0022-538X/78/0028-0905$02.00/0
Copyright©1978 American Society for Microbiology Printed inU.S.A.
Cell-Free Translation of Purified Virion-Associated
High-Molecular-Weight RNA Synthesized In Vitro by
Vaccinia
Virus
WALTER BOSSART,t ENZO PAOLETTI, AND DONALD L. NUSS*
Division of Laboratories and Research, New York State Department of Health, Albany, New York 12201
Received forpublication11May 1978
Virion-associated high-molecular-weight (HMW) RNA synthesized in vitro by
purified vaccinia virus particles has been translated in a wheat germ cell-free
proteinsynthesizing system. Purified HMW RNA directs the synthesis of
trans-lation products whichareidentical to the translation products made in response
toinvitro-synthesized,virion-released 8 to12SmRNA. The translation of HMW
RNA proceeds exclusively through a 5'-terminal cap-mediated initiation step.
Furthermore, only onecoding sequence is translated per HMW RNAmolecule,
and that sequence is probably located near the 5' end ofthe molecule. These
conclusions are based on the following results. (i) Sodium dodecyl
sulfate-polyacrylamide gel electrophoresispatternsoftranslation products
syn-thesized in response to HMW RNA and in response to 8 to 12S mRNAwere
qualitatively identical. (ii) Onanequalweightbasis, HMW RNA was 25 to 30%
as active as 8 to 12S mRNA in stimulating in vitro protein synthesis. (iii)
Unmethylated HMW RNAwastranslatedat10% theefficiency of themethylated
form of this RNA.
(iv)
m7pG
inhibited the translationof fullymethylated HMWRNAby 90%. (v) After the initiationstepoftranslationwasblockedby
aurintri-carboxylic acid, theratewithwhichaminoacidswereincorporatedintoindividual
polypeptides decreased in a similar mannerfor the translation of both HMW
RNAand8 to 12SmRNA. Virion-released8 to 12S mRNAderived from
virion-associated HMW RNA during a chase in the presence ofATP, GTP, and
S-adenosylmethionine
wasalso translated. Atlow RNAconcentrations, the derivedRNA appearedtostimulate amino acid incorporation more efficiently than the
HMWRNAprecursor. However,athigherconcentrations of this RNA,protein
synthesiswasseverely inhibited.
RNA synthesized and extruded by vaccinia
virusparticlesin vitro has an average
sedimen-tation value of between 8 and 12S (15, 35),
contains a
7-methylguanosine
cap at the 5'ter-minus (36),and ispolyadenylated atthe3' end
(16,24). Underappropriate conditions, vaccinia
virus particles
preferentially
synthesize ahigh-molecular-weight (HMW), 20 to 30S form of
RNA(26,29a). Theselarge transcriptsarefound
only
inassociation with the viruscoresandarenot released from the virion as HMW RNA
molecules (26, 27, 29a). HMW RNA is also
cappedatthe5'end(25),but it isnotextensively
polyadenylated at the 3' end (27, 29a). This
virion-associated HMW RNA can be chased
withinthe virionparticletoRNA in the8 to12S range, some of which is released from the virus (27). Thiscleavageprocess is notdependenton tPresentaddress: Institute forMicrobiologyandHygiene, University ofBasel, CH-4000, Basel, Switzerland.
continued RNA synthesis but does require
ri-bonucleosidetriphosphates and is
prevented by
theaddition of ethidium bromide
(27).
Recently,
an endoribonuclease whichappears
specific
forthe
cleavage
of the virion-associated HMWRNA has been solubilized from vaccinia virus
particles
(29). Furthermore, thetranscriptional
complexity of virion-released 8 to 12S mRNA
and virion-associated HMW RNA is identical
(28). These combined results suggesta
precur-sor-product
relationship
betweenvirion-associ-ated HMW RNA and virion-released 8 to 12S
mRNA.
Recently both we (7) and H. Pelham (30)
described the effect of UV irradiation on the
expression of vaccinia virus gene
products
syn-thesizedinacell-freesystem
coupling
transcrip-tion and translation. Although different
trans-lation systems were used in these two
studies,
similar resultswereobtained. These results
in-905
on November 10, 2019 by guest
http://jvi.asm.org/
906 BOSSART, PAOLETTI, AND NUSS
dicated that each translationally functional mRNA species produced in vitro by vaccinia virus cores is synthesized from individual pro-motersites.However,wecouldnotruleout the possibility that the polypeptides expressed in the coupled system are the products of se-quences found very near the 5' end of several long transcriptional units, whereas other se-quences that are expressed only after normal processingoftheselongtranscripts are transla-tionally silentin the invitro translationsystem. To investigate this possibility and to further characterize the virion-associated HMWRNA, we have determined a number of parameters with respecttothetranslation of this RNA in a wheat germ cell-free protein-synthesizing sys-tem.
MATERIALS AND METHODS
Synthesis and purification of vaccinia RNA. Vaccinia virus (WR)was grown in andpurifiedfrom
HeLa cells as described by Joklik (13). RNA was
synthesized in a standard in vitro RNA polymerase reactionconsistingof 50 mMTris-hydrochloride (pH 8.4), 10mMMgCl2, 10mMdithiothreitol,0.05%
Non-idetP-40, 2 mM eachATP, GTP, and CTP, and0.2 mM [:3H]UTP, (0.025 Ci/mmol). Purified virus was
addedto afinal concentration of4 to5optical density
units at 260nmperml.Incubationwasat37°C. For the preparation ofthemethylatedand unmeth-ylated forms of virion-released 8 to 12SmRNA, the
RNApolymerase reactionwasincubatedat37°C for
30minin thepresenceofeither10jimof S-adenosyl-methionine (AdoMet)or 100jim of S-adenosylhomo-cysteine (AdoHcy). Viruswas removed from the
re-action mixture atthe end of theincubation time by centrifugation at 15,000 rpm for 30 min. RNA was
phenol extracted from the virus-freereactionmixture and purified on sodium dodecyl sulfate
(SDS)-sucrose gradientsaspreviously described(25, 27).
For thepreparationofthemethylated and
unmeth-ylated forms ofHMW RNA, reaction mixtures
con-taining eitherAdoMetorAdoHcywerepreincubated for20min in the absence ofUTPtoreduce theATP
concentration(29a), andsynthesiswasthen startedby
theaddition of[3H]UTP (0.025Ci/mmol) to afinal concentration of0.2 mM. Afteranincubation period
of40min,RNA wasextracted from thevirusparticles asdescribedpreviously (26, 27, 29a), and then purified
by sedimentation through SDS-sucrose gradients. Bothtypesof RNA wererecovered from thegradients
byethanolprecipitation.Threeadditional ethanol
pre-cipitationswereperformedto removeall traces of SDS before the addition of RNA to thecell-free translation system.
