0022-538X/80/10-0109/06$02.00/0
Selective
Amplification of
Mouse
Mammary Tumor Virus in
Mammary
Tumors of GR Mice
THOMAS G. FANNING,JOHN P. PUMA,AND ROBERTD.CARDIFF*
DepartmentofPathology, SchoolofMedicine, UniversityofCalifornia,Davis,California 95616
DNAs extracted from the mammary tumors of GR mice were analyzed for mouse mammary tumor virus proviral sequences by the restriction enzyme-Southern blot procedure. The tumor DNAs contain more proviral copies of mouse mammary tumor virus than DNA from a
nonmalipant
tissue. The degree of proviral amplification issmall(ca. one to fiveadditionalcopies) andappears to be variable from tumor to tumor. The restriction patterns of the amplified proviral sequences suggestaclonalorigin forthe tumor mass. Inaddition, the restriction patterns observed after digestion with the enzymesBgl andSacI indicate that only one of theproviruses endogenous to GR mice is amplified. The amplified provirus found in GRmammar tumors is identical to the provirus thatis missinginGR-Mtv-2- mice,acongenic line exhibiting a low mammary tumor incidence.
Mouse mammary tumor virus (MuMTV) is
transmitted fromone mouse generation to the
nextby twomechanisms:vertically through the
germ cells, and horizontally through the milk
(3). Although all laboratorystrainsof mice ap-pear to contain several endogenous, vertically
transmittedMuMTVproviruses(11, 26), notall
strainstransmitamilk-bornevirus. Forexample, BALB/c and C57BL mice containnodetectable MuMTV in the milk of lactating females, whereasGRandC3Hmice contain appreciable
quantities ofthe milk-borne virus(14).
Mouse strainsconaining noMuMTV in the milk of females have alow incidence of
mam-marytumorsand,conversely,strains
exhibiting
the milk-borne virus have a high incidence of mammary tumors. Foster-nursing the pups of low-incidence strains on females from high-in-cidencestrains increases thetumorincidence of the former (2). The DNAs oftumorsobtained from the foster-nursed animals contain addi-tionalMuMTVproviruses (MuMTV amplifica-tion) which are the result of infection by the milk-borne virus (6,15).
Foster-nursing the pups of mice of strains
containing the milk-borne virus on those of
strains that donotcontain the virus in the milk
has,ingeneral,theeffectofreducingthetumor
incidence of the forner (1). However, the GR
mouse is an exceptional case in which
foster-nursinghas noapparenteffect on themammary
tumorincidence.GRmicefoster-nursedonmice
of strains conaining no milk-borne virus (e.g.,
C57BL) have a97% tumor incidence,which is
identicaltothatof GRmice ingeneral(4).This
observation, together with the finding that no
increaseinMuMTV proviral sequences canbe
detectedin GR mammarytumorsby liquid
hy-bridization (13, 15), has led to the
speculation
thatneitherinfectionnorMuMTV
proviral
am-plification isrequired for mammarytumor
de-velopment inGRmice (3;P.Bentvelzenand J.
Hilgers, inG.Klein, ed., Virological
Oncology,
inpress).
We haveused restriction enzymes, in conjunc-tion with the blotting technique of Southern (22), to examine the question of MuMTV pro-viralamplificationinGRmammarytumors.Our
findingsindicate that
proviral
amplificationdoesindeedoccur. In
addition,
theproviral
integra-tion patternswe observe
imply
that the tumormassis dominated bythe progenyofaunique
celloruniquesubsets ofcells,ashas been found
forBALB/cmiceinfected with the milk-borne
virus ofC3H animals (6). Further, our results indicate that only one of the endogenous MuMTV proviruses of GR mice is amplified.
Thisprovirusisnotpresent inGR-Mtv-2 mice,
a congenic line which lacks the genetic locus
responsibleforearlyGRmammarytumors(25).
MATERIALS AND METHODS
Mice, cells, and virus. GR mice were obtained
from the Cancer ResearchLaboratory,Berkeley,Calif. GR3A,acell line derived fromaGRmammary tumor (20), wasmaintainedinDulbecco-modifiedEagle
me-dium with 5%fetal calfserumand 5jLgof
dexameth-asoneperml.MuMTVvirions (from the Mm5mt/cl cellline) weresupplied by the Frederick Cancer
Re-searchCenter, Frederick, Md.,throughtheResearch Resources, Biological Carcinogenesis Branch,
Na-tional Cancer Institute.GR-Mtv-2 micewerekindly
provided by Jo Hilgers, TheNetherlands Cancer
In-stitute,Amsterdam, The Netherlands.
