions0.5 and0.6 (e.g., Fig. 2, top
panel). Since these loopsarelocatedinaregion
codingforhexon mRNA (28), the significance of
thisfinding is unknown.
Withinthe areas of the major loops, i.e., at
positions 0.95, 0.80, and 0.03, the quantity of
earlyRNAappearedtoincreasesteadily during
the first 6hofinfection, whereas surprisingly
fewloops formedatposition 0.65, which
corre-spondstothe RNAcodingfor the 72,000
molec-ularweightpolypeptide (26),anearly Ad2
poly-peptide givingriseto averyprominentbandon
1001
80 2 Hr P.I.
60-40L
80r 4HrP. I.
60~
-uj 40 D
uJ
-i
201-0
U- 0
0
o 6 rP.1
LU
LUI
60-0 0.2 0.4 0.6 0.8
FRACTIONAL LENGTH
FIG. 2. Timecourseof Ad2 mRNA production in
infected cells as determined by R-loop analysis.
Poly(A)-containing RNA wasisolated from infected
cellsatvarious timesafter infection. Each RNA
prep-arationwashybridizedtowhole Ad2 DNA for 16 hat 520C.Hybrid molecules were examined in the
elec-tron microscope, andthepositions of R loops were
determined. Thecombined datafor 50 molecules at eachtimepointarepresentedin theform ofa
histo-gram.Theabscissa shows the fractional length of the
adenovirusgenome,andthe ordinate gives the
per-centageofDNA molecules containing anR loopat thatposition.
SDS-polyacrylamide gels (i.e., Fig. 7). This
finding prompted us to repeat part of the
ex-periment, allowing more time for RNA:DNA
hybridizationtooccur.Andindeed, Fig. 3 shows
that,under theseconditions, the 8-hRNA,but
notthe 2-hRNA,formedaprominent peakof R
loopsatposition0.65. We cannotdecide, onthe
1.0 J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.501.283.439.58.452.2]MAPPING OF EARLY Ad2 mRNA 1023
basis of this experiment, whether the RNA specific for position 0.65 was in short supply
even 8hafterinfection or whether the higher
G+C content of DNA in thatregion of the
ge-nome (8, 29)causedadelay (28, 41) intherate
of DNA:RNA hybridization. The experiment
did show, however, that significantly more of
thisRNA was present at 8 h thanat 2 hafter
infection.
Orientation of R-loop positionsto the con-ventional Ad2 map. Cuts introduced by the
restriction endonucleaseBamHIinto Ad2 DNA
were utilized as markers for the orientation of
R loops. The enzyme cleaves at positions 0.3,
0.43, and 0.6 of theconventionalAd2 DNA map
(Mulder and Greene, personal communication), therebygeneratingfourfragments in theorder
B,
D, C, A. The lengthdistribution of allfrag-ments contained in the complete Ad2 DNA
di-gest is shown in Fig. 4. The two larger
frag-ments, A and B, corresponding to the ends of
the molecule, showed distinct length distribu-tionsand thuscould easilybeidentifiedin elec-tron micrographs of a mixture of fragments. This task was more difficult with the small internal fragments, C and D, which displayed overlappinglength distributions.We were nev-ertheless ableto arrive at a correctly oriented mapofRloops generated by early mRNA,since wecould correlateRloopsindistinctfragments towhole Ad2DNAmoleculescarryingmultiple loops. The electron micrograph of Fig. 5depicts aBamHI A-fragment containing twoR loops.
In Fig. 6, line drawings offragmented DNA
carryingRloops have beenarrangedinsucha waythat they reflect the distribution of loopsin
intact DNA. This methodof R-loop orientation
was used to determine the accuracy of
subjec-tive orientation based upon the position of a
100
2Hr RNA
80 16HrHYB
8Hr RNA 16 Hr HYB
60 I
-i
40--i20
0
0
2802HrRNA 8HrRNA
Z 4DayHYB ayHYB
60.
40iIll
-
11~i1
h.0 0.2 0.4 0.6 0.8 1.00 0.2 FRACTIONAL LENGTH
0.4 0.6 0.8 1.0
FIG. 3. R loops generated by Ad2 early mRNA after differentperiods of hybridization. Poly(A)-con-taining RNA wasprepared from infectedcells at2 and8 hafter infection,andhybridizedtoAd2 DNA at52Cfor16hor4days (96 h).