Cell-free translation. A cell-free
protein-synthe-sizingsystem was prepared from Niblack brand wheat germ by the method of Roberts and Paterson (32),
excluding the preincubation step. Reaction mixtures for cell-free translation contained wheat germ extract
at a finalconcentration of 25optical density units at
260 nm perml;20 mMN-2-hydroxyethyl piperazine-N'-ethanesulfuric acid (pH 7.5); 135 mM KOAc; 3.5
mMMg(OAc)2; 1.25mMATP;0.25mMGTP;15mM
creatine phosphate (Sigma Chemical Co.); 75 jg of creatine phosphokinase (Sigma) perml.; 3.6mM di-thiothreitol; 0.3 mM spermidine-hydrochloride; 50
,uMeach amino acid minus either leucine or methio-nine; and either 200jiCi of [3H]leucine perml (54.6 Ci/mmol; NewEngland NuclearCorp.) or500jiCiof
[35S]methionine perml (573 Ci/mmol;NewEngland Nuclear). Insomereactions, AdoHcywasincludedat aconcentrationof 0.3 mMtoinhibitmethyltransferase activitypresent in the wheat germextract.
RNA samples were heated at60°C for 1 min and subsequently quenchedonicejust before additionto the translation system to reduce any RNA-RNA aggregation. Protein synthesis was monitored as de-scribed byMansand Novelli(21).
Polyacrylamide gel electrophoresis. Samples from translation reaction mixtureswerepreparedand
analyzed by electrophoresis on 15% polyacrylamide gels followed byautoradiographyas described previ-ously (7). Radioactivepolypeptideswere detectedon
somegels bythefluorographicmethodofBonnerand Laskey (4).
RESULTS
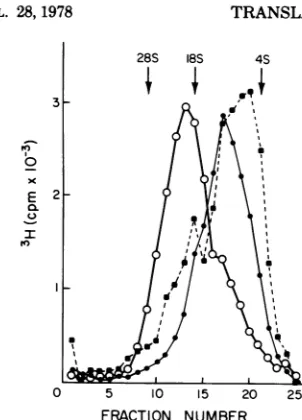
Stimulation ofprotein synthesis by
vir-ion-associatedHMW RNA. Purified vaccinia
virion-associated HMW RNA, synthesized
un-derrecentlyestablishedconditionswhich permit
the preferential synthesis of these large
tran-scripts (29a),wastested for theabilityto stim-ulateprotein synthesis inawheat germcell-free translation system. The level of amino acid in-corporation into protein which was directed by virion-released 8 to 12S mRNA or virion-asso-ciated HMW RNA is shown in Fig. 1A and B, respectively, as a function of RNA concentra-tion. Notice that both RNA populations stimu-late protein synthesistothe same extent at the concentration of RNA which issaturating. How-ever,the concentration of RNA whichgivesthis maximal stimulation is considerably lower for the 8 to 12S mRNAthan for the HMW RNA. The concentration of 8 to 12S mRNA which gave maximal stimulation ofprotein synthesis
wasconsistently foundtobe between three- and fourfold lower, on an equal weight basis, than the concentration of HMW RNA which gave maximal protein synthesis under identical con-ditions. In this regard, sedimentation analysis has shown that the HMW RNA is
approxi-mately three- to fourfold larger than 8 to 12S
mRNA (25-27). Furthermore, a comparison of
theincorporationofmethyl groupsatthe5' end
with theincorporation of UMPatinternal sites of these RNAs indicatedthat,on anequalweight
basis, 8 to 12S mRNA has three- to fourfold morecapped 5'-terminal ends thanHMWRNA
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
VACCINIA
synthesized under identical conditions (25).
Analysis by SDS-gel electrophoresis of
poly-peptidessynthesized inresponse to
virion-asso-ciated HMW RNA andvirion-released 8 to 12S
mRNA revealed thatthese two populations of
RNA directed the synthesis ofsimilar
transla-tionproducts. (Compare lanesAand B in Fig.
2.)Thereare someobservabledifferencesin the
relative synthesis of the larger (greater than
50,000 molecular weight) polypeptides when
comparing thetranslation products coded forby
HMWRNA with thetranslation products
pro-grammed by 8 to 12S mRNA; the larger
poly-peptides are moreprominent in translation
re-actions programmed by HMW RNA. Longer
exposure of the 8 to 12S mRNA translation
products doesreveal,however, that the
synthe-sis ofpolypeptides aslarge as90,000daltons is
directedby this population of RNA. These
quan-titative differencesareprobably duetothe sizing
ofthese RNAs on sucrose gradients as partof
the
purification
scheme. Itis importantto notethatthesynthesis of large polyproteins didnot
occurduring the translation of the HMW RNA.
This result suggests that normal termination
E250
LO) ' 150
0
E
Z 50
0
o 25
0 0
z
LZ 15
z
z
0 I
LJ 5
C
Po C 10 20 30 0 30 60 90
RNA (,ug/ml)
FIG. 1. Stimulation ofpolypeptide synthesis in a
wheatgermcell-freetranslation systeminresponse tothemethylated andunmethylated forms ofHMW RNA and 8to12S mRNA. Themethylatedand
un-methylated forms ofHMWRNA and 8to12SmRNA
were synthesized andpurified as described in the text. Translation was in the presence of0.3 mM
AdoHcy for30minat22°C. The levelofamino acid
incorporation directed byeachtypeofRNA is pre-sentedasafunction ofRNAconcentration.(A)
Meth-ylated8to12S mRNA; (B)methylatedHMWRNA; (C) unmethylated8to12SmRNA; (D) unmethylated
HMW RNA.
N
sm1mA
BC
:_ 66,000
o
47,000
:_ 30,000
AMaf
40wfk
_IvA
i_p 40
..
4=m
17,
000
FIG. 2. Autoradiograph of an SDS-polyacryl-amidegel after electrophoresis oftranslation
prod-ucts synthesizedin acoupled transcription-transla-tion system or in response to exogenous 8 to 12S mRNA or HMW RNA. [3S]methionine-labeled translationproducts synthesized in thecoupled
sys-tem(laneC)aspreviouslydescribed(3)orsynthesized under standard translation conditions in responseto purified8 to12S mRNA (lane A) orpurifiedHMW RNA(laneB) wereanalyzedby SDS-gel electropho-resis. Each well received 500,000 acid-precipitable cpm. Themigrationpositions ofmarkerproteins of
known molecular weight are indicated on the left margin. Theexposure timewas 4days.
signals arerecognized byribosomestranslating
thelarge transcripts.
We recently described a coupled
transcrip-tion-translationsystem in which we determined
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.505.253.447.83.466.2] [image:3.505.76.218.350.536.2]908 BOSSART, PAOLETTI, AND NUSS
theeffect of UV irradiationontheexpressionof
vacciniageneproductsinawheatgermcell-free system (7). The results of thatstudy suggested that eachtranslationallyfunctional mRNA
spe-cies producedin vitrobyvaccinia virus coresis synthesized from individual promoter sites. A comparison of the translation products synthe-sizedinthe coupledsystem(Fig. 2,laneC)with the translation products directed by 8 to 12S mRNA (lane A) and HMW RNA (lane B) also showed some quantitative differences in the
translation products synthesized in all three
cases. However, close examination of the gel
patterns revealed no qualitative differences, at this level ofsensitivity, between the translation products synthesizedin thecoupledsystemand thosesynthesizedinthe translation system
pro-grammed by either HMW RNA or 8 to 12S
mRNA.