Extraction of nucleic acids. GRmammary
tu-109
on November 10, 2019 by guest
http://jvi.asm.org/
110 FANNING, PUMA, AND CARDIFF
mors were minced and thendisruptedby
homogeniz-ing with a Douncehomogenizerin abuffercontaining
10mMTris-hydrochloride(pH 7.4)-10mMEDTA-10
mM NaCl(TEN buffer).The cellswerecollectedby centrifugation andresuspended in TEN buffer. GR 3A cellswerescraped fromtissuecultureplates, pelleted, andresuspendedinTENbuffer. Protease Kand
so-dium dodecyl sulfatewere added to thecell
suspen-sions to 100yg/ml and0.5%, respectively. The
mix-tures were incubated for 30 min at 37°C and then
extracted twice withphenol(9). The extracted nucleic acidsweredialyzedexhaustivelyat4°C against10mM
Tris-hydrochloride (pH8.0)-i mM EDTAtoremove
residualphenol and storedat4°C.
MuMTV virionsweredisruptedbytheaddition of
sodium dodecyl sulfate to 1%, and the RNAwas
ex-tracted twice with phenol. The MuMTV RNA was
heatedto90°C for3minand thencentrifuged ina 5 to 20% sucrosegradient (17). The materialsedimenting
in the 23-35S region ofthe gradient was collected,
ethanolprecipitated, anddissolvedinasmall volume of10 mM Tris-hydrochloride (pH 7.4)-50mMKC1-1
mMEDTA,andstoredat-20'C.
Preparation of MuMTV 32P-cDNA. Radiolabeled MuMTV-specific complementary DNA (32P-cDNA) wasprepared in a50-M1reaction mixturecontaining:
50 mM Tris-hydrochloride, pH 8.1; 75 mM KCI; 10
mMMgCl2;2mMdithiothreitol; 0.2mMeachdGTP, dCTP, and dTTP; 50 ILCi of[32P]dATP (2,000 Ci/
mmol; NewEngland Nuclear Corp., Boston, Mass.);
25,ugofactinomycinDperml;0.5ygofMuMTV
23-35S RNA;600ytgofcalfDNAprimers (24); and50U of avian myeloblastosis virus reverse transcriptase (providedbyJ.Beard, LifeSciences, Inc., St. Peters-burg,Fla.,through the Research Resources, Biological Carcinogenesis Branch, National Cancer Institute).
Themixturewasincubated for30min at37'C. EDTA, sodiumdodecyl sulfate, and NaOH werethen added
to final concentrations of0.05 M, 0.5%, and 0.3 M, respectively, and the mixturewas incubatedat65°C for 3 to4h tohydrolyzethe RNA.The32P-cDNAwas
separated from othercomponents ofthe mixtureby chromatography onSephadex G-50, ethanol precipi-tated, dissolved in 10 mM Tris-hydrochloride (pH
8.0)-imMEDTA,and stored at-40°C.
Enzyme digests and blotting. Restriction
en-zymes were purchased from New England Biolabs, Beverly, Mass. Digests were done in 100 mM Tris-hydrochloride (pH 7.4)-10 mM NaCl-5mM MgC12-100Mgoflysozymeper ml at 37°C. Approximately9
Mgofcellular DNAwasmixed with 1 Mgof bacterio-phage lambda DNA before enzyme digestion. The
completeness ofthe digestion reaction was verified, afterelectrophoresisin1%agarose gels, by the restric-tion pattern of the added lambda DNA. The transfer of DNAfragmentsfromtheagarose gels to
nitrocel-lulose filters was done essentially as described by
Southern(22). After transfer, the filters were treated
according to published procedures (6).
Autoradi-ograms were scanned with anSDC 300 density com-puter (Schoeffel Instruments, Westwood,N.J.).
RESULTS
We extracted thenucleic acids of four
mam-mary tumors and three livers obtained from
J. VIROL.
three female GR mice. Using the restriction
enzymes EcoRI, BamHI, PstI, BglI, BglHI and
Sac, we analyzed the three liver DNAs for
MuMTV proviral sequences by the Southern
blotting procedure (22).Eachenzymegenerated
a distinct restriction pattern, and for each
en-zyme the three liver DNAs had identical
pat-terns(not shown).Since the liver is not infected
byMuMTV (6, 15),these patterns were
consid-ered characteristic of the endogenous,
geneti-callytransmitted MuMTV proviruses (7).