70
C50-°z,
~40
0
~30
z~~~~~~~
10-1 2 3 4 5 6
LENGTHINMICROMETERS
FIG. 4. Length distribution of fragments gener-ated by digestion of whole Ad2 DNA with BamII restriction endonuclease. BamHI (0.4 unit, Be-thesda Research Laboratories, Inc.) was added to 3
Mg
of Ad2 DNA in10pJ
of 6 mMMgCl2, 6 mM,3-mercaptoethanol, 6mMTris-hydrochloride (pH 7.5), and incubated for 2 h at 370C (20). The digested DNA wasdiluted into R-loop buffer and examined in the electron microscope. Electron micrographs of 437 BamHIfragments were measured and the distribu-tion waspresented as a histogram. Since the proba-bilityof larger fragments extending off the electron micrograph, and thus not beingscored, was greater than that of smaller fragments, the number counted wasinversely proportional to the size offragments.
single loopinrelationto oneof the positionson a multilooped molecule (Table 1). Maps ofR loops were established either with the help of
specific
fragments of DNA carrying Rloops
(Fig. 6) or with the help of intact molecules
containing multiple Rloops (Fig. 2). Ineither
case,theratioofRloops occurringtothe left of
position0.07andtotheright of position0.9was
close to 1:2 in hybridizations using RNA ex-tracted 2 h after infection, and 1:1 in those
using RNA extracted8hafter infection.
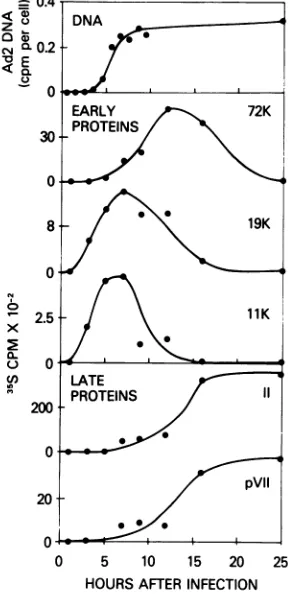
Timecourseof Ad2polypeptidesynthesisin
vivo. Inaneffort tocorrelate thetime courseof early Ad2 RNA synthesis to that of the
corre-sponding
viralgeneproducts,
cellswerelabeled with [35S]methionine atvarious timesafter in-fection, and thepolypeptides containedinfrac-tionated cell extracts wereexamined.
Autoradi-ograms of35S-labeled polypeptides resolved by
SDS-polyacrylamide gel electrophoresis are
shown in Fig. 7. Five of theearly
Ad2-specific
polypeptides-of 72,000, 19,000, 17,000, 14,000
and 11,000 molecular
weight-could clearly
bediscerned from the
background
of host cellpoly-peptides present at the initial stages of
infec-0 u-p ...
VOL. 21, 1977
D
c
I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.501.276.441.51.259.2] [image:5.501.55.245.477.611.2]1024 NEUWALD ET AL.
At X
e,'-,,- , Cb : e a
-.. k. H- - "'.
FIG. 5. BamHIA-fragment containingtwoRloops formed by hybridizationwithRNA isolatedfromcells2 hafter infection with Ad2. The DNAwasdigested withBamHIenzyme asdescribed in thelegendtoFig. 4, andhybridizedtotheRNAfor16hat520C. The barindicates1 ,um.
B D C A
0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0
;
I-0
z
0
co
FIG. 6. PositionsofRloopsonBamHIfragments
ofAd2 DNA as described inFig.5. Allfragments
containing one or more loops were photographed,
andthepositionsoftheRloopsweredetermined. The
fractional length ofthe conventional Ad2 map
cov-ered by each fragment is given at the top ofeach column. Upperset:Rloops formed by hybridization to RNA isolated 2 h after infection. Lower set: R loopsformed by hybridization toRNA isolated8 h afterinfection.
tion. Some of these virus-specific polypeptides
were abundant in the nucleus, others in the
cytoplasm.Asurveyofearlyvirus-specific
poly-peptides described in the literature (Table 2)
shows that, in addition to the early products
seen in Fig. 7, one additional polypeptide,
around40,000to50,000 molecular weight, was
commonly observed by other laboratories. The
highbackground of host cell polypeptide bands
intheareaof the gelcorrespondingtoaproduct
ofthis sizemayberesponsible forourfailureto
TABLE 1. Analysis of endloops
Timeafter . Bamfragmentsa Whole DNAb
Timeaftern
Positionofinfection N. Fe o
(h) loop No Fre- No
Fre-quency quency
2 0-0.07 24 0.36 22 0.37
0.9-1.0 42 0.64 38 0.63
8 0-0.07 38 0.49 23 0.48
0.9-1.0 40 0.51 25 0.52
aAsdescribedinFig.6.
bWhole Ad2 DNA hybridized to cytoplasmic mRNA at520C for 16 h and oriented according to the relative positions ofmultiple loops occurring within individual molecules (Fig. 2).
detect the 40,000 to 50,000 molecular weight
polypeptide. Polypeptides of comparable
elec-trophoretic mobilitieswereobserved ininfected
cells pretreated with 25pug of cycloheximideper
ml (16). Thedistribution of early virus-specific
polypeptides among subcellularfractions (Fig.