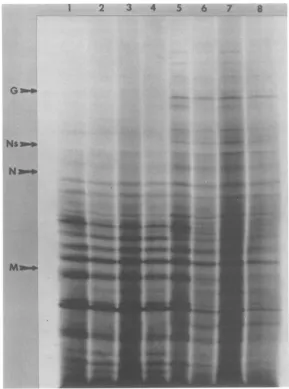
Requirementofacapped5' terminus for
efficient translation of HMW RNA. It is known that virion-associated HMW RNA is cappedatthe5'-terminal end(25).Todetermine whether the translation of this RNA proceeds exclusivelyviaa5'-terminalcap-mediated
initi-ationstep, thefollowing experimentswere
per-formed. Virion-released 8 to 12S mRNA and virion-associated HMW RNA were purified
from invitrotranscriptionreactions which
con-tained AdoHcy to prevent methylation of the transcription products. The stimulation of
pro-teinsynthesiswith respecttothe RNA
concen-trationwasthendeterminedfor thefully meth-ylatedandunmethylatedformsof both 8 to 12S mRNA and HMW RNA. In thisexperiment,the wheatgermtranslation systemwasalso
supple-mented with 0.3 mMAdoHcytoprevent meth-ylation ofanyunmethylated RNA by enzymes presentinthe cell-freesystem. Themethylated andunmethylated forms of each type ofRNA exhibited maximal stimulation of protein
syn-thesisatthesameRNAconcentration, i.e.,12to
17 .tgofbothmethylated and unmethylated8to
12S mRNAperml(Fig. 1A and C)and55to65 tigof bothforms of theHMWRNAperml(Fig.
1Band D).However,the unmethylated form of
both 8 to 12S mRNA and HMW RNA
stimu-latedprotein synthesis only 10%asefficientlyas
the methylated forms of these RNAs. The
re-quirement ofa 5'-terminal capfor the efficient
translation ofvaccinia8to12S mRNAhas
pre-viously been reported (34). The data presented inFig. 1suggestthat the efficienttranslation of HMW RNA also depends onthe presence ofa
5'-terminalcap.
The translation ofmRNA's which normally requireacapforefficient translationis inhibited
bytheaddition ofm7pG (11). The translationof
mRNA's which do not require the cap for effi-cienttranslation, e.g., EMC RNA (33) and, pre-sumably, the translation of RNA sequences which is initiated at internal initiation sites, is not affectedby the addition of m7pG at a con-centration which inhibits the translation of capped mRNA (14). The effect of m7pG on the translation of themethylated andunmethylated forms of both 8 to 12S and HMW RNA was tested (Table 1). The effect of m7pG on the translation of both the 8 to 12S mRNAand the HMW RNA populations was identical. The translation of the methylated forms of both RNAs was inhibited by 90% when 1 mMm7pG wasadded tothe cell-free system, whereas this concentration of
m7pG
had no effect on the translation of the unmethylated forms of these twoRNAs. Indetermining the effect of m7pGon thetranslation ofunmethylated RNA,onemust becognizant of the observation that the addition of anyRNA, e.g., rRNA (Table 1), results ina stimulation ofendogenous wheat germ protein synthesis in translation systems which are not preincubated. Thisendogenous protein synthe-sis is completely inhibited by the addition ofTABLE 1. Effect ofm7pGonthe translationof the methylated andunmethylated forms of HMW RNA
and 8 to12SmRNA
["S]methionineincorporation Reactionmixture' after
(min):'
0 10 20 30 60
-RNA
-m7pG 8.7 11.4 13.9 16.1 19.5
+m7pG 8.5 8.1 8.5 8.3 7.1
rRNA(10jig/ml)
-m7pG 7.9 15.7 22.1 25.3 33.5
+m7pG 7.6 9.2 9.2 10.0 9.1
Methylated 8 to 12S mRNA (10jig/ml)
-m7pG 70.4 163.9 230.6 276.8
+m7pG 6.3 12.3 16.7 22.7
Unmethylated8 to12S mRNA (10 jig/ml)
-m7pG 6.4 13.2 18.4 21.5
+m7pG 6.7 14.5 20.4 22.9
Methylated HMW RNA(40jug/ml)
-m7pG 89.7 186.9 247.0 280.0
+m7pG 7.3 15.1 20.6 23.3
Unmethylated HMW RNA(40jg/ml)
-m'pG 3.7 9.5 13.1 19.4
+m7pG 4.4 10.9 19.2 22.9
'Themethylatedandunmethylatedforms of HMW RNA and 8 to 12S mRNA were translated under standard conditions in the presence or absence of 1 mM m7pG. AdoHcy was present at a concentration of 0.3 mM in all reaction mixtures topreventmethylationofunmethylatedRNAbythe wheat germmethyltransferase.
bResults are presented as countsper minute x 10-" of
[C3S]methionine
incorporationinto hotacid-precipitableradio-activityper 5jilof reaction mixture.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
m7pG.
Thus,the stimulationofendogenouspro-tein synthesis by the addition of unmethylated
RNA and theinhibition of that endogenous
pro-tein synthesisby
m7pG
mustbe taken intoac-count when calculating the effect of m7pG on
the actual translation of unmethylated RNA.
Theresults presented here withrespect to the
relative translation of the methylated and
un-methylated forms of the 8 to 12S mRNA and
the HMW RNA, aswell asthe effect of
m7pG
on the translation of these RNAs, are in total
agreementwithpreviously published results for
8 to 12S mRNA (34) and stronglysuggest that
the translation of HMW RNA also proceeds
exclusively
through
a5'-terminal cap-mediatedinitiationstep.
Asafurthertestof this conclusion, the
trans-lationproductsdirected by both methylated and
unmethylated HMW RNA and8 to12SmRNA
in thepresenceand absence of
m7pG
werecom-pared by
SDS-gel
electrophoresis. Ifanyof theHMW RNA sequences are translated from an
internal initiation site, then the corresponding
polypeptide would beveryprominent inthegel
analysis of translation products directedbythe
unmethylated form of the HMW RNA. Also, the
synthesis of these polypeptides would not be
inhibited after the additionof
m7pG.
AsshowninFig. 3, the
gel
profiles
of the translationprod-uctsof8 to 12SmRNA, which are synthesized
only from5'-terminal initiation sites,are
similar
tothe
proffies
of HMW RNA translationprod-ucts synthesized under each set of conditions.
Thus, we can conclude that the translation of
HMW RNAproceeds througha5'-terminal
cap-mediated initiation step and can rule out the
possibility
that there is asignificant
level ofinitiation of HMW RNA translation atinternal
initiation sites.
Number and probable location of
trans-lated sequences on the
BMW
RNAmole-cule.
Although
the datapresented
inFig.
1and2andinTable1ruleoutthepossibility that the
translationofHMW RNA starts atinternal
ini-tiation
sites,
it ispossible
that multiplese-quenceswell downstream from the5'end of the
HMW RNA are translated after translation is
initiated atthe 5' end of the molecule, as has
been determined for the translation of some
coliphage
mRNA (18). Toinvestigate thispos-sibility,
thefollowing experimentwasperformed.
