Domains required for assembly of adenovirus type 2 fiber trimers.
Full text
Figure
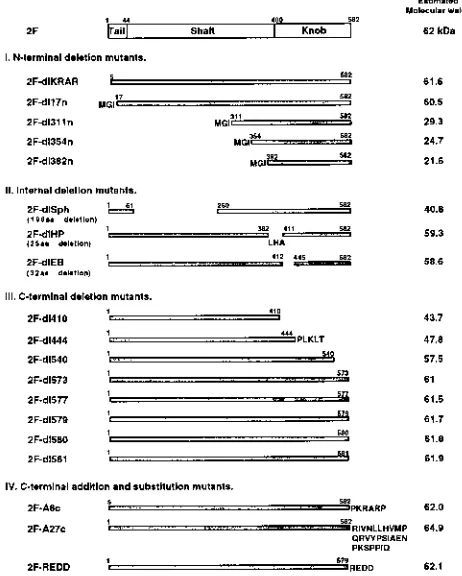
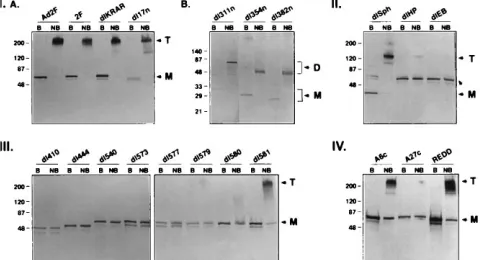
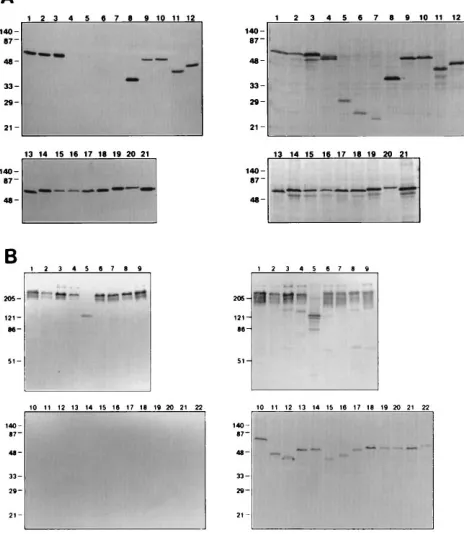
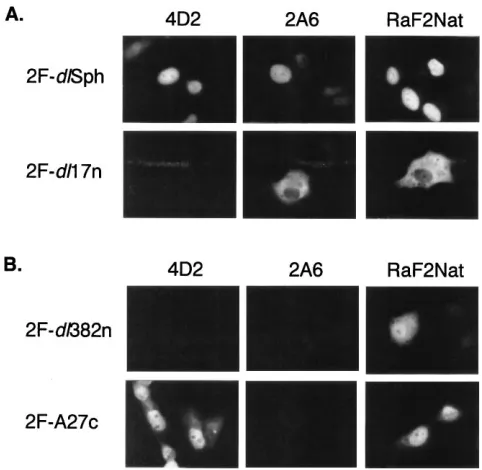
Related documents
The large proportion of amino acid changes in the NAs and internal gene products of the Hong Kong H5N1 isolates indi- cates that these viruses are evolving rapidly and therefore
Within the last 30 years the technical emphasis has focused on the use of tissue expanders with implants, latissimus dorsi myocutaneous transfer, and the transverse rectus abdominis
Experiments in which BFA was used to block transport from the ER illustrate that class II molecules do not associate intracellularly with ER-resident vSag1, and the rapid
These data reveal a novel class of HBV variants with HNF1 binding sites in the core promoter which are characterized by a defect in hepatitis B e antigen expression,
Lanes 1 to 7, HIV-1 virions from peripheral blood of HIV-1-infected individuals; lane 8, HIV-1-seronegative blood plasma; lane 9, HIV-1-seronegative sample and total cellular DNA
In the most rapid mode, this can be accomplished as follows: day 1, transform donor plasmid into competent cells harboring the shuttle vector and helper plasmid; day 2,
upstream elements that are required for efficient use of the downstream poly(A) addition site have been identified in the adenovirus Li site (19), the simian virus 40 late site
Comparison of the polymerase activity in P15 fractions from Sindbis virus (SIN)- and Semliki Forest virus (SFV)-infected cells indicated that both had similar kinetics of viral