7)compared well with similar data in the
litera-ture (Table 3). Minor variations were most
likely dueto differences in the methods of cell
fractionation used by the individual groups of
investigators.
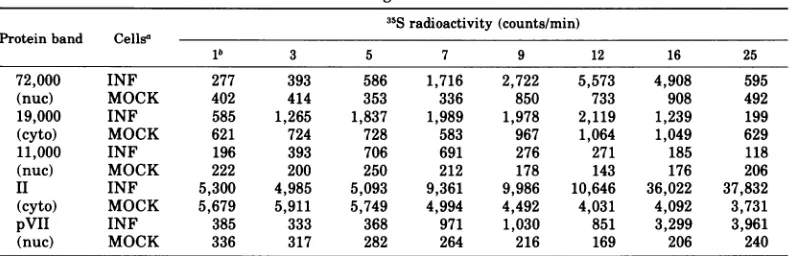
To quantitate our observations, we
deter-mined theradioactivity containedinindividual
polypeptide bands, subtracted the correspond-ing mock values, and related these data to the time course of Ad2 DNA synthesis in the in-fected cells. Time points between 7 and 25 h
after infection were taken from a gel (not
shown) similar to that ofFig. 7. Only
virus-specific polypeptides that could be cleanly cut
fromthe gels were utilized in thisanalysis. The
results (Fig. 8 and Table 4), indicated that
19,000 and 11,000 molecular weight
polypep-tides wereamong the earliest of the
virus-spe-J.- VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.501.77.444.72.223.2] [image:6.501.59.254.269.448.2] [image:6.501.262.458.273.385.2]MAPPING OF EARLY Ad2 mRNA
1025
NUCLEUS
CYTOPLASM
INF
1 3 5 7 24 1
MOCK
3 5 7 24
INF MOCK
1 3 5 7 24 1 3 5 7 24 HRS
_
w-iiAr
a_.
11.5K loo
FIG. 7. SDS-polyacrylamide gel electrophoresis of nuclear and cytoplasmic polypeptides synthesized at various times after infection.Ad2-infectedormock-infected KB cellswerepulse-labeledwith[35S]methionine
for1h priortoharvesting. Cellswerefractionated into nuclear and cytoplasmicfractions and analyzedon13%
polyacrylamide slab gels. Sampling times (hours after infection) are shown at the top of each column.-Apparent molecular weights (calibrated by co-electrophoresis of the24-hINFsample with known marker proteins) aregiven in thecenter, with virion polypeptides (24) showninparentheses. Abbreivations:INF, samples preparedfrom Ad2-infectedcells; MOCK, samples prepared from mock-infectedcells.
TABLE 2. Comparison of early virus-specific polypeptides observed by various groupsofinvestigators Results of Walter and Chin and Saborio etal. (37) SaborioandOberg (36) Atkins etal.
this study Maizel (44) Maizel (4) (2) (invitro)
(in vivo) (in vivo) (in vivo) Invivo In vitro Invivo Invitro
72a 70 70 70 70 74 74 72b
67 67
60 60
45 45 44-55b
42 42-50 42
40
35 35
26.5
19 19 19 19 19 19 19 19b
18.5 18.5
17 17 17 17 17.5 17.5
14.5-16 15.5 15.5b
15 15b
14 14 14.5 14.5
12.5 12.5
11 10 11 10 10 10.5 10.5 11
aMolecularweight x103.
bShowntobevirus-coded byLewis etal. (26). VOL. 21, 1977
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.501.47.444.63.359.2] [image:7.501.47.444.457.640.2]TABLE 3. Intracellular location ofearly viral-specific polypeptides
Location Polypeptide (mol
wt x 103)a Results of this Walter and Mai- Chin and Ma- Saborio et al. Saborio and
Oh-study zel(44) izel (4) (37) erg(36)
72 N(C)b C N,C C C
40-50 C C C
19 C(N) C C C(N) C(N)
17 C C C
14 C C C
11 N N N N N
aApparent molecularweightsvaryslightlyamongobservationsbydifferent laboratories.
bAbbreviations: N,polypeptidefound innucleus; C,polypeptidefoundincytoplasm.Lettersin
parenthe-sesindicate location ofaminorfraction ofpolypeptide.
= 0.4
0a
z L0
CNcl0.2 E
< a
30j
0
0.
