0022-538X/80/03-1013/13$02.00/0
Number and Location of Mouse Mammary Tumor Virus
Proviral
DNA in
Mouse DNA of Normal Tissue and of
Mammary
Tumors
BERNDGRONERAND NANCY E. HYNES*
Swiss Institutefor ExperimentalCancerResearch, Lausanne,Switzerland
The Southern DNA filter transfer technique was used to characterize the
genomiclocation of themousemammary tumorproviral DNA in different inbred
strainsof mice. Twoof the strains (C3H and CBA) arosefrom a crossofaBagg
albino (BALB/c) mouseandaDBAmouse.Themousemammary tumor
virus-containing restriction enzyme DNAfragments ofthese strains had similar
pat-terns,suggesting that the proviruses of these miceareinsimilar genomic locations.
Conversely, the pattern arising from the DNA of the GR mouse, a strain
genetically unrelatedtotheothers, appeared different, suggesting that itsmouse
mammary tumorprovirusesarelocatedindifferent genomic sites. Thestructure
of another gene, thatcoding for
fl-globin,
wasalso compared. The micestrainswhichwestudiedcanbecategorizedintotwoclasses,expressing eitherone or two
,8-globin
proteins. The macroenvironment of thefl-globin
geneappeared similaramongthe mice strainsbelongingto onegenetic class. Female mice of the C3H
strainexogenously transmitmousemammary tumorvirus viathemilk, and their
offspring have a high incidence ofmammary tumor occurrence. DNA isolated
from individual mammary tumorstaken from C3Hmice or from BALB/c mice
foster nursedonC3H motherswasanalyzed by theDNAfilter transfertechnique.
Additionalmousemammary tumorvirus-containing fragmentswere found in the
DNAisolated from eachmammary tumor. These proviral sequences were
inte-gratedintodifferent genomic sitesin each tumor.
Mouse mammary tumor virus(MMTV) is an
RNA-containing virus which causes mammary
tumorsinmice.Incertaininbred strains of mice
the virus isexogenouslytransmitted tothe
off-spring via the milk. Animals which receive this
milk-bornevirus haveahigh incidence of
mam-mary tumor occurrence.Inaddition, mice
trans-mit MMTV-specific information to their
off-spring via the germ cell. This endogenously
transmittedMMTVispresent asproviralDNA
integrated into the genomicDNAofallcells of
themouse (2,23,27).
All tested inbred strains of mice contain
en-dogenously transmitted MMTV proviral
se-quences (22, 25, 35, 44). The origin and the
possiblefunction of proviralDNA remain
spec-ulative. Inallbut the GRmouse strain (3) the
endogenously transmitted MMTV proviral
se-quences appear to have norolein early mam-marytumoroccurrence. One hypothesisis that these proviral sequences arise from the viral
infectionof the germcell ofthe animal (43). A
comparative study ofthe genomic locations of
the endogenous MMTV proviruses in different
inbred mouse strains may be informative in
de-termining their origins.
Mammary tumors arising in mice that were
exogenously infected by milk-borne virus have
been shownbyhybridizationanalysestocontain
not only the endogenous proviruses but also
additional proviral copies (22, 25). These are
assumedtohave arisen from theexogenousviral
infection ofmammary gland cells, followed by
reverse transcription and viral integration into
genomicDNA.These mammarytumor-specific
proviralsequences arethoughttoberelatedto
mammary tumorformation. The control of their
expressionmay bebased,in part,ontheprimary
nucleotidesequencewithin and around the
pro-viralintegration site.Therefore,wehavebegun
acharacterizationof theintegration sitesof the
exogenously acquired MMTV proviruses of
mammarytumorDNA.
We used the Southern DNA filter transfer
technique (37) tostudyboth the genomic
loca-tionsofendogenousMMTVprovirusesand the
genomicintegration sitesoftheexogenously
ac-quired provirusesinmammarytumorDNA.We
comparedtheendogenous provirusespresent in
the liverDNAof seven differentinbred mouse
strains. Six of the strains have been bred in the UnitedStates,and ofthese,two(CBAandC3H)
1013
on November 10, 2019 by guest
http://jvi.asm.org/
1014 GRONER AND HYNES
canbedirectly traced from a BALB/candDBA mating (19, 38). The seventh strain, GR, has beenbred in Europe (26, 38). The resultssuggest
that the genomiclocations of the MMTV
pro-virusesin mouse strains of related geneticorigins
are similar. Conversely, the MMTV proviral
DNA in mousestrains of unrelated origins ap-pears to be located in different genomic sites. We determined theintegrationsites of the newly acquired MMTV proviruses in mammarytumor
DNAisolated fromindividualC3H orBALB/c mammary tumors. C3Hfemales produce milk-borne MMTV, and their offspring have a high incidence of mammary tumor occurrence. BALB/c mice transmit no milk-borne MMTV,
but when foster nursed on C3H mothers
(BALB/cfC3H), these animals also have a high
incidence of mammary tumor occurrence.
Ad-ditional MMTV-containing sequences were
identifiedin the mammary tumor DNA of both
strains. Theadditionalcopieswereintegrated at
differentgenomic sites in eachtumor.
MATERLALS AND METHODS Mouse strains and genetic origins. BALB/ cfC3H micebearingmammary tumorsandGR mice wereobtained from the Netherlands Cancer Institute, Amsterdam, The Netherlands. C3H/HeJ, BALB/c, CBA/H-T6,C57BL/6J, DBA/2J, and A/J micewere
obtained from colonies maintainedby the animalcare centerof the Swiss Institute forExperimentalCancer Research. All of the inbredstrains,with theexception
ofGR, have been bred in theUnited States. Approxi-mately60years agoaBagg albino femalewasmated with a DBA male. A number of different strains, includingC3H andCBA,canbe traceddirectlyfrom thismating. Inaddition,theBALB/cstrainoriginated
directly from the Bagg albinostock,and the A strain descended fromacrossofaBagg albino with another
mousestock. The C57BLstraincannotbetracedback directly from the above-mentionedcrossesbutit de-scends from stocks whichwereusedforbreedingother American strains (19, 38). Thus,five of these strains
areknowntobegenetically related,and thepossibility
that theC57BL strain is also relatedtoother Ameri-can-bred strains isnotunlikely.TheGR strain
origi-natedand has been bred inEurope(26, 38).
Purificationof MMTV viral RNA. GR cells from
a mousemammarytumorcelllineobtainedfrom K. Yamamoto weregrown in roller bottlesin Dulbecco-modifiedEaglemediumcontaining 10%fetal calf se-rumand 10-6M dexamethasone. The medium from confluent cultureswascollectedat24-hintervals and
clarifiedbycentrifugation,andthe viruswaspelleted byasecondcentrifugationfor45minat25,000 rpmin aBeckmanSW27rotor.The pelletedviruswas sus-pendedinTNE (0.1 MNaCl, 5mMEDTA,10mM
Tris-hydrochloride[pH7.5])andrepelletedina
Beck-manSW65rotorbycentrifugationfor30minat35,000
rpm. The viralpelletwasresuspendedinTNE,and proteinaseKandsodiumdodecylsulfate(SDS)were added to,respectively, 100,ug/ml and 0.5%. After
in-cubation for1hat370C,the viral RNA was extracted withphenol-chloroform and chloroform and precipi-tated by the addition of 2 volumes of ethanol. The RNA was dissolved in 10 mMTris-hydrochloride (pH 7.5)-5 mMEDTA and layered onto a sucrose gradient (10 to30%sucrose in 0.1MTris acetate[pH8.0],0.2% SDS) which was centrifuged for 2 h at 50,000 rpm at
20C
inaBeckman SW65 rotor. The 70S region of the gradient wascollected,theRNA wasprecipitated by theaddition of2volumes of ethanol, and the precipi-tate was collected bycentrifugation. The RNA was redissolved in 10 mM Tris-hydrochloride (pH 7.5)-5 mM EDTA and heat denatured for 2 min at700C.