We cleaved samples ofone of the GR
mam-marytumorDNAs with the restrictionenzymes
listed above and compared the digestion
prod-uctswithsamples of GR liver DNA cleaved with
the same enzymes. The autoradiograph of one
comparison, with EcoRI, BamHI, and PstI, is
presentedin Fig. 1. There were no major
differ-ences between the mammary tumor DNA and
liver DNA restriction patterns.
The restriction patterns of another GR tumor
DNA showed major differences in theform of
manyadditionalfragments when digested with
several of the enzymes (Fig. 2A, slot 2, andFig.
2B, slot 2). These additional restriction
frag-ments probably contained both MuMTV and
15o0+
*imui
6.4 -_
4.3 -_ -i
2.9
_4-1.44
0
1
2
3
4
5
6
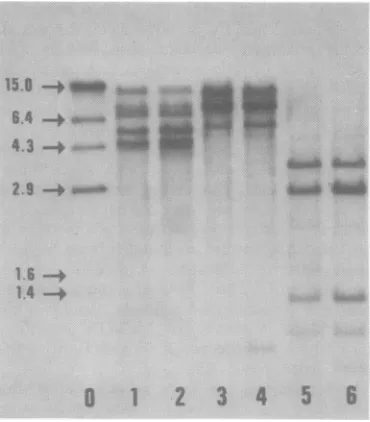
FIG. 1. Comparison ofMuMTVproviralsequences in GR liver and GR mammary tumor DNAs. Both the
tumor and liverDNAs used in this example were
from animal 2 (see Fig. 2). Each DNA (9 MLg) was
digestedwith EcoRI(slots1 and 2),BamHI (slots 3 and4), orPstI(slots5and6) andanalyzed by the Southern blottingprocedure and autoradiography. The liver DNA wasrun inslots 1, 3, and 5; tumor DNAwas runin slots 2, 4, and6.Slot0is aHindIII
digestof32P-labeled bacteriophagelambda DNA (19). Themolecularweights(x106) ofthelambda markers
aregivenontheleft(16).
:. 10., k ddk--.Ak
.i -1:
am-l&
own mpmw
is'
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.510.273.458.338.549.2]MuMTV AMPLIFICATION IN GR TUMORS 111
mousesequences, thus suggesting a clonal origin for the tumor mass (see below). Thus, amplifi-cation of MuMTV proviral sequences was easily detected insome GR mammary tumors, but was not readily identified in others. However, we could show that each tumor DNA contained unique MuMTV proviral fragments provided the appropriate restriction enzyme was used. Thus, tumors 2, 3, and 4 could be distinguished from oneanother by the appearance of unique
BglIfragments (Fig. 2B, slots 2, 3, and 4), and
tumor 1 could be distinguished from the other threeby a unique BamHI fragment (not shown). During the course of this investigation we observed that someautoradiographic bands in the GR mammary tumor DNAs were consis-tently darker (i.e., the fragments bound more cDNA) than comparable fragments from GR liver DNA; that is, specific MuMTV proviral sequences appeared to be amplified. The phe-nomenon was best demonstrated with theBglII orSaclrestriction enzyme (Fig. 3).
Of the eight major MuMTV DNA fragments found in BglII digests of GR mammary tumor DNA, one fragment of 2.6 x 106 molecular weight had bound more cDNA than the
com-parable liver fragment (Fig. 3A). Of the eight
major MuMTV-specific proviral fragments found inSacl digests oftu
bound more cDNA than t
A
'^F
*::.>...-v...:....3
*,. ,.2Af;."*_ __ - i S :.
__ __ _ a a_
a = ;^s
s_ _= s_
_w.
*-lrB...f,.
.:::::B;:r
S... ...:: :.
B
1
2
3
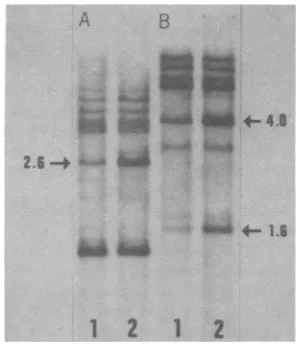
4FIG. 2. EcoRI and BglI MIMTVproviralsequencesi
DNAs. Each tumor DNA (9
either EcoRI (A) orBglI (B,
Southern blottingprocedure
Slot 1, Tumor1fromanimal
animal1;slot3,tumorfromi from animal3.