C,) (n
qn
2.5
0
200
0
20-0 5 10 15 20 25
HOURS AFTER INFECTION
FIG. 8. Time ofappearance ofAd2-specific
pro-teins and of viral DNA in infected cells. DNA synthe-sis was measured by DNA:DNA hybridization on
nitrocellulose filters as described in Materials and
Methods. Levels of both early and late polypeptides synthesized ateach timepoint were determined by
scintillation counting of 35S radioactivity contained in individual polypeptide bands of the SDS-poly-acrylamide gels. Radioactivitypresent in the same
regions of the corresponding mock samplewas
sub-tracted from each value and the net amount was
plotted. Totalradioactivity recovered from the
appro-priate regionsofeach gelwasasshown in Table 4.
cific polypeptides, followed by the 72,000
mo-lecular weight polypeptide, which appeared
concomitant with the start of Ad2 DNA
repli-cation. Polypeptides II (hexon) and pVII, both
late Ad2 gene products located in the virion particle, appeared within a few hours after the
onset of Ad2 DNA synthesis, and continued
tobe produced while the rate of early
polypep-tide synthesis decreased. Thehighbackground
ofhost cell polypeptides present in the 17,000
and14,000 molecular weight regions of thegels
prevented quantitation of these early,
virus-specific polypeptides. Distinct bands were not observed at these locations in the
autoradi-ograms until5 to 7 h after infection. However,
thesepolypeptides may exist prior to that time
atlevels undetectablebyourassay method.
DISCUSSION
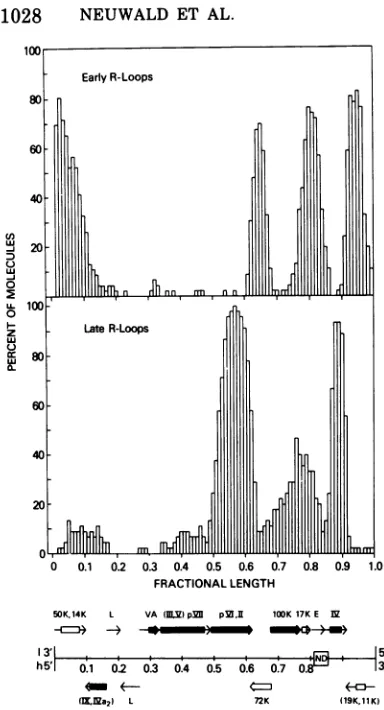
Agenetic map ofadenovirustype 2 may be
constructed from the data contained in this
study, inthe accompanyingreport (28), andin
thepublished accounts of otherlaboratories. In
Fig. 9, we present two histograms which are representative for the spectrum ofRloops
ob-servedinhybridizations of Ad2 DNA with early
orlate Ad2 mRNA. Below thesehistograms we
have drawn a map of the genetic elements of
Ad2 which, in our opinion, represents the best
fit ofall available data. Transcriptional units
are drawn as arrows. Their extent and
posi-tionsaredefined by the sizes and positions of R
loops, bythemap positions of strand switches of transcriptional units (28, 32, 38), by the map positions of certain size classes of early RNA (5, 7), and by the map positions of the special
markers VA and ND, i.e., thevirus-associated
RNAs (27, 30, 31, 34, 35, 39, 45), andthe Ad2
DNA sequences deleted in nondefective (ND)
Ad2-SV40 hybrid viruses (see reference 28 for details). Bars within the arrows indicate the
minimumsequences needed to code for the
var-1026
NEUWALD ET AL. J. VIROL.on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.501.57.457.84.186.2] [image:8.501.81.225.230.528.2]MAPPING OF EARLY Ad2 mRNA 1027
TABLE 4. Totalradioactivity recovered from the appropriate regions of each gel mentioned in the legend to Fig. 8
35Sradioactivity (counts/min) Protein band Cellsa
lb 3 5 7 9 12 16 25
72,000 INF 277 393 586 1,716 2,722 5,573 4,908 595
(nuc) MOCK 402 414 353 336 850 733 908 492
19,000 INF 585 1,265 1,837 1,989 1,978 2,119 1,239 199
(cyto) MOCK 621 724 728 583 967 1,064 1,049 629
11,000 INF 196 393 706 691 276 271 185 118
(nuc) MOCK 222 200 250 212 178 143 176 206
II INF 5,300 4,985 5,093 9,361 9,986 10,646 36,022 37,832
(cyto) MOCK 5,679 5,911 5,749 4,994 4,492 4,031 4,092 3,731
pVII INF 385 333 368 971 1,030 851 3,299 3,961
(nuc) MOCK 336 317 282 264 216 169 206 240
aAbbreviations: INF, samples prepared from Ad2-infected cells;MOCK, samplesprepared from mock-infected cells.
bHoursafterinfection.
ious early and late polypeptides listed on the
map. Thesepolypeptides havepreviously been
allocatedtorestriction fragments of Ad2 DNA (2, 25,26), andthus we wereabletoalign them with the individualRloops.