Thesolution was layered onto a second sucrose gra-dient madeasdescribed above and centrifuged for 4.5 hat50,000 rpm inaBeckman SW65 rotor. The RNA sedimenting in the 20 to 40S region was collected, precipitated by the addition of2volumes of ethanol, and isolated by centrifugation. The RNA was dis-solved in sterilewaterand storedat-20°C.Isolation of DNA from mouse tissues. Mam-mary tumorsand organs were removed from the ani-mals, frozen inliquid nitrogen, and stored at-700C. One gram of tissuewasthawed in10mlof homogeni-zation buffer (0.3 M sucrose, 10 mM NaCl, 1.5 mM magnesium acetate, 10 mM Tris-hydrochloride [pH
8.0], 1 mM dithiothreitol, 0.1% Nonidet P-40) and
homogenized. The nuclei were sedimented by centrif-ugation for5min at3,000xg, thepelletwaswashed
3 times with 10 ml of homogenization buffer and suspended in 10 ml ofTNE, and proteinase K and SDSwereadded to,respectively,100,ug/ml and 0.5%. After incubation for 12 h at 370C, the DNA was extracted withphenol-chloroformandchloroform and
precipitatedby the addition of2volumes of ethanol. The DNA was collected by centrifugation and
sus-pendedin1mM EDTAto aconcentration of
approx-imately500,ug/ml.
Samples of DNA used in hybridization analyses
werebroughtto0.3NNaOH and incubatedat
1000C
for 10min todenature and decrease the size of the
DNA and to digest the nuclear RNA. The solution
wasneutralized, and the DNAwasprecipitatedby the addition of 2 volumes of ethanol and collected by
centrifugation. The pellet was dissolved in 1 mM EDTA, and the DNA concentrationwasdetermined by thediphenylamine analysis(6).
Preparation ofcDNA. MMTV-specific
comple-mentaryDNA(cDNA)wassynthesizedwith20 to40S
viralRNAasthetemplateinareaction mixture (0.1
ml) containing 0.05 M Tris-hydrochloride (pH 8.3);
0.05MKCI;8mMMgCl2;5mMdithiothreitol;1mM
dATP,1mMdGTP,1mMdTTP,0.1mM[3H]dCTP
(25.5Ci/mmol),or0.05mM[32P]dCTP(40Ci/mmol);
actinomycin D (100,ug/ml); RNA (15to 20,ug/ml); and avian myeloblastosisvirusreverse transcriptase
(100 U/ml).Thesynthesiswasprimed bydenatured fragments ofDNApreparedfrom Micrococcusluteus
DNA treated with DNase I asdescribed previously (40). TheDNAfragmentsat afinal concentration of
260 ,ug/ml were used to prime the synthesis. The
mixture wasincubated for2hat
250C,
andthe cDNAwasisolatedasdescribedpreviously (15, 16).
For determining the extent towhich the MMTV RNAwasreversetranscribedintocDNA, increasing
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
amounts of cDNA were hybridized with a fixed amountof "25I-labeledMMTV RNAprepared as de-scribedpreviously (41).AtacDNA/RNAratio of2:1,
50% of thehybridizable RNAwas ina cDNA-RNA
hybrid, suggestingthatthe cDNA isafairly homoge-neouscopyof the templateRNA. Ata 10:1 cDNA/
RNAratio,amaximum of 80% of the RNA became
resistanttoRNase A(datanotshown).Thepurityof the cDNAwasassessedby analyzingforthepresence
ofsequencescomplementarytorRNA,themostlikely
contaminant of the viral RNApreparation. Withthe RNA ifiter transfer technique (1) total polysomal
RNA isolated fromaGR cell linewasboundtopaper
andhybridized with MMTV cDNA. There was no
evidence ofhybridizationin theregioncorresponding tomouserRNA(see Fig.3 ofreference12).
cDNAhybridizationto total cellular DNA. The number of MMTV proviral genomes in thevarious
preparations ofcellular DNA was determined by a
cDNAexcesshybridizationtechnique (4).TheDNA
(1.6to
10.ug)
waslyophilized togetherwithamounts of 3H-labeled MMTV cDNA(specific activity,19cpm/ pg) rangingfrom 30to 300 pgtoachieve theratios indicatedin thefigures.Thelyophilizednucleic acidsweredissolvedin 0.24 Msodiumphosphatebuffer(pH 6.8)containing1mM EDTAataconcentration of 0.5
mg/ml,denatured at 100°Cfor 10mm,andannealed underparaffinoil at69°Cfor 48 h.Thetotal content of each tubewasremovedinto 0.5 ml of SInuclease
assaybuffer(0.3MNaCl,0.03 Msodiumacetate[pH 4.5],3mMZnSO4, denaturedM.luteusDNA[10,g/
ml]) andthe proportion of cDNA which hybridized
wasdeterminedasdescribedpreviously (15, 16).
With thistechnique (4)it is alsopossibleto
deter-mine thekineticcomplexity ofthecDNA, providing
anadditional measure-forthe representativeness of theprobe.Formeasuringthekinetics ofhybridization, samples containing 1,000cpmofcDNAwereremoved at various time intervals into 0.5 ml of Si nuclease
assaybuffer, andtheproportionofhybridizedcDNA
wasdetermined.Figure1shows thekineticsof
hybrid-ization of MMTVcDNA with the total DNAisolated fromaC3Hmouseliver.Fromtheinitial slope of the curvetheDot1/2value of 6.9x 10-4ml-Iswas
calcu-lated. The substitution of this valueinto Bishop and
Freeman'sequation3(4) yieldedavalue forthekinetic
complexity of theMMTV cDNA. Afteracorrection wasmadefor the saltconcentration (5), this valuewas
foundto be 2.35 x 10', a number whichrepresents approximately80%of thereportedmolecular weight
of MMTVviralRNA(10).Theseresultsalsosuggest thatthe cDNAproberepresents mostof theMMTV genome.
Restriction endonuclease digestion of DNA.
Restriction endonucleases HindIII and BamHIwere
purchasedfrom NewEnglandBiolabs,Beverly,Mass., EcoRI was purchased from Boehringer Mannheim
Corp.,WestGermany,and PstIwasagift of G. Fey.
DNA (20,ug) wasdigestedwith atwofoldenzyme
excessin the buffersspecified bythemanufacturers.
Thedigestion mixtureswereincubatedfor6hat37°C
andthen foranadditional 30 minat370C with RNase
(50 ug/ml) toremoveRNA.Finally,the reactionwas
terminatedbytheadditionofSDSto0.2%. Toensure
complete digestion, in some cases the mixture was a
6-m
4 4-z a u
z
w
2-CL
z
-500 a
0 0
300 S
-100 a
Q
a.
4 8 12 16 20
HOURS
FIG. 1. Kinetics of hybridization of 3H-labeled MMTVcDNA with C3H liver DNA. C3H liver DNA
(15pg) washybridizedwith 1.27ngofMMTV
[3H]-cDNA in 30itlof hybridizationbufferat69°C.Atthe indicated times2-,ul samples were diluted into Si
nucleasedigestion buffer, and thepercentcDNA hy-bridized was determined as described in the text.
Approximately1%ofthe cDNAwasresistantto
diges-tionwithS1nuclease,and this valuewassubtracted
fromeachpoint.
extracted withchloroform,the DNAwasprecipitated
bythe addition of 2 volumes ofethanol,thepelletwas
resuspended in the appropriatebuffer,andthe DNA
wasredigestedwithatwofoldexcessofenzyme.After
the digestionreaction,eachsampleof DNAwas pre-cipitated bythe addition of 2 volumes ofethanol.The
precipitateswerecollected andsuspendedin 20pl of
10%Ficoll-0.1%SDS-1mM EDTA-0.02%
bromophe-nolblue.
Electrophoresis, transfer,andhybridizationof
enzyme-digestedDNA.Restrictionenzyme-digested
DNA samples were electrophoresed on 0.8 or 1.0%
agaroseslabgels (slotsurface,5by10mm)madein36 mM Tris-30 mM sodium phosphate (pH 7.2)-i mM EDTA at 18 V for 18 h at roomtemperature. The
digestedDNA and the molecularweightmarkerswere
visualizedbyethidiumbromidestainingand photog-raphyunder UVlight.The DNAfragmentsinthegel
weretransferredtoanitrocellulose filter sheetbythe
blotting technique ofSouthern (37) as modified by
Ketner andKelly (18).Beforehybridization,the
mem-brane filter (Millipore Corp., Bedford, Mass.) was
washedtwice in4xSSC(lx SSC is0.15MNaCl plus
0.015 M sodiumcitrate) plusDenhardtsolution(9)at 65°C,with the second washcontainingdenatured M. luteusDNA(20ug/ml) and 3.3%(vol/vol) phosphate
buffer (0.5 M sodiumphosphate, 1.5% sodium
pyro-phosphate [pH 7.3]). Thefilterwasthen hybridized
by saturatingit withasolution of32P-labeled MMTV cDNA (specific activity, 50 x 106 cpm//ug), SSC, EDTA, SDS, anddenatured M. luteusDNAsothat
the final concentrations were,respectively, 25ng/ml,
4x, 10 mM, 0.1%, and 20 uLg/ml. The hybridization
mixture and the filter were incubated in asealable
plasticbagbetweentwoglass platesfor 20 hat65°C.