Translationof8 to12S mRNA and HMW RNA
wasallowed to initiate andproceedfor 4minin theabsence ofisotope, atwhichtimethe
inhib-itorofpolypeptidechain initiation,
aurintricar-boxylicacid(ATA),wasaddedtoafinal
concen-tration of 75 ,M. The ATA-inhibited reaction mixtures were then supplementedwith the
la-beled amino acid either at the time of ATA
additionorat1-min intervals after the addition
of ATA, andelongation wasallowedtoproceed
for a totalof30 min.Thisexperiment measures
the time it takes for ribosomes to finish the
translation of the mRNA molecules on which
they had initiated beforeATAaddition, i.e.,the
runoff
time.
Arunofftime
for HMWRNAlast-ing three to four times longer than the runoff
time for 8 to 12S mRNA would suggest that
sequences all along the HMW RNA molecule
arebeingtranslated after initiationatthe 5' end.
Identical runoff times for HMW RNA and8 to
12S
mRNA
wouldsuggestthat onlyonecodingsequence per HMW RNA wastranslated. The
runoff kinetics wereidentical for both 8 to 12S
mRNAand HMWRNA (Fig. 4).
As afinaltest,thetranslation products labeled
during the runoff of both8 to 12S mRNA and
HMWRNAwereanalyzed bySDS-gel
electro-phoresis. Thetime required for runoff,as
deter-mined by the decrease in the level of
radioactiv-ity incorporated into polypeptides with
time
after ATAaddition, increases
progressively
withincreasing molecular weight of thepolypeptides
coded forby the8 to12SmRNA(Fig. 5A).From
these measurements, aribosometransittime of
approximately
40codonspermin wascalculated.Acomparison of thegelpatternsinFig. 5A and
Breveals that the individualpolypeptides coded
forby both the8 to 12SmRNA andtheHMW
RNA have identicalrunoff kinetics.
Witharibosome transittime of40codonsper
min
(calculated
from data inFig. 5),aribosomewhich initiates at zero time can proceed
only
about160codons from the ribosomebinding site
by4min,the time of ATA
addition;
160codonsrepresentsabout 70% of the
coding region
ofanmRNA coding for a
polypeptide
of molecularweight 30,000. Since the initiation of HMW RNA
translation,
like the initiation of8to 12SmRNA translation, proceeds through a
5'-ter-minal
cap-mediated
step, the similar runoffki-netics forpolypeptides codedforby HMWRNA
and 8 to 12S mRNA suggests that the one
se-quencetranslated from eachHMW RNA
mole-culeislocatednearthe 5' end ofthatmolecule.
However,itisconceivable, though
unlikely,
thatthesecondary structure of the
large transcripts
is such that the 5' terminus is in an unusually
close functional association with the initiator codon for acoding sequence nearthe 3' end of
the HMWRNA.
Inpreliminary experimentswedeterminedthe
stability of both the HMW RNA and8 to 12S
mRNA in the wheat germ system
by
sucrosegradientanalysis of theRNA's recovered from
the translation system with
increasing
time of 28,1978on November 10, 2019 by guest
http://jvi.asm.org/
910 BOSSART, PAOLETTI, AND NUSS
1 2 3 4
J. VIROL.
5
6
7 8Gum.
Nsa_
Na..
[image:6.505.118.407.69.458.2]M_..
FIG. 3. Autoradiograph ofaSDS-polyacrylamidegel afterelectrophoresis of translationproducts synthe-sized inresponsetomethylatedandunmethylatedHMWRNA and8 to12S mRNA in the presenceorabsence ofImMm7pG.Each well received120,000
acid-precipitable
cpm.Samples werenormalized with respecttoprotein concentration byadding appropriate amounts of non-radioactive wheat germextract. (Lanes1 and 2)Methylated8to12S mRNA in the absence and presenceofm7pG,respectively; (lanes3and4)unmethylated
8 to12S mRNA in the absence and presenceof m7pG respectively; (lanes5and6)methylatedHMW RNA in the absence and presenceof m7pG, respectively, (lanes7and8)unmethylatedHMWRNA in the absence and presence ofm7pG,respectively. The positions of molecular weight markersare shown in themargins.Exposure
timeforthisautoradiographwas 3weeks.
incubation. Both HMW RNA and 8 to 12S mRNA'sweredegradedatsimilar rates, and the sourceofthenuclease activitywastracedtothe creatine phosphokinase preparation. Several lots of creatinephosphokinaseweretested,and
allwerefoundtocontainnucleaseactivity(12).
Althoughsomedegradationof HMW RNAand
8to12S mRNAwasobserved in the translation system,over50%of the HMW RNAsedimented
atgreaterthan 18Swithin15minofincubation.
The time for initiation and runoff after ATA additionwas10min(Fig. 4); consequently,under these conditions if thereweretranslation of in-ternal sequence near the 3' end of the HMW RNAafter the initiation of translation occursat the 5' end it would be reflected in both the kinetics of runoff(Fig. 4) and in thegel analysis
ofpolypeptides labeled duringthe runoff (Fig.
5).Furthermnore,allpolypeptideswhichare
nor-mally synthesized in response to HMW RNA
on November 10, 2019 by guest
http://jvi.asm.org/
TRANSLATION TRANSCRIPTS
z w
w
40-Q
20-0 5 10 20 30
[image:7.505.63.218.60.264.2]MINUTES
FIG. 4. Incorporation ofamino acidsintoprotein
with time afteraddition ofATAto translation sys-temsprogrammed byeither HMW RNA or8to 12S mRNA. The translation of either HMW RNA (40
pg/ml)or8to 12S (10pg/ml) mRNA wasinitiated and allowedtoproceed for4minin the absence of added leucine, atwhichtime ATA wasaddedtoa
finalconcentration of75pM. [3HJleucinewasthen
addedtoportions oftheATA-inhibited reactionsat zerotime,i.e., with the ATA,oratthetimesindicated
in the graph afterATA addition. Elongation was allowedto continuefor a total of30 min after the addition of[3H]leucine for each timepoint.
[3H]-leucineincorporationintoproteinwasdeterminedby the methodofMans and Novelli (21). The levelof [3H]leucine incorporation resulting when
[3H]leu-cinewasaddedwithATA (zero time) isconsidered 100%o incorporation aspresented on the ordinate. Symbols: (0) runoffkineticsfor8to12S mRNA; (0) runoffkineticsforHMWRNA.
when initiation of translation isnotlimited are
also synthesized when the initiation step is in-hibited4min after thestartoftranslation (Fig. 5B). Given the kinetics ofrunoff (Fig. 4), this result strongly suggests that none of the
ex-pressed coding sequences in the population of HMW RNA arelocated exclusively atinternal
sites ofapolycistronicRNA molecule. From the
combined results, we conclude that only one
codingsequenceistranslated from each HMW RNAandthat thatsequenceisprobablylocated
near the 5'-terminal end of the HMW RNA molecule.