LateAd2gene expression isdiscussedinthe
accompanying report (28). Here we will touch
on somefeaturesofearlyAd2 geneexpression.
The early region to the left of position 0.1 is of chiefinterest because of its primary
involve-ment in celltransformation (14). The
polypep-tidemostlikelyspecified bythemRNAforming the left-mostRloopisofuncertainsize.
Accord-ing tocell-freetranslation, itssize is near50,000
molecularweight (26). However,thecoding ca-pacityof the correspondingmRNA, judged, by the R-loop size,iscertainlygreater. It is
tempt-ing tospeculate thata58,000 molecularweight
polypeptide which is specific for
Ad2-trans-formedhamster cells(Chin, Lewis, and Maizel,
personalcommunication) isidenticaltothe pol-ypeptide seen by Lewis et al. (26), and thus specified bythe left-mostearlyAd2 mRNA.
According to Atkins et al. (2), the 14,000 molecularweightpolypeptidemapstothe
right
of the 50,000 molecular
weight
polypeptide.ThiscorrespondstotheR-loop peak observedat
position 0.09 intheupper
histogram
ofFig.
9.Although
Rloops
werenotobservedinthisre-gion of the DNA when hybridized with RNA isolated 2 hafter infection (Fig. 2 and 6), the limitations of the
R-loop
technique
inregard
toquantitation of individual mRNAspeciesdonot
allow us to ruleoutthepresence of this message
atveryearlytimes after infection.
The Rloopcorrespondingtothe well
charac-terized72,000 molecularweightpolypeptide
ap-pearedintheregion between position 0.61and
0.68, an area matching itsminimal codon size
andflankedbystrand switchpoints(28,32,38).
In keeping with the proposed function of this
early Ad2 geneproduct during Ad2 DNA
repli-cation (43), itmay be interesting to note, from
the results ofour study, that both the 72,000
molecular weight product and its mRNA
ap-peared concomitant with Ad2 DNA synthesis,
i.e., later thanthe earliest Ad2 polypeptides. A
similar result was obtained by Gilead et al.
(13).
Thethird early region of Ad2 mapped within
positions 0.77 to 0.84 and included the ND
dele-tion.Judging from the size of the corresponding
Rloop, a polypeptide of up to 80,000 molecular
weight could be encoded here. Yet, the only product known to map in this area of the
ge-nome is the 15,500 molecular weight
polypep-tideof Lewis et al. (26), which probably
corre-spondsto our17,000molecularweight polypep-tide.InourAd2 map, we have tentatively posi-tioned this product to the left of the ND deletion becauseitseemedunlikely to us that the 15,500
to 17,000 molecular weight product is encoded
byanareaof thegenome which is not required forlytic infectionofhuman cells in culture (23).
As yet, the presence of the 15,500 to 17,000
molecular weight polypeptide in Ad2+ND4 in-fected cells hasnotbeenreported.
A rather large and quite abundant R loop generated by early Ad2 mRNA waslocated to the right of position 0.9. The corresponding
19,000 and 11,000 molecular weight
polypep-tides (26) require only afraction of the coding capacity of an RNA of this size. We have no evidencethat morethan one RNA is
responsi-ble for theirsynthesis. Both polypeptides may,
infact, be breakdown products of some larger,
yetunstable, Ad2 geneproduct. The19,000 and
11,000 molecular weightpolypeptides appear to
be the earliest viral gene products synthesized
after infection.