The filterwas thenwashed at65°C with0.1%SDS and 3.3% (vol/vol) phosphate buffer containing 2x
SSC, followedbysecond and third washes in which the 2x SSCwasreplaced bylxSSC and thenby0.1x SSC foratotalperiodof3h. Thedryfilterwasthen
* 0
I
e.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.504.254.451.55.187.2]1016 GRONER AND HYNES
exposed at-70°C to Kodak XR-5 film with an inten-sifying screen.
Preparation of and hybridization with nick-translatedmouse8-globin plasmid DNA. Plasmid pCR1containing themouse,B-globinstructural DNA sequences (34) was provided by B. Mach. The plasmid wasnick-translated with[32P]dCTP to a specific activ-ity of 3x107cpm/,ugasdescribed previously (46). The filters were prepared and hybridized as described abovewith theexception that the plasmid DNA was included at a final concentration of 100 ng/ml.
Isolation ofunintegrated MMTV viral DNA. HTC-M1 cells from a rat hepatoma cell line infected withMMTV(GR) (31) were obtained from K. Yama-moto. The cells were grown in Dulbecco-modified
Eagle medium, 10% fetal calf serum, and 10' dexa-methasone. The cells have been shown to contain unintegratedMMTV viral DNA (33) which was iso-lated as follows. Confluentcells wereremovedfrom
petridishes bytrypsinizationandwashed3times with cold phosphate-buffered saline. Approximately 109 cells weresuspendedin 50 ml of10mM
Tris-hydro-chloride(pH 8.0)-5 mM EDTA and disrupted witha
Potter homogenizer. The nuclei were collected by centrifugation, and thepelletwasreextracted in the samebuffer. Thesupernatantfractionscontaining the cytoplasmic, linear DNA molecules were pooled, and proteinase K andSDS were added to, respectively, 100 ,tg/ml and 0.5%. After extraction with phenol-chloroform and chloroform, the nucleic acids were precipitatedby the addition ofethanol,collected by centrifugation, and dissolved in2ml ofwater. Dena-tured M. luteus DNA (50 ,tg) was added, and the solutionwasbroughtto 0.1MNaCI-5mMEDTA-10 mM Tris-hydrochloride (pH 7.5). RNase, incubated for 10minat100°Cin 50 mM sodium citrate(pH5.0)
to removeDNase, wasaddedto 100,tg/ml, and the RNAwasdigested for2hat37°C.SDSwasaddedto
0.5%, and thedigestwasextracted with phenol-chlo-roform and chloroform. The aqueousphasewas
ap-pliedto aSephadexG-50column (0.5 by40cm)in10
mMTris-hydrochloride (pH 7.5),theexcluded volume waspooled,and thenucleic acidswereprecipitatedby the addition of2volumesofethanol. The DNAwas
collectedby centrifugationandsuspendedin 250
pi
of1 mM EDTA. The content of MMTV-specific
se-quences was determined by hybridizing increasing
amountsof this solution withanexcessof 3H-labeled MMTV cDNA under the conditions described above. The hybridization was begun after incubating each reaction for10minat100°C.
RESULTS
MMTVproviral number inmouse DNA.
The DNA of all tested inbred strains ofmice
containsMMTVproviralsequences (22,25, 35,
44). Beforedoingthe genomicmappingstudies
of the MMTV proviruses of different mouse
strains, we determined, by hybridization, the
number of MMTV proviruses endogenous to
BALB/c,C3H, andGR mice.BALB/cmice do
nottransmitMMTV in themilkandhavealow
incidence ofmammarytumor occurrence. C3H
mice have ahigh incidence ofearlymammary
tumor occurrence and transmit viral particles in
the milk. In the GR mouse a single dominant
gene, the Mtv-2 locus, controls the high
inci-dence ofmammary tumor occurrence (3). This
strain isan exceptionas the endogenous
provi-rusesof other testedmouse strains seem to play
norole in earlymammary tumor occurrence. We
also determined, by hybridization, the number
of MMTVproviruses inC3HandGR mammary
tumors.
Otherinvestigators havedetermined the
num-ber of MMTV proviruses in organ DNA and
mammary tumor DNA isolated from various strains of mice by measuring the kinetics of
hybridization of radioactivecDNA to an excess
of cellular DNA (22, 25, 44). We employed a
cDNA excesshybridizationtechnique developed
by Bishop and Freeman (4) to determine the
amount of MMTV proviral genes in the DNA
isolated from livers and mammary tumors of
different mice. With this method both the
com-plexity of the cDNA probe and the comcom-plexity
of thehybridizable DNAcanbedetermined.
Thenumber of proviral genesinthe DNA of
eachmousestrainwascalculatedbydetermining the amount of cDNAhybridized at increasing
ratios of cDNAtoDNA. Thisvaluewas
repre-sentedaspicogramsof cDNAhybridper 100,tg
of DNA. The plateauvalue reached when the
cDNA-to-provirus ratio was about 10 to 20:1
reflects theamountofprovirusin the total
cel-lular DNA. This method has previously been
describedfordeterminingthe number ofglobin
genes in various cellular DNAs (4, 28). The
results of suchhybridizationsare showninFig.
2. Knowing thehybridizationplateauvalue and
the molecularweightofonegenomeequivalent
of MMTVproviralDNAandcorrectingfor the
fact that approximately 80% of the viral RNA
complexity is present in the cDNA (as
deter-mined above), we were able to calculate the
number of MMTVproviral copies. These
num-bers and the calculationsarepresentedinTable
1.C3H andBALB/cliver DNA andC3H
mam-mary gland DNA contained approximately 7
copiesof MMTVproviralDNAper
diploid
ge-nome, whereasC3HmammarytumorDNA
con-tained8.5 to 9copies. GR liverDNAand
mam-mary tumor DNA contained approximately 10
copiesof MMTVproviralDNAperdiploid
ge-nome. These data agree with previously
pub-lished work which shows that C3H mammary
tumors contain increased amounts of MMTV
proviralDNA(21, 22,25).
Restriction enzyme analysis of the
loca-tionof theendogenous MMTV
proviruses.
The digestionof the total genomic DNA with
restrictionenzymes,the separationof the
frag-ments by agarose gel electrophoresis, and the J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
MMTV PROVIRAL DNA: GENOMIC NUMBER AND LOCATION
AB
0 ,
. . I I
0 00
@
4O.2-648-220 16 2
1017
4 8 12 16 20 4 8 12 16 20
ngcDNA/100pg DNA
FIG. 2. Saturationhybridization ofMMTV[3H]cDNAwithmouseDNA. DNA(1.6to10pg)waslyophilized
with amountsofMMTV[3HJcDNA ranging from30 to300pgtoachieve the indicated ratios. Thenuckeic acidsweresuspendedinhybridization bufferataDNAconcentrationof0.5mg/mLAfterincubationat69°C for48h, the entire mixturewasdilutedinto Si nuclease digestion buffer,andthepercent cDNAhybridized
wasdetermined.The countsperminutehybridizedwereconverted topicograms of hybridper100pgof input
DNA. The DNAwasisolatedfrom(A) C3Hliver(0), C3Hmammarygland (0),BALB/cliver(x); (B)C3H
mammarytumor;(C)GRliver; (D)GRmammarytumor.