Translation of8 to12SRNAderived from BMWRNA. Since normallyreleased 8to 12S mRNAwasfoundtobeapproximatelythree- to fourfoldmoreefficienton aweightbasis in stim-ulating protein synthesis than was purified
HMW RNA, it was considered important to
determinewhether 8to12SRNA derived from
corescontaining HMWRNA during incubation
in the presence of ATP, GTP, andAdoMet is
translated
with a similar efficiency as HMW RNA or asnormallyreleased 8 to 12S mRNA.Virion-released8 to 12S mRNA and
virion-as-sociated HMW RNA were synthesized as
de-scribed above. Cores containing HMW RNA
synthesizedat limiting ATPconcentrations were
washed twice, and the RNA in one-half of the
cores was chased for60min in the presence of
ATP, GTP, and AdoMet. The three RNA
spe-cies were then prepared for translation by
phenol extraction, chromatography on G-25
Sephadex to remove nucleoside triphosphates
and,finally, ethanol precipitation and
resuspen-sion inwater.Itmust be stressed that
prepara-tionof these RNAs didnotincludeapurification
step on sucrose gradients to remove traces of
virion-associated 8 to 12S RNA in the HMW
RNA preparation as was done for the other
experiments reported in this paper. Since the
contaminating 8 to 12S RNA is three to four
times more efficiently translated than true
HMW RNA, even a 10% contamination will
significantly
decrease theapparentdifferenceintranslational efficiencies of thesetwoRNA
pop-ulations. Sedimentation patterns of the three
RNA
preparations
wereexaminedtodeterminewhether the chase was efficient. There was a
slight contamination of the HMW RNA with
RNA whichsedimentsinthe
region
ofreleased8to12SmRNA(Fig. 6).Asinmostexperiments,
HMW RNA was chasedto 8 to 12S RNA and
released from the virion with an efficiency of
approximately 65%.
Shown inFig. 7A, B, and Carethesaturation
curves for the translation of the three RNA
species.Asexpected, the difference in saturation
curvesforHMWRNA andnormally released8
to 12S mRNA is reduced from that shown in
Fig. 1. Atlow concentrations ofRNA, the level
of
protein
synthesis
stimulatedby
derived8to12S RNA issimilartothat of
normally
released8 to 12S mRNA rather than the
virion-associ-ated HMW RNA.Thisincreased
specific
trans-latability
suggeststheprocessing of HMWRNAintoactive 8 to12S mRNAmolecules. The
sat-uration curve is unusual, however, in that the
level ofproteinsynthesis atthe apparent
satu-ration levelof RNA is reducedfrom thelevels
observed for both the
normally
released 8to12S mRNA and HMW RNA.Furthermore,athigherRNA concentrations, the level ofprotein
syn-thesis isradicallyreduced.
This reduction in proteinsynthesisathigher
RNAconcentrationssuggeststhepresenceofan
inhibitor.
Recently,
thesynthesis
ofapolyade-nylated and capped small
(4S)
RNAspecies
VOL. 28,1978
on November 10, 2019 by guest
http://jvi.asm.org/
912 BOSSART, PAOLETTI, AND NUSS
0 1 2 3 4 5 6 7
_
* ,zbF
.S . u.. ..
w X yX
rs
1E
illiL
Fs-s--e
k _E 39 =. .b.; :..l'S | s ':'
_ Xb |
.kw. w ,,
i ,
^g
..
4 4
>
A
B
FIG. 5. Autoradiograph ofaSDS-polyacrylamide gel afterelectrophoresis ofpolypeptide labeled duringa
runoff experiment. Conditions for the runoff experimentwereidenticalto those describedfor Fig.4, except
that [3H]leucine wasreplaced by [35S]methionine. Each well received anequal volume of the respective
reaction mixture, with thezero time sample (lane1) containing80,000cpm. (A)8to12S mRNA; (B) HMW
RNA. The numbersatthetopofeachlaneindicate the timeafterATAaddition thatthe labeled amino acid
wasadded. Thefluorographic procedure ofBonner andLaskey (4)wasusedfortheprocessing of these gels.
(polyadenylated leader sequences [PALS]) by
vacciniacoresunderavariety of conditions has
been observed (Paoletti and Lipinskas,
manu-script in preparation). If the distortion of the saturation curve forthe derived 8to 12S RNA
results from the presence ofthese small RNA
species actingasinhibitors oftranslation, then
addition ofa purified fraction of this RNA to
released 8to12S mRNA shouldgeneratea curve
similartothatobserved inFig.7C. As shown in Fig. 7D, E, and F, addition of increasingamounts ofPALS did indeed result inanalteration of the
saturation curve for 8 to 12S RNA to curves
approaching that shown in Fig.7C.
PALS are synthesized in the ATP-depleted
reaction butarereleasedonly after restoration
of the ATPconcentration in the reaction mix-ture, as wasdone for the chase in Fig. 6.Thus,
PALS are nornallypresent in the virion-asso-ciated HMW RNApopulationas well asinthe
population of derived 8 to 12S RNA yet, inex-plicably, high concentrations of HMW RNA do
notshow theinhibition ofprotein synthesis ob-served withhigh concentrations of derived 8to 12S RNA.
Analysis of the translation products synthe-sized inresponseto the derived 8to 12S RNA showed the appearance of no new bands, no
increase in thesynthesis ofasubset of
polypep-tides also synthesized in responseto virion-as-sociated HMW RNA, andno synthesis of
low-molecular-weight protein duetothe translation ofPALS(datanotshown). The translation prod-uct directed by virion-associated RNA and de-rived 8 to 12S RNA were identical and again
similartotheproduct synthesized inresponseto normally released8to 12S mRNA and synthe-sizedinthecoupledsystem.
DISCUSSION
Purified vaccinia virion-associated HMW RNA directs polypeptide synthesis in a wheat
germ cell-free translation system. Translation
Ns_
N_
J. VIROL.
6-.ML. .%-in.
Ga--*.
.w
X.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.505.70.463.72.373.2]TRANSLATION OF LARGE TRANSCRIPTS OF VACCINIA
C-'
0 5 10 15 20 25
[image:9.505.63.214.53.263.2]FRACTION NUMBER
FIG. 6. Sedimentation profiles of normally
re-leased8to 12SmRNA, virion-associated RNA
syn-thesizedatlimiting concentrationsof ATP, and re-leased 8to12S RNA derivedfrom the
virion-associ-ated RNA duringa chase in thepresenceof ATP,
GTP, and AdoMet. Virion-released 8 to 12S RNA (0) waspurified fromavirus-freesupernatantafter
40min of synthesis. Virion-associated HMW RNA (0) wassynthesizedinanATP-depleted reactionas
described(29a) from virus pelleted fromapolymerase reaction.Released8to12S mRNA (a) derived from the virion-associated RNA during a chase in the
presence of ATP, GTP, and AdoMet waspurified
fromavirus-freesupernatantafter60minof chase.