VOL. 21, 1977
on November 10, 2019 by guest
http://jvi.asm.org/
1028 NEUWALD ET AL.
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7
FRACTIONAL LENGTH
0.8 0.9 1.0
50K,14K L VA(UIL)p3M pMI 1K 17K E
- _ )->
0.1 02
(1 j2)a, L
0.3 0.4 0.5 0.6 0.7 0.8
72K
5' 13
(-c1-1
(09K,1K)
FIG. 9. Comparison oftheR-loop patterns
gener-atedby early and late Ad2 RNA andproposed
ge-neticmapof adenovirustype2.Theupperhistogram
isrepresentative of early R loops and is derived from Fig. 3 (lower right panel). The lower histogram is representative ofmost late R loops and is derived from Fig. 6, panel three, of Meyeret al. (28). The
mapshown below thehistograms is baseduponthe
accumulateddata of several laboratories,andis dis-cussed in detail in thetext. Thepolarity of both the heavy (h) and light (1) strands of theDNA molecule is indicated (38).NDreferstotheregion ofdeletion of Ad2 DNA sequences in nondefective Ad2-SV40 hybrids. Thearrowsgive the direction of
transcrip-tion and size of mRNA species produced both early and lateininfection. Boxes insertedinthesearrows
indicate the minimal length of mRNA required to code for each polypeptide. The locations of boxes within arrows isarbitrary. Early (open boxes) and
late (closed boxes) polypeptides mostlikely encoded by these messages are indicated. Thegeneorder is unknownfor those polypeptides within parentheses. Abbreviations: L, unknown late products; E,
un-knownearly products. VAshows themapposition of
virus-associated, low-molecular-weight RNA.
Because of the differential kinetics of loop
formation in various regions of the Ad2
ge-nome, it is difficult to correlate the relative
frequencies ofR loops with the proportions of
mRNA's present ininfected cells(28).However,
in this report we have compared the
frequen-ciesofRloopsgenerated bymRNA'sisolatedat
various times after infection. In this case, we feel itisvalidto correlateR-loop
frequency
atany one position to the relative amount of
mRNA present at different times ofinfection,
since dissociation of the DNA duplex in this
regionwouldremain constantforeach
hybridi-zation. Thus, although one cannot
accurately
comparethe frequencies of mRNA'sproducing
Rloops at,for example,positions 0.65and0.95,
one cansaythat theRNAwhichgeneratesthe
R loop at position 0.65 is considerably more
abundant at 8hthan at2 hafter infection.
The R-loop technique certainly has not yet
been fully exploited by the experiments dis-cussed here. Further studies are expected to detect additional, less abundant classes of Ad2
mRNA. Itshouldalso be interesting to mapthe
Ad2 mRNA's present in various
Ad2-trans-formed cells, as well as the viral transcripts
isolated from nuclei of both virus-infected and
transformed cells. Since, at the same time,
work on the Ad2 polypeptides is expected to
reveal further details of thevarious viralgene functions, all thepiecesof the puzzlemay even-tually be elucidatedso as to construct a gratify-ing picture of the molecular genetics of Ad2 gene expression.
ACKNOWLEDGMENTS
The expert technical assistance of Sing Ping Lai and Margery Sullivan and the editorial assistance of Cecilia Levi aregratefullyacknowledged. We wishtothank
Wil-liam Chin and Andrew Lewis, Jr., Raymond White and David Hogness, andCarel Mulder and Ronni Greene for transmittingtheir results priortopublication. Wearealso grateful to Jose Saborio and Bo Oberg, and to Marjorie Thomas, Raymond White, and Ronald Davis for making their manuscripts available beforepublication.JurgMeyer was supported by a fellowship from the Swiss National Science Foundation andbyaPublic HealthService Interna-tional ResearchFellowship (no. F05TW2256-01).
LITERATURE CITED
1. Anderson, C. W., P. R. Baum,and R. F.Gesteland. 1973.Processing ofadenovirus 2-induced proteins.J.
Virol. 12:241-252.
2. Atkins, J. F., J. B. Lewis, C. W. Anderson, P. R.
Baum,and R. F.Gesteland.1975.Mapping of
adeno-virus 2genesby translationof RNAselectedby hy-bridization, p.293-298. In A. L.HaenniandG.Beard (ed.),InstitutNational delaSanteetde la Recherche
Mddicale Symposium no. 47. Editions INSERM, Paris.
3. Aviv, H.,and P. Leder. 1972.Purification ofbiologically activeglobin messengerRNAbychromatographyon
(n IL
C-0 0 z
0U
U
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:10.501.62.254.55.410.2]MAPPING OF EARLY Ad2 mRNA 1029
oligothymidylic acid-cellulose. Proc. Natl. Acad. Sci. U.S.A. 69:1408-1412.
4. Chin,W.W., and J. V. Maizel, Jr. 1976. The polypep-tides of adenovirus. VII. Further studies of early polypeptides in vivo and localization of E2 and E2A to the cellplasma membrane. Virology 71:518-530.
5. Craig, E. A., M. McGrogan, C. Mulder, and H. J. Raskas. 1975.Identification of early adenovirus type 2RNAspecies transcribed from the left-hand end of the genome. J. Virol. 16:905-912.
6. Craig, E. A., and H. J. Raskas. 1974. Effect of cyclohex-imide on RNA metabolism early in productive infec-tion withadenovirus 2. J. Virol. 14:26-32.