TABLE 1. AmountofMMTV-specific DNA in mouse tissuesasdetermined byhybridization with MMTV
cDNA
pgof MT
cDNA MMTV
Mouse Tissue hybrid gene
nor
strain per 0
gene
nipo.d
1tg
of per diploidDNA genome
C3H Liver 550 6.9a,6.3b
Mammary gland 550 6.9,6*0b Mammary tumor 680 8.6
BALB/c Liver 550 6.9
GR Liver 790 10.0
Mammary tumor 860 10.7
aDetermined as
follows:
the molecular weight ofoneproviral equivalentis6.0 x10'and one mouse cell has DNAwithamolecular weight of 3 x 1012;
there-fore, oneMMTV genome is 2 x 10-4% of the total genome. 100 ugof DNA containing one MMTV
ge-nomehas 200 pg of genebut only 100 pg of + strand. Aplateauvalue of550pg of cDNAhybrid per 100ug of DNA isequivalent to 687 pg of gene per 100
iLg
of DNA asthe cDNAprobe represents approximately80%ofthe viralgenome. 687 pg of gene is equivalent
toapproximately6.9copies of MMTV per diploid cell.
bDetermined from the kineticsof MMTV cDNA
hybridization asdescribed in the text and in Fig. 2.
transfer of the DNA to anitrocellulosefilter (the Southern DNA filter transfer technique [37])
followed by hybridization with radioactive
cDNA allowed an analysis of the restriction
enzymesitesinproviralDNA and theflanking
host DNAsequences.We used thesetechniques
to analyzethe location of the MMTVprovirus in DNA isolatedfrom the livers andmammary tumorsof different inbred strains of mice. The
restriction enzymecuttingsites of the MMTV
viral DNAhave been determined.Unintegrated viral DNA hasbeen isolated from rathepatoma cells infected with MMTV and digested with
various restrictionenzymes(36;N. E.Hynes,B.
Groner, H. Diggelmann, R. Van Nie, and R.
Michalides, Cold SpringHarbor Symp. Quant.
Biol.,inpress).EcoRI andHindmcut the
pro-virus once close to the center of the genome,
whereas BamHI cuts it twice and yields an
internalfragmentofapproximately1.0kilobases
(kb) alongwith two endfragments.Upon diges-tion of the cellular DNA with EcoRI or with
HindIIl, each provirus yields two fragments
whosemolecularweights correspondto the dis-tancebetween the restrictionenzymesiteonthe
provirus and the nearest site in the cellular
DNA. This is also the case for digestion with
BamHI. An additional fragment of approxi-mately1.0kbcontaining only proviralDNAmay
alsoarise fromaBamHIdigestionof the
endog-enousproviruses.
For analyzing the genomic locations of the
endogenous MMTV proviruses, DNA isolated
from theliver,anorganwhich containsonlythe
endogenous proviruses, was digested with
EcoRI, HinduII, and BamHI. The DNA
frag-mentsresulting from the digestionofthe liver VOL. 33, 1980
800-
600-4c
z
a
400-co
0
200-a
0
=
800-z
4
Z
600-f
400-
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.504.52.244.322.484.2]1018 GRONER AND HYNES
DNAof various strainsof micewerefractionated
by electrophoresis and transferred to
nitrocel-lulose filters,andprovirus-containingfragments
wereidentified. Theresults of this analysisare
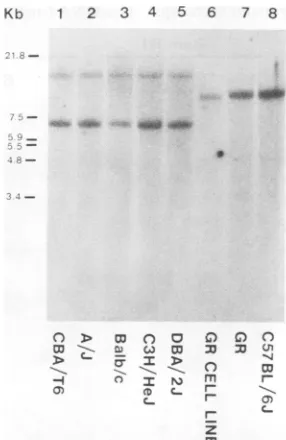
shown in Fig. 3.
For facilitating the comparison, the sizes of
theMMTV-containing EcoRIfragments of the
different strains are listed in Table 2. Such a
comparisonreveals thatamongtheseven
differ-ent strains, the BALB/c, CBA, and C57BL
strains had almost identical EcoRI fragments,
the C3H strain shared fragments with the
BALB/c mouse and the DBA mouse, andthe
DNA from the A mouse also had fragments commontothe BALB/cmouseDNAandtothe DNAofotherexaminedmice. Thesimilarityin
the EcoRIpatternsoftheCBA, BALB/c, and
Hind 3
C57BL DNA ledustodigestthese DNAs with
anotherenzyme. Lanes 2 and 4 of Fig.3reveal
thatthe MMTV-containing HindIII fragments
of CBA and BALB/c DNA were identical,
whereas the C57BL DNA yielded thesame
pat-ternwithtwoadditionalbands. Thepatternof
MMTV-containing EcoRI fragments arising
from theDNAof theGRmouseappearedtobe
different from that ofthe other strains tested.
Seven ofnine fragmentsidentifiedin Fig.3and
listed in Table 2 did not correspond to those
found in either the DBA DNAorBALB/c DNA.
The limits of resolution ofthe method donot
allow usatthis pointtoruleoutthepresenceof
additional EcoRIfragments,especiallythose of
thesize of about 7kb.This leavesthepossibility
thatadditionalEcoRIfragments ofsimilar
mo-Bam Hi Eco RI Eco Rl
1 2 Kb 3 4 5 Kb 6 7 8 9 10 11 Kb 12 13 14 15 16
...
-ivr-fteidctd resrit\'ion
bromide.ie.
... _~~~~~~~~~~~~~~~~~~~W5_ Q
[image:6.504.69.458.241.453.2]7 ;
FIG. 3. AnalysisoftheendogenousMMTVprovirus in theDNAof differentstrainsofmice. DNA was isolatedfromtheliversoftheindicated strainsofmice anddigestedwithdifferentrestriction endonucleases.
Thesampleswereelectrophoresedinan0.8%agarosegel, transferredtoanitrocellulosefilter,andannealed
with32P-labeled MMTV cDNA asdescribed in the text. The markers indicate theposition in this and in
subsequentfigures ofthefragments ofEcoRI-cut lambdaphageDNAdetectedbystainingwith ethidium bromide.
TABLE 2. MMTV-containingDNA
fragmentsa
resultingfromtherestrictionenzymedigestion ofthe liverDNAof differentmousestrains
Strain Enzyme Kboffragments
DBA/2J EcoRI 14.5 13.5 11.5 9.7 8.7 7.7 7.4 6.7 6.2 5.9 5.4 4.2
BALB/c EcoRI 14.5 9.6 7.5 7.15 6.7 6.1
C3H/HeJ EcoRI 14.5 7.5 6.7 6.2 5.9 5.4 4.2
CBA/H-T6 EcoRI 9.6 7.5 7.15 6.7 6.1
C57BL/6J EcoRI 10.3 9.6 7.7 7.5 7.15 6.7 6.1
A/J EcoRI 14.5 8.85 7.7 6.7 6.1 5.4 4.3
GR EcoRI 22.8 16.0 11.5 11.0 10.0 8.1 7.8 6.9 6.7
C3H/HeJ HindIII 22.0 16.5 12.0 8.2 7.0 6.0 4.3 4.15 3.7
C3H/HeJ BamHI 21.8 10.5 7.7 7.0 6.1 4.8 4.6 4.2
aOnlythefragmentslargerthan 3 kbarelisted.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.504.66.463.544.640.2]MMTV PROVIRAL DNA: GENOMIC NUMBER AND LOCATION
lecular weightwere presentin the DNA ofGR
and DBA miceorofBALB/c mice.
Theresultscanbestbeinterpreted in lightof
the genetic origins of the different mice. As
previously noted,theC3HandCBA strainscan
betracedtothe cross of aBALB/cmousewith
aDBA mouse.EachMMTV-containingEcoRI
bandwascorrelated withaband presentinone
of the parents. The DNA from A mice, which
are related to the BALB/c strain, also had
MMTV bands in common with the BALB/c
DNAaswell aswiththeDNAofDBAandC3H
mice. The DNA from C57BL mice, which can
also betraced backtorelatedstocks,had, with
theexception oftwoadditionalbands, thesame
pattern of MMTV-containing fragmentsas the
BALB/cmouse.Theresultssuggestthatinmice
of relatedgeneticoriginsthepatternsof
MMTV-containing EcoRI fragments are similar,
sug-gesting thattheprovirusesarelocatedinsimilar
genomic environments. The pattern of
frag-ments arising from the GR mouse, which is
genetically unrelated to the others, appeared
different. Inaddition, the patterns of
MMTV-containing EcoRI fragments arising from two
other European bred strains (RIII and 020)
have also been identified and differ from each
other and from theGR strain (Hyneset al.,in
press).Thissuggeststhat the MMTVproviruses
in thesestrainsarelocatedindifferentgenomic
environments.