/2HJUTPspecific activity wasmaintainedconstant
inallreactionstomonitor thesynthesis of RNA. productssynthesized inresponsetoHMWRNA
are qualitatively identical and quantitatively
similar to the translation products synthesized in responseto virion-released 8to 12S mRNA. Efficient initiation of HMW RNA translation requires that these largetranscripts be capped at the 5' terminus. Initiation of translation at internal initiation sitesonthe HMWRNA
mol-ecules seems not to occur. Finally, only one
codingsequenceoneachHMW RNAmolecule
istranslated, and thatsequence isprobably
lo-catedverynearthe 5'-end of themolecule.
Aninterpretationof theseresults intermsof
theorigin andbiological significance of the
vac-cinia HMW RNA is somewhat limited by the
sheer complexity of this large DNA virus. The size of the vacciniagenomeis estimated at123
X 106daltons (10). Basedonthecalculation that
vaccinia mRNA hasan averagelength of 1,100
nucleotides (5) and the assumption that only
non-complementary regions of the DNA are
transcribed, there isaminiimal potentialwithin
the vaccinia genome for the expression of
be-.2 A D
100 /
o50 ** r
50
Q,o 20 E0o lo 2
0
w
10 _E_
U 0
0 20 40 0 2
RNA (,ug/ml)
FIG. 7. Stimulation ofpolypeptide synthesis in a wheatgerm cell-free translation system in response to normallyreleased 8 to 12S mRNA, virion-associ-atedRNA made at limiting concentration of ATP, and released 8 to 12SRNA derived from the virion-associated RNA during a chase in the presence of ATP, GTP, and AdoMet. Translation of each species of RNAsynthesized as described in the legend to Fig. 6 was performed asdescribed for Fig. 1. (A) normally released 8 to 12S mRNA; (B) virion-associated RNA made at limiting ATPconcentrations; (C) released 8 to 12S RNA derived from virion-associated RNA during a chase in the presence of ATP, GTP, and AdoMet. (D), (E), and (F) show the effect of adding increasing concentrations ofsmallpolyadenylated and cappedRNAs (PALS) on the translation of nor-mallyreleased 8 to 12S mRNA. The final concentra-tions of PALS were 0, 5, and 10 pg/ml, respectively, for (D), (E), and (F). PALS were synthesized in the presence of2Opgof actinomycin Dper ml andpurified byphenol extraction and selection on poly(U)-Seph-arose. Details of thesynthesis and characterization of PALS will be presented in afuture communication.
tween 160 and180 differentgene products.
Al-though transcriptional complexity studies
indi-cate that essentially all of the viral genome is
transcribedinvitro (6, 28),abundancy
measure-ments do suggestthat asmall proportion of the
genome is represented bythe majority of the in
vitro-synthesized RNA (6). In this regard, the
translationproductssynthesizedin the coupled
transcription-translation system programmed
byvaccinia virusparticles (7, 30, 31) or in
cell-free systems programmed by purified 8 to 12S mRNA (7) (Fig.2)havebeenresolved into 30 to
40 discrete polypeptides on one-dimensional
polyacrylamidegels and approximately 60
poly-peptidesontwo-dimensional gels.
Considerable evidence hasaccumulatedwhich 913
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.505.277.429.59.263.2]914 BOSSART, PAOLETTI, AND NUSS
isconsistent with aprecursor-product relation-ship between the vaccinia virion-associated HMW RNA and the virion-released 8 to 12S mRNA(25-29a). Nevertheless,thereareseveral observations which are, at present, difficult to reconcile with thishypothesis.Forexample, only
transcripts having5'-di-ortriphosphate termini,
butnot5'-hydroxylor-monophosphate termini,
whichwould beproducedbycleavageofaHMW RNA precursor, are capped by the virion-asso-ciatedguanylytransferase(2, 9, 22,23). Although the elucidation of the mechanismby which caps are formed allows the elimination of several possible pathways which could be envisioned for processing ofalarge transcript, several alterna-tive pathways, e.g., a mechanism analogous to
gene splicing inadenovirus (3, 8, 17) orsimian
virus 40 (1) have not yet beenruledout. Asecond observation which isdifficultto un-derstand in terms of a processing mechanism comesfromrecent studies whichexamined the
effect of UV irradiation on the expression of
vaccinia gene products in a coupled transcrip-tion-translation system (7, 30). The results of these studies indicate that eachtranslationally
functionalmRNA producedinvitroby vaccinia
cores is synthesized from individual promotor sites and isnotderived fromlarge polycistronic
transcripts. In this case, it is conceivable that
thepolypeptidessynthesized in thecoupled
sys-tem arecodedforbyRNA sequenceslocated at ornearthe 5' terminus ofafewlarge transcrip-tionalunitsand that sequencesdownsteamfrom the 5' end of these large transcripts are not
expressed because of a lesion in the normal
processingmechanism under in vitro conditions.
Consistentwiththispossibilityarethe data
pre-sented inFig.2, 4,and5which suggest thatonly onesequence perHMW RNAmoleculeis
trans-lated, that this sequence is probably locatedat
ornearthe5' end of the HMW RNAmolecule, and that the sequences expressed in response to HMW RNA are the same as those expressed both in the cell-free translation system in re-sponse to 8 to 12S mRNA and in the coupled system.However, ifoneconsiders the possibility that.virion-released 8 to 12S mRNA isderived fromasetofpolycistronic HMW RNAs by an in vitro processing mechanism in which only the sequences derived from the 5' end of the large transcript are translationally functional, whereas the processed sequences derived from internal regions of the HMW RNA precursor
are translationally nonfunctional, then one
wouldpredict that the translational activity of the HMWRNA and the derived 8 to 12S RNA would be identical on anequal weight basis. As shown inFig. 1 and in Table 1, this is not the
case; 8to 12S mRNA is approximately three- to fourfold more active instimulating protein syn-thesis in the cell-freesystem than acomparable amount of HMW RNA. Thus, there is no evi-dence for the buildup oftranslationally inactive RNA sequence as a result offaulty processing. Furthermore, if only the 5'-terminal sequence of apolycistronic HMW RNA precursorwere proc-essed in vitro to yield a translationally active molecule, but several translationally active mRNA moleculeswerederivedfrom each HMW RNA precursor in the infectedcell, where proc-essing would proceed normally, then there should be major differences in the
polyacryl-amide gel patterns of the translation products synthesized in a cell-free system in response to eitherpolysomal mRNA from vaccinia-infected cells or in response to invitro-synthesized8 to 12SmRNA. Infact,polyacrylamide gelpatterns of the translationproducts directedbyeither in vitro-synthesized vaccinia mRNA or RNA iso-lated frompolysomes ofvaccinia-infected cells are quite similar (J. Cooper and B. Moss, per-sonalcommunication).
Perhaps each HMW RNA is the precursor of only one 8 to 12S mRNA molecule and the remainder of the HMW RNA sequences are eitherdegraded orremain virion associated. In
thisregard, the results ofexperimentsin which
HMW RNA was preferentially (27) or
exclu-sively (29a)labeled and thencleavedtoRNA in
the8 to12S size range suggeststhat themajority
of this labeled HMWRNA is conserved during
cleavage;i.e.,there isnoevidencesuggesting the
extensivedegradationofaportionof the HMW
RNA during the cleavage step. Indeed,
tran-scriptional complexity studies have indicated
thatthesamesequencesarerepresented in both
the virion-released 8 to 12S mRNA and the
virion-associated HMW RNA (28).