7. Craig, E. A., S. Zimmer, and H. J. Raskas. 1975. Anal-ysisof earlyadenovirus 2 RNA using Eco R * RI viral DNAfragments. J. Virol. 15:1202-1213.
8. Doerfler, W., and A. K.Kleinschmidt.1970. Denatura-tion pattern of the DNA ofadenovirus type 2 as determined by electron microscopy. J. Mol. Biol. 50:579-593.
9. Eron, L., R.Callahan, and H. Westphal. 1974. Cell-free synthesis ofadenovirus coat proteins. J. Biol. Chem. 249:6331-6338.
10. Eron, L., and H. Westphal. 1975. Cell-free translation products of early RNA from adenovirus type 2-in-fected KB cells, p. 299-304. In A. L. Haenni and G. Beard (ed.), Institut National de la Sante et de la Recherche M6dicale Symposium no. 47. Editions INSERM,Paris.
11. Eron, L., H. Westphal,and R. Callahan. 1974. In vitro synthesis of adenovirus core proteins. J. Virol. 14:375-383.
12. Flint, S. J., P. H. Gallimore, and P. A. Sharp. 1975. Comparison of viral RNA sequencesinadenovirus
2-transformed andlyricallyinfected cells. J. Mol. Biol. 96:47-68.
13. Gilead, Z., K. Sugawara, G. Shanmugam, and M.
Green.1976.Synthesis of theadenovirus-codedDNA bindingproteinininfected cells. J. Virol. 18:454-460.
14. Graham, F. L., A. J. vander Eb, and H.L.Heijneker. 1974.Sizeand location of the transforming regionin
human adenovirus type 5 DNA. Nature (London) 251:687-691.
15. Green, M., J. T. Parsons, M. Pifia, K. Fujinaga, H. Caffier,and I. Landgraf-Leurs. 1970.Transcription ofadenovirus genesin productively infected andin
transformed cells. Cold Spring HarborSymp. Quant. Biol.35:803-818.
16. Harter, M.L., G. Shanmugam, W. S. M.Wold, and M. Green.1976. Detectionof adenovirus type 2-induced earlypolypeptidesusingcycloheximidepretreatment toenhanced viral protein synthesis.J.Virol. 19:232-242.
17. Horwitz, M. S.1971.Intermediates in thesynthesisof type 2 adenovirus deoxyribonucleic acid. J. Virol. 8:675-683.
18. Horwitz, M. S., C. Brayton, and S. G. Baum. 1973. Synthesisof type2adenovirus DNAinthe presence ofcycloheximide.J.Virol. 11:544-551.
19. Ishibashi, M., and J. V. Maizel, Jr. 1974. The poly-peptidesofadenovirus. VI.Earlyand late glycopoly-peptides. Virology 58:345-361.
20. Ketner,G., andT.J.Kelly,Jr. 1976.Integratedsimian virus 40sequencesintransformed cellDNA:analysis using restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 73:1102-1106.
21. Levine, A.J., P.C. vanderVliet, B. Rosenwirth,J. Rabek, G.Frenkel, andM.Ensinger.1974. Adenovi-rus-infected, cell-specific, DNA-binding proteins. Cold Spring HarborSymp. Quant. Biol. 39:559-566.
22. Levinson,A.,A.J.Levine,S.Anderson,M.Osborn,B.
Rosenwirth, and K. Weber. 1976. The relationship between group Cadenovirus tumor antigen and the adenovirus single-strand DNA-binding protein. Cell 7:575-584.
23. Lewis, A. M., Jr., M. J. Levin, W. H. Wiese, C. S. Crumpacker, and P. H. Henry. 1969. A nondefective (competent) adenovirus-SV40 hybrid isolated from the ad2-SV40 hybrid population. Proc. Natl. Acad. Sci. U.S.A. 63:1128-1135.
24. Lewis, J. B.,C. W. Anderson, J. F. Atkins, and R. F. Gesteland. 1974. The origin and destiny of adenovirus proteins. Cold Spring Harbor Symp. Quant. Biol. 39:581-590.
25. Lewis, J. B., J. F. Atkins, C. W. Anderson, P. R. Baum,and R. F. Gesteland. 1975. Mapping of late adenovirus genes by cell-free translation of RNA se-lected by hybridization to specific DNAfragments. Proc. Natl.Acad.Sci. U.S.A. 72:1344-1348. 26. Lewis, J. B., J. F. Atkins, P. R.Baum, R. Solem, R. F.
Gesteland, and C. W. Anderson. 1976. Locationand identification of the genes for adenovirus type 2 early polypeptides. Cell 7:141-151.