The interpretation of the preceding results
dependsupon two conditions. Onecondition is
thatthe integrated viral DNA is colinearwith
the unintegratedspecies. Thiscanbe testedat
least fornewlyintegratedmilkvirus-specific
pro-viral DNA. The resultssuggesting that this is
true arepresented below. The second condition
isthat whereastherestrictionenzymedigestion
patternof the viralDNAsynthesizedfrom
milk-borne MMTVisknown(36), the restriction
en-zyme digestion pattern for the MMTV
provi-ruses endogenous to different mouse strains is
notknown. Atthepresenttime thisproblem is
difficulttosolveasthere isnoreadilyavailable
sourceof viralRNA or DNAfromstrains, such
asBALB/c orC57BL/6J, withalow incidence
ofmammary tumor occurrence. Some evidence
forthe relatednessoftheendogenous proviruses
canbeobtainedfromthe resultsofthe BamHI
digestionpatterns. Figure 3, lanes 6 through 8,
shows that at least some of theproviruses
en-dogenousto GR, BALB/c, and C3H/HeJ mice
containedthe internalBamHI fragment of
ap-proximately1.0kb which also arose from a
diges-tion of the isolated viral DNA.However, another smaller fragment also arose from the BamHI
digestion ofGRaswell asC3H DNA. In
addi-tion, Cohen et al. (7) have published a pattern
ofMMTV-containing fragments resulting from
thedigestion of theBALB/c DNA with BamHI
in which the 1.0-kb fragment is absent. These
results suggest that these may be endogenous
proviruses with different BamHI sites. These
resultswill befurtherdiscussedbelow.
Analysis of the location of the
/8-globin
gene indifferent strains of mice.Inthe
pre-ceding section we analyzed the genomic
loca-tions ofthe MMTV provirusesin anumber of
differentmice. Weextended thiscomparisonto
another gene, that coding for
f3-globin.
LiverDNA isolated from the different mice was
di-gested with EcoRI, an enzyme which doesnot
cutwithin the coding orintervening sequences
of the
fi-globin
gene (34, 42),and,followinggelelectrophoresis, the ,B-globin-specific fragments
wereidentified. The resultsareshowninFig.4.
C3H/HeJ, BALB/c, A/J, CBA/H-T6, and
DBA/2J mice exhibit the hemoglobin diffuse
gene product (38) and contain two linked
,B-globingenes(29).Lane 3shows thetwo
,B-globin-containing restrictionfragments of the BALB/c
DNAwhichmigrateatapproximately 14and7
kb, as previously identified (42). This pattern
appears identical to that obtained with DNA
isolated from the livers of CBA/H-T6, A/J,
Kb 1 2 3 4 5 6 7 8
WXc; w ;
Co > CcoI> MUl) -4
cr
~
0 coz
[image:7.504.277.420.347.567.2]m
FIG. 4. Analysisofthe
fl-globin
gene in the DNAof different strains ofmice. DNA wasisolated from the liversof the indicated strains of mice, digested
withEcoRI,andanalyzedasdescribedin the legend toFig. 3, except that thefilters wereannealed with
32P-labelednick-translatedpCR1 plasmidDNA
con-taining ,B-globin-specific sequences. Thesizes of the
fragmentsinlanes 1 through 5 are 14.0 and7.0kb; in lanes 6through8thefragmentis10 kb.
1019 VOL. 33,1980
on November 10, 2019 by guest
http://jvi.asm.org/
1020 GRONER AND HYNES
C3H/HeJ andDBA/2J mice (Fig. 4; lanes 1, 2,
4,and 5). TheC57BL/6J andGR mouse strains
contain only one
fl-globin
chain (38). Lanes 7and 8 show that the
f,-globin
gene from bothstrains was contained inan EcoRI DNA
frag-mentofapproximately10kb. Ourresults suggest
that in strainsof different genetic origins such
asGR and C57BL/6J themacroenvironmnentof
the
f,-globin
geneissimilar.Analysis of MMTV proviral integration
sites in mammary tumor DNA. C3H mice
transmit MMTV in the milk and have a high
incidenceof earlymammary tumor occurrence.
BALB/c mice transmit no milk-borne MMTV
and have alow incidence oftumorformation.
However,whenBALB/c micearefosternursed
on C3H mothers (BALB/cfC3H), their
mam-mary tumor occurrencebecomes high.The
hy-bridization ofmammary tumorDNA has
dem-onstrated that there isanincreasein theamount
of MMTVproviruses. The extra copies are
as-sumed to derive from the exogenous-virus
ac-quired via the milk. It may be possible that
mammarytumorformation is relatedtothe site
ofintegration of the exogenous provirus in the
cellular DNA.Onecanpartiallycharacterize the
integration sites byanalyzing the DNA isolated
from differentmammary tumorswiththe DNA
filter transfer technique. The DNA from
individ-Bam Hi Eco Rl
ual C3H mammary tumors was isolated and
digestedwithdifferent restrictionenzymes.The
fragmentscontainingMMTV-specificsequences
weredetected, and the resultsareshowninFig.
5. It can be seen that the DNA of individual
C3Hmammary tumorscontained notonly the
restrictionfragmentspresentin the liver DNA,
but for each tumor it was possible to identify
additional MMTV fragments. Thesenewbands
areindicatedby the linesonthe left side of the
individual lanes. Itwas notpossible in allcases
toidentifynewMMTV-specific bands from the
digestion with one enzyme. This might have
beenduetothe coelectrophoresis of
tumor-spe-cificbands with those deriving from the
endog-enousproviralsequences.Inthe EcoRI digest of
tumor 3DNA (lane 6)nonewbandswereseen.
However, a new band was evident when the
same DNAwas digestedwithBamHI (lane 2).
If one copy of the provirus was additionally
integrated in the mammary tumor DNA, two
new MMTV-specific bands wereexpected. We
observed this for theEcoRI digestion oftumor
1DNA(lane8) and for theHindIIIdigestionof
tumor 4 DNA (lane 11). Other tumor-specific
bands in tumors 2, 3, and 5 might have been
maskedby theendogenousproviruses.
The DNAisolated from fourseparate tumors
removed fromoneBALB/cfC3Hmouse was
di-Hind 3
Kb 1 2 3 4 5 6 7 8 9 10 11
5
e-*P
,-;i
Kb 12 13
r.
4is -_
_E
w...*=_
. .* ; t t A,
'. '* .
.' _
_w--'Xt' F
.*.
FIG. 5. AnalysisofMMTVproviralsequences in theDNAofindividual C3H mammarytumors.DNAwas isolated from five individual C3H mammary tumors (tl through t5), digested with different restriction endonucleases, andanalyzedasdescribed in thelegendtoFig.3.The markerstotheleft oflanes2, 3,5, 7, 8, 9, 11, 12,and13indicatethepositionof tumor-specific MMTV-containing fragmentsidentifiedinthe DNA isolatedfromthedifferenttumors.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
gested with EcoRI, andthe ing MMTV-specific sequen
These results are shown inF
to the right, the bandsarisir
nous proviruses are indica
whereas the tumor-specific
cated by dotted lines. As w
C3H
tumors, eachBALB/cI
mor contained additional M ies.
It should be noted that
DNA was obtained from t]
kept in the Netherlands
whereas the BALB/cDNAu
13 of Fig. 3 was isolated fron
of the Swiss Institute for
E:
Research. TheMMTV DNA
arising from the SwissBALB
ing from the DNA of the A colony. Cohen et al. (7) ha
pattern
of MMTV-containin ments generated from BAL missing the 6.7-kb band andterns
shown in Fig. 6.Possil
these results could be that the original BALB/c stock w
that the Amsterdam colony(
sequences or that the BALI
zerland gained an MMTV
observation has also been i
and Varmus (8), who compa
Kb
21.8
-
7.5-5.9_
5.5-
4.8-
3.4-1 2 3 4
U."i
.,s__o
FIG. 6. Analysis of MMTVI the DNA of individual BALB/
mors. DNA isolated fromfour j
tumors removedfromone mou.
EcoRIand analyzed as descri
Fig. 3. In the schematic diagr bands arising from endogenous cated by solid lines, whereasti
specific fragmentsareindicatea
fragments contain- MMTV provirusesindifferent colonies ofC3H/
Ices were detected. St mice.
Pig.
6. In the scheme The results shown in Fig. 5 and 6 can beig from the endoge- summarized asfollows. First, although each
tu-Lted
by solid lines, mor contained at least one additional copy offragmnents are indi- MMTV proviral DNA, the extra bands in the
ras the case for the different tumor DNAs did not coelectrophorese.
FC3H
mammary tu- This suggests that exogenous proviralDNA can[MTV proviral cop- integrate into differentsitesinmammarygland
cell DNA. Second, in most cases theadditional
the BALB/cfC3H bands in the tumor DNAs were clearly visible,
he BALB/c colony although they were not of thesameintensity as
Cancer Institute, those specific for the endogenous proviruses.
ised for lanes 10 and Therefore, it can be concluded thatahigh
per-amiceof the colony centage of the cells ineach tumorcontained the
Kperimental Cancer additional MMTV provirusesintegrated into the
v fragment of 6.7 kb same site in the genomic DNA. The results also
B/c
colony wasmiss- suggest that each tumor originatedfromasmallmsterdam
BALB/c number of cells or from one singleMMTV-in-ve also obtained a fected cell.