Given the difference in efficiencywith which
equal weights of released 8 to 12S RNA and
purified virion-associatedHMW RNA direct in
vitro proteinsynthesis (Fig. 1), a determination
of whether HMW RNA isprocessed into func-tional messenger RNA should be possible by comparing the efficiency of translation of de-rived8 to12S RNA withvirion-associated HMW RNA from which it was derived and normally released8 to 12S mRNA. However, this
experi-ment is plagued by two problems. First, the
virion-associated RNA synthesized under
limit-ing ATP concentrations is contaminated with some 8 to12S RNAwhich is released upon chase in the presence of ATP and thusbecomes part of the population of derived 8 to 12S RNA. While this virion-associated 8 to 12S RNA makes uponlyafraction of the totalbulk ofthe
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
915
virion-associated RNA, it contributes more to
the translational efficiency of the total RNA
than toitsbulkbyafactor of3 to 4.Thus, the
differenceinefficiency of the normally released
8 to12S mRNAand the unpurifiedtotal
virion-associated RNAwill be on the order of 1.5- to
2.5-fold rather than 3- to 4-fold. Second, the
presenceofinhibitory PALS released under the
chase conditions (Paoletti and Lipinskas,
man-uscriptinpreparation) addsanadditional
com-plication to the interpretation of these data.
However, based on the level of incorporation
observed atlow concentrations ofderived 8 to
12S RNA, it wouldseemthat thetranslational
efficiency of the virion-associatedRNAdoes
in-creaseduring the chase. This result is consistent
with theproposal that the presynthesized HMW
RNA isprocessedquantitatively within the
vir-iontofunctional mRNA.
An alternative explanation for the origin of
the HMW RNA is that this RNAresults froma
lesion in the termination oftranscription, i.e.,a
read-through mechanism. It is possible that
faulty termination, which mightoccur atlimiting
ATP concentrationsinvitro (19,20,29a), could
result inthe synthesis of large transcripts with
properties similar to those described for the
vaccinia virion-associated HMW RNA. Such
large transcriptswould be cappedat the 5' end
(25). The ratio of molecules having either
m7G(5')ppp(5')Gm
orm7G(5')ppp(5')Am
terminiwould bethe same for the largetranscripts
re-sulting from read through and for the correctly
terminated8 to 12S mRNA (25). The 8 to 12S
mRNA and the
large
read-through transcriptswould have identicalsequencecomplexity (28).
Translation products synthesized in cell-free
translation systems inresponse tothe
large
tran-scripts would be identical to the translation
products synthesized in response to 8 to 12S
mRNA (Fig. 2), and the sequences that are
translatedonthelarge transcripts would be
lo-catedatthe 5' endofthose molecules(Fig.4and
5).
Finally,
themechanismofcapformation andtheobserved effectof UVirradiationonvaccinia
gene expression in the coupled
transcription-translation systems arenotinconsistent with a
read-through mechanism. At the same time, a
read-through mechanism for the generation of
HMW RNA does not
explain
the consistentobservation thatprelabeled HMWRNA can be
cleavedwithin thevirion core, underappropriate
conditions,toRNA in the size range of 8 to 12S
(27, 29a) which appears to be
translationally
functional (Fig. 7). Also,the detection ofa vac-cinia virion-associated endoribonucleaseactivity(29) whichacts
specifically
on the HMW RNA but notthe8 to 12S mRNAsupportsaprocess-ing mechanism rather than a read-through
mechanism. Additional studies arerequired to
determine whether the vaccinia virion-associ-atedHMWtranscriptsresult from faulty termi-nationof transcription or, a more exciting pros-pect,theyrepresent precursortranscripts to
ma-turevaccinia mRNA.
Independent of the mechanismbywhich
vac-cinia HMW RNA isgenerated, it is likely that
these large transcripts contain more than one
gene sequence
and,
consequently,
morethanonepotential ribosome binding sitepermolecule. It
isinteresting, therefore, that translation of these
large transcripts is initiated only through a
5'-terminal cap-mediated step and that initiation
at internal sites does not occur. These large
transcripts shouldproveuseful for studies aimed
at understanding the molecular basis of
site-specific recognition of mRNA by eucaryotic ri-bosomes.
ACKNOWLEDGMENTS
We aregratefultoBernard Lipinskasand Andrew Peterson forexcellenttechnical assistance.
This work wassupported in partby Public Health Service grant1R01 GM23853-01 from the National Institute of Gen-eralMedical Sciences. W.B.was arecipientofaSwiss National Science Foundationpostdoctorialfellowship.
LITERATURE CITED
1. Aloni, Y., R.Dhar, 0. Laub, M. Horowitz, and G. Khoury. 1977. Novel mechanism for RNA maturation: theleader sequences of simian virus40mRNAare not transcribed adjacent to the coding sequences. Proc. Natl. Acad. Sci. U.S.A. 74:3686-3690.
2. Ball, L. A., and C. N. White.1976.Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 73:442-446.
3. Berget, S.M., C. Moore, and P. A. Sharp. 1977.Spliced
segmentsatthe 5'-terminus of adenovirus2late mRNA. Proc.Natl. Acad. Sci. U.S.A. 74:3171-3175.
4. Bonner, W. M., and R. A.Laskey.1974. Afilmdetection method fortritium-labeledproteins and nucleic acids in
polyacrylamide gels.Eur. J. Biochem.46:83-88. 5. Boone, R.F., and B. Moss.1977.Methylated 5'-terminal
sequences ofvaccinia virus mRNA species made invivo at early and late times after infection. Virology 79:67-80.
6. Boone,R.F., and B. Moss.1978.Sequencecomplexity andrelative abundance of vacciniavirusmnRNA's syn-thesized in vivo and in vitro. J. Virol. 26:554-569. 7.Bossart,W.,D.L.Nuss,and E. Paoletti.1978.Effect
of UV irradiationon theexpressionofvaccinia virus geneproductssynthesizedinacell-free systemcoupling
transcription and translation. J. Virol. 26:673-680. 8. Chow,L.T.,R.E.Gelinas,T. R.Broker, and R. J.
Roberts. 1977. An amazing sequence arrangementat the 5'-ends of adenovirus 2 messenger RNA. Cell 12:1-8.
9. Ensinger, M. J., S.A. Martin, E. Paoletti, and B. Moss. 1975. Modification of the5'-terminusof mRNA by solubleguanylylandmethyltransferasesfrom vac-ciniavirus.Proc.Natl.Acad.Sci.U.S.A. 72:2525-2529. 10.Geshelin, P., and K. L. Berns. 1974. Characterization andlocalization ofthenaturallyoccurring cross-linksin vaccinia virus DNA. J.Mol.Biol. 88:785-796.
VOL. 28,1978
on November 10, 2019 by guest
http://jvi.asm.org/
916 BOSSART, PAOLETTI, AND NUSS
11. Hickey, E. D., L. A. Weber, and C.Baglioni. 1976. Inhibition of initiation of protein synthesis by 7-meth-ylguanosine-5'-monophosphate. Proc. Natl. Acad. Sci. U.S.A. 73:19-23.