27. Mathews, M. B. 1975. Genes for VA-RNAinadenovirus 2. Cell 6:223-229.
28. Meyer, J., P.D. Neuwald, S. P. Lai, J.V.Maizel,Jr., and H. Westphal. 1977. Electron microscopy of late adenovirus type 2 mRNA hybridized to double-stranded viral DNA. J. Virol. 21:1010-1018.
29. Mulder, C., J. R. Arrand, H. Delius, W. Keller, U. Pettersson, R. J. Roberts, and P. A. Sharp. 1974. Cleavage maps of DNA from adenovirus types2and5
by restriction endonucleases Eco RI and Hpa I. Cold Spring Harbor Symp. Quant. Biol. 39:397-400.
30. Ohe, K. 1972. Virus-coded origin of a low molecular weight RNA from KB cellsinfected with adenovirus 2.Virology 47:726-733.
31. Pettersson, U., and L. Philipson. 1975. Location of sequences ontheadenovirus genomecoding for the 5.5S RNA. Cell 6:1-4.
32. Pettersson, U., C. Tibbetts, and L. Philipson. 1976.
Hybridization maps of early and late messenger RNA sequences onthe adenovirus type2genome.J. Mol. Biol. 101:479-501.
33. Philipson, L., R. Wall, G. Glickman, and J.E.Darnell.
1971.Addition ofpolyadenylatesequences to virus-specific RNA during adenovirus replication. Proc. Natl.Acad.Sci. U.S.A. 68:2806-2809.
34. Raska, K., L. M. Sehulster, and F. Varricchio. 1976.
Three new virus-specific low molecularweight RNAs
inadenovirus type2infectedKBcells. Biochem. Bio-phys. Res. Commun. 69:79-84.
35. Reich, P. R.,B.G. Forget, S. W. Weissman, and J.A.
Rose.1966.RNA of low molecularweightinKBcells infected with adenovirus type2.J. Mol. Biol. 17:428-439.
36. Saborio,J., and B. Oberg. 1976.In vivoand invitro synthesis of adenovirus type2earlyproteins. J. Vi-rol. 17:865-875.
37. Saborio,J., B.Oberg,and L.Philipson.1975.In vitro synthesisofadenovirustype2early proteins,p.
325-330. In A. L. Haenni and G. Beard (ed.), Institut National de la Sante et de la Recherche M6dicale Symposiumno. 47.EditionsINSERM,Paris. 38. Sharp, P. A., P. H. Gallimore, and S.J. Flint. 1974.
Mapping of adenovirus2RNA sequencesinlyrically infected cells andtransformed cell lines. Cold Spring HarborSymp. Quant. Biol.39:457-474.
39. Soderlund,H., U.Pettersson, B.Vennstrom,L.Philip. son, and M. B. Mathews. 1976. A new species of
virus-coded low molecular weight RNA from cells infected with adenovirus type2.Cell7:585-593. VOL. 21, 1977
on November 10, 2019 by guest
http://jvi.asm.org/
1030 NEUWALD ET AL.
40. Thomas, D. C., and M. Green. 1969.Biochemical
stud-iesonadenovirus multiplication. XV. Transcription
of theadenovirus type 2genome duringproductive
infection. Virology 39:205-210.
41. Thomas, M., R. L. White, and R. W. Davis. 1976.
Hybridization of RNAtodouble stranded DNA: for-mation of R-loops. Proc. Natl. Acad. Sci. U.S.A. 73:2294-2298.
42. van derVliet, P. C., and A. J. Levine. 1973. DNA-bindingproteinsspecificforcells infectedby
adenovi-rus.Nature (London) New Biol.246:170-174. 43. vander Vliet, P.C., A. J. Levine, M. J.Ensinger,and
H. S. Ginsberg. 1975. Thermolabile DNA binding
proteins from cells infected witha
temperature-sensi-tive mutant of adenovirus defective in viral DNA
synthesis. J. Virol. 15:348-354.
44. Walter, G.,and J. V.Maizel, Jr.1974.Thepolypeptides of adenovirus.IV. Detection ofearly and late virus-induced polypeptides and their distributionin subcel-lular fractions. Virology57:402-408.
45. Weinmann, R.,T.G.Brendler,H.J.Raskas,and R.G. Roeder. 1976. Low molecular weight viral RNAs transcribed by RNA polymeraseIIIduring
adenovi-rus2infection. Cell7:557-566.
46. Westphal, H., J. Meyer, and J. V. Maizel, Jr. 1976.
Mapping ofadenovirus messengerRNA by electron
microscopy. Proc. Natl. Acad. Sci. U.S.A. 73:2069-2071.
J. VIROL.