.g EcoRI DNA frag- A comparison of the genomic locations of the
.B/c
DNA which is newly acquiredproviral copies inmammarytu-Iresembles the pat- mor DNA depends upon the assumption that
ble explanations for the integrated proviralDNAiscolinear with the
the proviral loci in unintegrated viral DNA. We tested this by
di-rere heterozygous or gesting the DNA isolated from a C3H mammary
of mice lost proviral tumor containing additionalMMTV proviral
se-B/c colony in Swit- quences with PstI, anenzyme which cleaves the
provirus. A similar proviral genome infiveplaces to yield six
frag-reported by Cohen ments of DNA which have beenorientated with
red the endogenous respect to the viral RNA genome (36). One end
fragment could not be detected because one
1 2 3 4 enzyme
recognition
sitewastoocloseto the endof the proviral DNA. If the newly integrated
MMTV is colinear with the unintegrated viral
DNA, then the digestion of mammary tumor
DNA with PstI should yield bands which
cor-respond to the internal PstI cutting sites,
whereas the fragment which is joined to the
- -
=-
- cellular DNAshould have analtered molecularweight.
The DNA froma C3H mammary tumor and
a C3H liver was digested with PstI, and the
fragments of DNA-containing MMTV
se-quences were identified. These were compared
to fragments arising from a PstI digestion of
unintegrated viral DNA isolated from
MMTV-infected cells. The results are shown in Fig. 7.
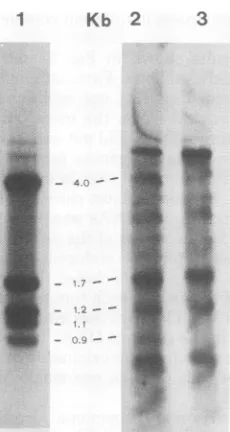
C3H mammary tumor DNA (lane 2) yielded
four fragments of 4.0, 1.7, 1.2, and 0.9 kb which
arose from internal PstI cutting siteson the viral
Droviral
sequences in DNA,whereas the1.1-kb
fragment
arising from.cfC3H mammary tu- the 3' end of the unintegrated MMTV viral
indiidual
mammary
DNA was notevident (compare lanes 1 and2).se wasdigested with
'bed
in the legend toThis
suggests that the newly acquired mammaryam on the
rlght,
the tumor-specific provirus is attached to thecellu-provirues
areindi-
lar DNA somewhere within this endfragment.
he additional tumor- Othermoredetailed restrictionenzymemapping
iby dottedlines. experiments ofnewly integrated mammary
tu-1021 VOL. 33,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.504.47.242.395.571.2]1022 GRONER AND HYNES
1 Kb 2 3
[image:10.504.104.219.65.281.2]§-4 -- F
FIG. 7. Analysis of MMTV(GR) viral DNA and C3HDNAdigestedwith PstI. Linear viral DNA (lane 1), C3H mammary tumor DNA (lane 2), and C3H liver DNA(lane 3) were digested with PstI and ana-lyzed asdescribed in the legend to Fig. 3, except that
a 1.0% agarosegel was used. The markers in the center indicatethe sizes of thefragmentsresulting
fromthePstI digestion ofMMTV(GR) viral DNA.
mor-specific proviruses (7) show that the
provi-rus is attached to the cellular DNA within a
region a few hundred base pairs from the ends
of the unintegrated viral DNA. These results
suggestthat newlyacquired proviruses are
co-linearly integrated.
Lane 2 reveals that C3H mammary tumor
DNAcontainedtwoadditionalfragments at 5.2
and 0.7 kb which strongly hybridized with
MMTV.These fragmentsarosefromthe
endog-enous proviruses as they were also present in
C3H liverDNA (lane 3). The intensityofthese
fragments suggests that they may have arisen
from intemalPstIsiteson theC3Hendogenous
proviruses. This leads to the conclusion that
C3Hendogenousproviruseshave differentPstI
cutting sites than doesviral DNA arising from
a milk-borne viral infection. In a recent
publi-cation (8),Cohen and Varmus havecome to a
similar conclusion fromastudyofthe
endoge-nous proviruses of the C3H strain and related
strains.
Digestion with PstI yielded fragments
com-prised of viral and cellular DNA andtherefore
shouldbe usefulin characterizing the genomic
location of proviral DNA. However, the bands
representing these fragments will be weaker than those representing the internal proviral
fragmentsastheyarisefrom only one provirus.
InFig. 7,lanes 2 and 3,itis possible to seefaint
bands which mightrepresent the endfragments,
but digestion with EcoRI, HindIII, or BamHI
yielded patterns whicharemore useful for
char-acterizing the genomiclocation of proviralDNA.
DISCUSSION
Inthispaper wepresentedthe results of
stud-ies of theMMTVproviralnumber and its
orga-nization within thecellularDNAisolatedfrom
organsand mammarytumorsof differentstrains
ofmice.
All strains of mice contain MMTV-specific
sequences intheirgenomicDNA(22, 25, 35, 44). By a cDNA excesshybridization technique (4),
wedetermined that C3H liverDNA,C3H
mam-marygland DNA,andBALB/cliver DNA
con-tainapproximately7copies each of MMTVper
diploidgenome,whereasGR liverDNAcontains
approximately 10. These numbers are in close
agreement with published values (22, 25). As
previously observed (22, 25),wefound that C3H
mammary tumor DNAcontains additional
cop-ies of the MMTVprovirus.
We used the DNA filter transfer technique
(37) to compare the locations of the MMTV proviruses endogenous to different mouse strains.The results reveal thatin mousestrains
ofunrelatedorigins, suchasBALB/c,
DBA/H-T6, andGR,the patternsofMMTV-containing
restriction enzyme fragments arising from the
digestion of liverDNAs are different.In mouse
strains ofrelated genetic background the pat-terns ofMMTV-specific fragmentsare similar.
One interpretation of these results is that the
endogenous MMTV proviruses of unrelated
mousestrainsareindifferentgenomiclocations,
whereas inrelatedmice theirgenomic locations
aresimilar. Onehypothesiswhichcouldexplain
the results is that proviral DNA arose from a
number of independent germ cell infection
eventsin which viralDNAwasabletointegrate
intodifferentgenomic locations.However,once
the viral DNA was integrated it remained a
stablegeneticelement.Hence,we areabletosee
similarities in thegenomiclocations of the pro-viruses ofgenetically related mice. Cohen and
Varmus (8) have recently published a similar
analysis and have come to similar conclusions.
They also saw that different inbred mouse
strainsresulting from the Baggalbino x DBA
matingcarried MMTVproviruses insimilar
ge-nomic locations. In addition, they determined
that thegenomiclocations of MMTVproviruses
inferal micearedifferent, resultswhichcanbe
compared to our findings on inbred mouse
strains which are genetically unrelated. In
an-J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
otherpublication (24), the authors determined
that the MMTVproviruses endogenous tothe
A/HeJmouse arelocatedon atleast three
sep-aratechromosomalpairs. This result alsofavors
theidea thatseparate viral germ line infection
events ledtoproviral DNA. Analternative
in-terpretation of the differences seen inthe
pat-terns of MMTV-containing fragments in, for
example, the GR and C57BL DNA is that the
sequencessurrounding theprovirusesinthe
dif-ferentmice have diverged. To analyze this
pos-sibilityweused theSouthem DNA filter transfer
technique (37) to analyze the/3-globin gene in
the different mousestrains. C57BL/6J and GR
micehave theHb-S locus and code for onlyone
f8-globinprotein. Both strains contain the
,8-glo-bingenewithina10-kb EcoRI fragmentofDNA.
Recently ithasbeenreported that theC57BL/
10 mouse has its fl-globin gene within a 10-kb
EcoRI fragment (45). Upon further analysisby
RPC5chromatography, thisfragmentwasfound
to be composed oftwo different 10-kb pieces
whichcould bedifferentiated by digestion with
restriction enzymes. Whether the 10-kb band
arising from the digestion of GR DNA with
EcoRI also contains twoseparate fragments is
not known. The 83-globin gene in these two
mousestrains ofdifferentgeneticoriginsappears
tobein asimilar macroenvironment. Thesame
analysis has been carriedoutfor the a-amylase
gene. Arecombinantplasmid containing
a-am-ylase mRNA-specific sequences recognizes at
least four genomicDNAEcoRIfragments.The
DNA from the seven different mouse strains
whichweanalyzedgaverisetothesame pattern
of a-amylase-containing EcoRI fragments (B.