12. Hickey, E. D., L. A. Weber, and C.Baglioni. 1978. Nuclease activity in preparations of creatine phospho-kinase: effect on mRNA stability. Biochem. Biophys. Res. Commun. 80:377-383.
13. Joklik, W. K. 1962. The preparation and characteristics of highly purified radioactivity labeled poxvirus, Biochim. Biophys. Acta 61:290-301.
14. Kaempfer, R., H. Rosen, and R. Israeli. 1978. Trans-lational control: recognition of the methylated 5' end andaninternal sequence in eukaryotic mRNA by the initiation factor that bindsmethionyl-tRNAf"e'. Proc. Natl. Acad. Sci. U.S.A. 75:650-654.
15. Kates, J., and J. Beeson. 1970.Ribonucleic acid synthe-sis invaccinia virus. I. The mechanism of synthesis and release of RNA in vaccinia cores. J. Mol. Biol. 50:1-18. 16.Kates, J.,and J. Beeson.1970.Ribonucleic acid synthe-sis invaccinia virus. II. Synthesis of polyriboadenylic acid. J.Mol. Biol. 50:19-33.
17. Klessig, D. F. 1977. Two adenovirus mRNAs have a common5'-terminalleader sequence encoded at least 10Kb upstream from their main coding regions. Cell 12:9-21.
18. Lodish, H.F. 1976. Translational control of protein syn-thesis. Annu. Rev. Biochem. 45:39-72.
19.Lowery,C., and J. P. Richardson. 1977. Characteriza-tionof the nucleosidetriphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor P.I. Enzymatic properties and effects of inhibitors. J. Biol. Chem. 252:1375-1380.
20. Lowery,C., and J. P. Richardson. 1977. Characteriza-tion ofthe nucleoside triphosphate phosphohydrolase (ATPase). Activity of RNA synthesis termination factor P.II.Influence of synthetic RNA homopolymersand random copolymers on the reaction. J. Biol. Chem. 252:1381-1385.
21. Mans, J. R.,and G. D. Novelli. 1961. Measurement of theincorporation of radioactive amino acids into pro-tein by a filter-paper disc method. Arch. Biochem. Biophys.,94:48-53.
22. Martin,S. A., and B. Moss. 1975. Modification of RNA by mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J. Biol. Chem. 250:9322-9329.
23. Moss, B., A. Gershowitz, C. M. Wei, and R. Boone. 1976.Formation ofthe guanylylated and methylated 5'-terminus ofvaccinia virus mRNA. Virology 72:341-351. 24. Nevins, J. R., and W. K. Joklik. 1975. Poly(A)
se-quences ofvacciniavirusmessenger RNA: nature, mode ofaddition and functionduring translation in vitro and in vivo.Virology 63:1-14.
25. Nuss, D.L., and E.Paoletti.1977.Methylgroupanalysis
ofvirion-associatedhigh-molecular-weight RNA syn-thesized in vitro by purified vaccinia virus. J. Virol. 23:110-116.
26. Paoletti,E.1977.Invitrosynthesisofa
high-molecular-weight virion-associated RNA by vaccinia. J. Biol. Chem. 252:866-871.
27. Paoletti, E. 1977.The high-molecular-weight virion-as-sociated RNA of vaccinia:apossibleprecursorto8-12S mRNA. J. Biol. Chem. 252:872-877.
28. Paoletti, E., and L. J.Grady. 1977. Transcriptional
complexity of vaccinia virus in vivo and in vitro. J. Virol. 23:608-615.
29. Paoletti, E.,and B. R.Lipinskas. 1978.Soluble endo-ribonucleaseactivity from vaccinia virus: specific cleav-ageofvirion-associatedhigh-molecular-weightRNA. J. Virol. 26:822-824.
29a.Paoletti, E.,and B. R.Lipinskas.1978.Therole of ATP in the biogenesis of vaccinia virus mRNA in vitro. Virology 87:317-325.
30. Pelham, H. R. B.1977.Use ofcoupled transcriptionand translation to study mRNA production by vaccinia cores.Nature(London) 269:532-534.
31. Pelham,H. R.B., J. M. M.Sykes,and T.Hunt.1978. Characteristics ofacoupledcell-freetranscriptionand translation system directed by vacciniacores.Eur.J. Biochem. 82:199-209.
32. Roberts, B. E., and B. M. Paterson. 1973. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA inacell-free system from commercial wheatgerm.Proc.Natl.Acad.Sci. U.S.A. 70:2330-2334. 33. Shafritz,D.A.,J.A.Weinstein,B.Safer,W.Merrick,
L.A.Weber,E. D.Hickey,andC.Baglioni.1976. Evidence for the role ofm7G-phosphategroupin rec-ognition ofeukaryoticmRNAbyinitiation factor IF-M3. Nature(London)261:291-294.
34. Weber,L.A.,E.D.Hickey,D.L.Nuss,andC.
Bag-Honi.1977.5'-terminal7-methylguanosineand mRNA function: influence ofpotassiumconcentrationon trans-lation in vitro. Proc. Natl. Acad. Sci. U.S.A. 74:3254-3258.
35. Wei, C. M., and B. Moss. 1974.Methylationofnewly synthesized viral messenger RNA by an enzyme in vaccinia virus. Proc. Natl. Acad. Sci. U.S.A. 71:3014-3018.
36. Wei,C. M., and B. Moss.1975.Methylatednucleotides block 5'-terminus of vaccinia virus messenger RNA. Proc.Natl.Acad. Sci. U.S.A.72:318-322.
J. VIROL.
![FIG. 2.amidetranslationtemundertionpurifiedcpm.uctsmRNAmargin.RNAresis.known AutoradiographofanSDS-polyacryl- gel after electrophoresis of translation prod- synthesized in a coupled transcription-transla- system or in response to exogenous 8 to 12Sor HMW RNA.[3S]methionine-labeled products synthesized in the coupled sys- (lane C) aspreviously described (3) or synthesized standard translation conditions in response to 8 to 12S mRNA (lane A) or purified HMW (lane B) were analyzed by SDS-gel electropho- Each well received 500,000 acid-precipitable The migration positions of marker proteins of molecular weight are indicated on the left The exposure time was 4 days.](https://thumb-us.123doks.com/thumbv2/123dok_us/1522305.104819/3.505.76.218.350.536/amidetranslationtemundertionpurifiedcpm-uctsmrnamargin-autoradiographofansds-electrophoresis-transcription-aspreviously-synthesized-precipitable.webp)
![FIG. 4.pg/ml)finalmRNA.withtems[3H]leucinezeroandaddedaddedSymbols:inallowedadditionleucinetherunoffcine100%o the Incorporation of amino acids into protein time after addition of ATA to translation sys- programmed by either HMW RNA or 8 to 12S The translat](https://thumb-us.123doks.com/thumbv2/123dok_us/1522305.104819/7.505.63.218.60.264/finalmrna-withtems-leucinezeroandaddedaddedsymbols-inallowedadditionleucinetherunoffcine-incorporation-translation-programmed-translat.webp)