Groner, N. E. Hynes, and U. Schibler,
unpub-lishedobservations). These resultssuggestthat
these structural genes in the different mouse
strains aresimilarlyorganized,whereas MMTV
proviral DNA is located in different genomic
sites. This couldreflectseparate germ line
inte-gration events into a relatively similar
back-ground of structural genes in the different
strains. The validityofthis idea awaits further
comparative studies with probes specific for
otherstructural genes.
One difficulty in comparing the endogenous
proviruses ofdifferent strains is that the
restric-tion enzyme sites in these proviruses have not been mapped and may vary from the pattern
determinedforisolated viral DNA. For most of
themousestrains itappears that theendogenous
proviruses never yield viral particles. Thus,
pu-rified genomic DNA may be the best substrate
for suchananalysis. Previouspublications sug-gest thatthereis a highdegree of cross hybrid-izationbetween the endogenous MMTV
provi-ruses of different strains (20, 30). However,
RNase T1 analyses (11) andhybridization
anal-yses (20) have shown that differences do exist.
Aspointedoutin theresults,wehave evidence
that atleastsomeof the restrictionenzymesites
ofendogenousproviruses correspondtosites in
isolated viral DNA. The viral internal BamHI
fragment of 1.0 kb also appears in a BamHI
digestion ofliverDNA fromC3H, BALB/c, and
GR mice. However, the digestion ofC3H liver
DNA withPstIreveals thatonlytwoof the four
internalPstI fragments obtained from a digest
of viral DNAareidentical. This resultsuggests
thatsomeof the restrictionenzyme sites of the
endogenous proviruses vary from those
deter-mined for isolated viral DNA.
An additional observation which we have
made in the courseofthesecomparativestudies
is that DNAisolated from different coloniesof
BALB/c mice yields different numbers of
MMTV-containing EcoRI fragments. There is
oneadditionalfragmentpresentin the DNA of
the Swisscolony. Weattemptedtoanalyzethis
difference further by digesting the DNA with
PstI. This enzyme has been used in this work
(Fig.7)and in others (7,8)todistinguish
endog-enous MMTV proviruses from those
corre-sponding to the exogenous milk-bome virus.
Upon digestion with PstI, DNA isolated from
mice of both colonies gave rise to the same
pattern (datanotshown),whichhaspreviously
beenpublished (8).Ifthe SwissBALB/ccolony
hasacquired a newMMTV provirus,itcannot
be distinguished from the others by PstI and
therefore does not correspond to the BALB/
cfC3Htumor-specific viral variant.
With theSouthemblottingtechniquewe
an-alyzed theDNAofmammary tumorstaken from
C3H miceorfromBALB/c mice foster nursed
on C3H mothers. The animals received exoge-nous, milk-borne MMTV, and we determined
that each tumor contained at least one
addi-tionalcopy of MMTVproviralDNA.It is
inter-esting to note thattumors fromBALB/cfC3H
animals contain higher amounts of newly
ac-quiredproviruses than those isolated from C3H
animals. We analyzed a number ofC3H
mam-mary tumorsand have detectedamaximum of
one new proviral copy, whereas one of the
BALB/cfC3HtumorDNAsshown inFig.6
con-tained at least four newproviral copies. Other
investigators (7) who looked at BALB/cfC3H
tumorsalso found highamounts ofnewly
inte-grated MMTV proviruses. Perhaps C3H mice
are more resistant to the C3H virus than are
BALB/c mice. Different types ofexperiments
haveled others to suggest this (13).
The comparison of the pattern of MMTV-33,
on November 10, 2019 by guest
http://jvi.asm.org/
1024 GRONER AND HYNES
containing restriction enzyme fragments
ob-tained with different tumor DNAs allows the
following conclusionsto be made. First, the
pat-temof the extra MMTV-containing DNA bands
is different in each tumor, suggesting that the
exogenous viral DNAcanintegrate at different
sites in the hostgenome. Second, the fact that
extrabands ofMMTV-containing DNA are ev-ident suggests that a high percentage of the
tumorcells containedthe exogenous viral DNA
integratedin thesame site and that the tumor
mayhave arisen fromone cell or from a small
number ofinfected mammary gland cells.
Intwo recentpublications(7, 32), other
inves-tigators have reported that MMTV proviral
DNA can integrate into a number ofgenomic
sites. In one case (7), the integration sites of
newly acquired, tumor-specific proviruses were
compared.Inthe otherreport(32), the genomic
locations of the proviral DNA in an
MMTV-infectedratcell line were determined. The
ex-ogenous infectionof cells with otherretroviruses,
including murine leukemia virus (39), spleen
ne-crosis virus (17), and aviansarcoma virus (14),
has yielded results which suggest that the
pro-viral DNAspecific for these virusescanintegrate
intoanumber ofgenomic sites.
The differentpatterns of MMTV-containing
sequences invariousmousestrainsand the
dif-ferentproviral integrationsitesfoundin
individ-ual mammary tumor DNAssuggestthat there
aremultiplesites onthe cellular genome
avail-ablefor proviral DNA integration. Conclusive
evidence as to whether the different MMTV
integration sites sharecommonfeaturescanonly
be drawn from a sequence analysis of the
ge-nomic DNAflanking the provirus. Suchan
anal-ysis may help to clarify the mode of proviral
DNAintegration andmayrevealsequences
re-latedtothe controlofMMTVproviral
expres-sion.
ACKNOWLEDGMENTS
The support and encouragement of H. Diggelmann, in whoselaboratory this workwasperformed, isgratefully ac-knowledged.Wethank J.Beard for the avianmyeloblastosis virus reversetranscriptase,B.Mach forplasmid pCR1DNA containingthe8-globinsequences,L. Bootand J.Hilgersfor supplyinguswithsomeof the mammarytumor-bearing mice, G.Fey for PstI, and K. Yamamoto for the GR cell line. We also thankR.Michalides forcritically readingthemanuscript. Finally wethank S. CherpillodandP. Dubied forhelpin preparing the manuscript.
Thisworkwassupported by grants 3.706-0.76SR and 3.273-0.78SR fromtheSwiss NationalScience Foundation. N.E.H. wassupported by a grant from the Swiss Cancer League.
LITERATURE CITED
1. Alwine,J.C., D. Kemp, and G. R. Stark.1977.Method for detectionofspecificRNA inagarosegelsby transfer todiazobenzyloxymethyl-paper and hybridizationwith DNA probes.Proc. Natl.Acad. Sci. U.S.A. 74:5350-5354.
2. Bentvelzen,P. 1974. Host-virus interactions in murine
mammarycarcinogenesis.Biochim. Biophys.Acta 355: 236-259.
3. Bentvelzen, P., and J. H. Daams. 1969. Hereditary infections with mammary tumor viruses in mice. J. Natl. Cancer Inst. 43:1025-1035.
4.Bishop, J. O., and K. B. Freeman. 1974. DNA sequences neighboring the duck hemoglobin genes. Cold Spring Harbor Symp. Quant. Biol. 38:707-716.
5. Britten, R. J., D. E. Graham, and B. R. Neufeld. 1974. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 29:363-418.
6. Burton, K. 1968. Determination of DNA concentration withdiphenylamine. MethodsEnzymol. 12B:163-166. 7. Cohen,J.C.,P.R. Shank, V.L. Morris, R. Cardiff, and H. E. Varmus. 1979. Integration of the DNA of mousemammary tumorvirus in virus-infectednormal andneoplastic tissue of the mouse. Cell 16:333-345. 8. Cohen,J. C., and H. E. Varmus. 1979. Endogenous
mammarytumor virusDNA varies among wild mice and segregates during inbreeding. Nature (London) 278:418 423.
9. Denhardt, D. T. 1966. A membrane-filter technique for the detection of complementary DNA. Biochem. Bio-phys. Res. Commun. 23:641-646.
10. Dion,A.S.,U.L.Heine,A.A. Pomenti, J. Korb, and G. H. Weber. 1977. Electrophoretic analysis of the molecular weight of murine mammary tumor virus RNA. J. Virol. 22:822-825.
11. Friedrich, R.,V. L.Morris,H. M.Goodman, J. M. Bishop, and H. E. Varmus. 1976. Differences between genomesof twostrainsof mouse mammary tumor virus asshownby partial RNA sequence analysis. Virology 72:330-340.
12. Groner, B.,N. E.Hynes,and H.Diggelmann. 1979. Identification of mouse mammary tumor virus-specific mRNA. J.Virol. 30:417-420.
13. Hilgers, J., and P.Bentvelzen. 1978. Interaction be-tweenviral and genetic factors in murine mammary cancer.Adv.Cancer Res. 26:143-195.
14. Hughes,S.H.,P. R.Shank,D.H.Spector,H. J.Kung, J. M.Bishop,H. E. Varmus, P. K. Vogt, and M. L. Breitman.1978.Proviruses of aviansarcomavirus are terminally redundant, co-extensive with unintegrated linearDNAandintegrated at many sites. Cell 15:1397-1410.
15. Hynes, N.E., B. Groner,A. E. Sippel, S. Jeep,T. Wurtz, M. C. Nguyen-Huu, K.Giesecke, and G. Schiitz.1979.Control of cellularcontentofchicken egg whiteprotein-specificRNAduringestrogen administra-tionand withdrawal.Biochemistry18:616-624. 16. Hynes,N.E.,B.Groner,A. E.Sippel,M. C.
Nguyen-Huu,andG.Schutz.1977.mRNAcomplexityand egg whiteproteinmRNAcontentinmatureand hormone-withdrawn oviduct.Cell11:923-932.
17.Keshet, E.,and H. M. Temin. 1978. Sites ofintegration ofreticuloendotheliosis virusDNA in chicken DNA. Proc.Natl. Acad. Sci. U.S.A. 75:3372-3376.
18.Ketner, G., andT. J. Kelly. 1976. Integrated simian virus40sequencesintransformed cell DNA: analysis
usingrestriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 73:1102-1106.
19. Klein,J. 1975.Biologyof themousehistocompatibility-2
complex: principles of immunogenetics applied to a
single system, p.26. Springer-VerlagNewYork, Inc., New York.
20. Michalides, R., and J. Schlom. 1975.Relationshipin nucleic acid sequencesbetweenmousemammarytumor
virusvariants.Proc.Natl. Acad.Sci. U.S.A. 72:4635-4639.
21. Michalides, R.,L.VanDeemter,R.Nusse,G.Ropcke,
and L. Boot. 1978.Involvement ofmousemammary tumor virus in spontaneous and hormone-induced mammary tumors in low-mammary-tumor mouse
strains.J. Virol. 27:551-559.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
22.Michalides, R.,G.Vlahakis, and J.Schlom.1976. A biochemical approach to the study of the transmission of mousemammary tumor viruses in mouse strainsRu andC3H. Int.J. Cancer 18:105-115.
23. Moore, D. H. 1975.Mammary tumor virus, p. 131-167. In F.F. Becker(ed.), Cancer:acomprehensive treatise, vol. 2. Plenum Publishing Corp., New York.
24. Morris, V. L, C.Kozak, J. C. Cohen, P. R. Shank, P. Jolicoeur,F.Ruddle,and H. E. Varmus. 1979. En-dogenous mouse mammary tumor virus DNA is distrib-uted amongmultiplemousechromosomes.Virology 92: 46-55.
25.Morris, V. L., E. Medeiros, G. M. Ringold, J. M. Bishop, and H. E. Varmus. 1977. Comparison of mousemammarytumorvirus-specific DNA in inbred, wild and Asian mice, and in tumors and normal organs from inbred mice. J. Mol. Biol. 114:73-91.
26.Miihlbock,0. 1965. Note on a new inbred mouse strain GR/A. Eur. J. Cancer 1:123-124.
27. Nandi, S.,andC. M. McGrath. 1973.Mammary neopla-sia in mice.Adv. Cancer Res. 17:353-414.
28. Old, J., J. B.Clegg,D. J.Weatherall, S. Ottelenghi, P. Comi,B. Giglioni,J.Mitchell, P. Tolstoshev, and R. Williamson. 1976. A directestimateof the number ofhuman,B-globin genes. Cell 8:13-18. 29. Popp,R.A.,and E. G. Bailiff.1973.Sequenceof amino
acids in themajor and minor , chains of the diffuse hemoglobin from BALB/c mice. Biochim. Biophys. Acta 303:61-67.
30.Ringold, G. M.,P. B.Blair,J. M.Bishop,and H. E. Varmus. 1976. Nucleotidesequence homologies among mousemammary tumor viruses.Virology 70:550-553. 31.Ringold,G.M.,R. D.Cardiff,H. E.Varmus,and K. R. Yamamoto. 1977. Infection ofcultured rat hepa-tomacellsby mouse mammary tumor virus.Cell 10:11-18.
32. Ringold, G.M., P. R.Shank,H. E.Varmus, J. Ring, and K. R. Yamamoto. 1979.Integration and transcrip-tionofmousemammarytumor virus DNA in rat hep-atomacells. Proc. Natl.Acad. Sci.U.S.A. 76:665-669. 33.Ringold, G. M.,K. R.Yamamoto, P. R. Shank, and H. E. Varmus.1977.Mouse mammary tumor virus DNA in infected rat cells: characterization of unintegrated forms.Cell 10:19-26.
34. Rougeon,F.,and B. Mach. 1977. Cloning and amplifi-cationof a and,8mouse globingene sequences
synthe-sized in vitro. Gene 1:229-239.
35.Scolnick, E. M., W. Parks, T.Kawakami,D.Kohne, H.Okabe, R. Gilden, and M. Hatanaka. 1974. Pri-mateandmurinetype-Cviral nucleic acid association kinetics:analysis ofmodel systems and natural tissues. J. Virol. 13:363-369.
36.Shank, P. R., J. C. Cohen, H. E. Varmus, K. R. Yamamoto, and G. M. Ringold. 1978. Mapping of linear and circular forms of mousemammary tumor virus DNA with restrictionendonucleases: evidence for alarge specific deletion occurring athigh frequency during circularization. Proc. Natl.Acad. Sci. U.S.A. 75: 2112-2116.
37. Southern, E. M. 1975. Detection ofspecific sequences among DNAfragments separatedby gel electrophore-sis. J. Mol. Biol. 98:503-517.
38.Staats, J. 1976. Standardizednomenclature for inbred strains of mice:sixthlisting.Cancer Res. 36:4333-4377. 39.Steffen,D., and R. A.Weinberg. 1978.Theintegrated genome of murine leukemia virus. Cell 15:1003-1010. 40.Taylor,J.M., R.Illmensee, and J.Summers. 1976. Efficient transcription of RNA into DNA by avian sarcomaviruspolymerase.Biochim.Biophys.Acta 442: 324-330.
41. Tereba, A.,and B. J.McCarthy.1973.Hybridizationof 5I-labeled RNA.Biochemistry12:4675-4679. 42. Tiemeier,D.C.,S. M.Tilghman,F. I.Polsky,J.G.
Seidman,A. Leder,M.H. Edgell, and P.Leder. 1978.Acomparisonof two cloned mouse,B-globingenes and theirsurroundingandinterveningsequences. Cell 14:237-245.
43. Todaro, G.J., R. E.Benveniste,R.Callahan,M. M. Leiber, andC.J.Sherr.1975. Endogenous primate and felinetype-C viruses. ColdSpringHarborSymp. Quant. Biol.39:1159-1168.
44. Varmus, H. E., J. M.Bishop,R.C.Nowinski,and N. H. Sarkar. 1972.Mammary tumor virus specific nu-cleotide sequences in mouseDNA. Nature (London) NewBiol. 238:189-191.
45. Weaver, S.,N.L.Haigwood,C. A.Hutchison,and M. H.Edgell.1979.DNAfragmentsof theMus musculus ,B-globin haplotypesHbb' and Hbbd. Proc.Natl.Acad. Sci.U.S.A.76:1385-1389.
46. Weinstock,R., R.Sweet,M.Weiss,H.Cedar,and R. Axel.1978.IntragenicDNAspacersinterruptthe oval-bumin gene. Proc. Natl. Acad. Sci.U.S.A. 75:1299-1303.
on November 10, 2019 by guest
http://jvi.asm.org/

![FIG. 2.forDNA.mammarywithacidswas Saturation hybridization ofMMTV[3H]cDNA with mouse DNA](https://thumb-us.123doks.com/thumbv2/123dok_us/1498310.102530/5.504.52.244.322.484/fig-fordna-mammarywithacidswas-saturation-hybridization-ofmmtv-cdna-mouse.webp)