A B
1212
FIG. 3. BglII and SadI restriction patterns of
MuMTVproviral sequences in OR liver and OR mammary tumor DNAs. Nine micrograms ofeach DNA (fr-omanimal2)wasdigestedwith eitherBglIII (A)orSadI(B)andanalyzedbytheSouthernblotting
procedureandautoradiography. Slot1, LiverDNA;
slot2,tumorDNA. Theamplified proviralsequences
in the tumor samples are indicated by the arrows. The numbersgive the molecularweights (x 106) of
thefragments.
nmorDNA, two had fragments (Fig. 3B). ThesetwoSacIfragments ;he comparable liver have molecular
weights
ofapproximately 4.0x106
and 1.6 x106,
the sum of which representsvirtuallythe entireMuMTV genome (6.0x 106)
(8). The otherGR mammarytumorDNAs also contained the amplified BgllI and SacI
frag-_
__
~~ments.
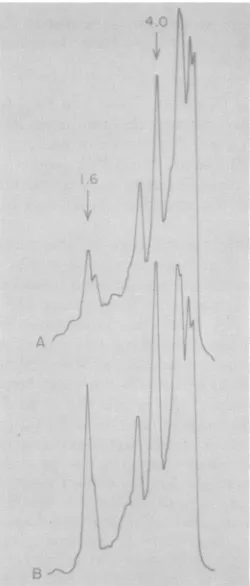
Densitometer tracings of the SacI digests shown in Fig. 3 are presented in Fig. 4. These confirm whatcanbe surmisedupon inspection.
Two SacI fragments from tumor DNA bound
more cDNA than the
comparable
liverfrag-ments. Integrationof the two curvessuggested
thattwo tothree additionalMuMTVproviruses
were present in this GR mammary tumor DNA.
Weinterpretthe results shown in Fig.3 and
4asfollows:oneof the GRMuMTVproviruses (or one class of provirus) contains two BglII restriction sites that give rise to a 2.6 x 106-- O *s_ daltonfragment and threeSacI restriction sites
generating
1.6x 106- and 4.0 x 106-daltonfrag-1
2~
3 4 ments. The other GRproviruses
donotcontainthe same sites. The GRprovirus inquestion is
restrctio pattes selectively amplified in GRmammary tumors.
restriction patterns of The degree of amplification can range from a
,nOR
maimmary
tumorismgle
additionalcopytofourorfiveadditional
pg was digested withsigead
)and
analyzed by thecopies.
In those cases where four or fiveaddi-and autoradiography. tionalcopiesarepresent,unique proviral
restric-1; slot 2, tumor 2from tion fragments can normally be detected with
animal2;slot4,tumor enzymesthatcleavethe
provirus
atasingle
site (Fig. 2A,slot2).Whenonlyoneadditionalpro-VOL. 36,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.510.273.422.72.244.2]112 FANNING, PUMA, AND CARDIFF
virus is present, it may be difficult toidentify.
Unique proviral fragments may beobscured by
comigration with other high-molecular-weight
restrictionfragmentsorrelativelysmallamounts
of MuMTV sequences may be present in the
provirus-mouse fragments. However, the GR
mammarytumorsweexaminedcouldbeshown
to have selective MuMTV amplification using
BglII and SacI, which cleave at several sites
within theamplifiedprovirus.
The amplified provirus we detected in GR
mammary tumors was not present inthe tissue
DNAsofGR-Mtv-2- mice(Fig. 5).This linewas derivedthroughabreedingprogramdesignedto
eliminate thegeneticlocusresponsible for early
mammary tumors in GR mice (25). Thus, our resultssuggestthat thisgenetic locus is,atleast
inpart, one ofthe GRproviruses.We will
hence-forth refer to this provirus as the GR-MTV-2 provirus.
The low degree ofMuMTVDNA
amplifica-tion detected in GR mammary tumors wasin
FIG. 4. Densitometertracings of the SacI restric-tion patterns shown inFig. 3. (A) GR liver; (B) GR mammarytumor.The 1.6x106-and 4.0x106-dalton
proviralsequencesamplified in thetumorsampleare
indicatedby thearrows.
A '.-...;A
~~~~~~~~~~~~~~~~4.~~~~~~~g
1.6-+ .. 4.
[image:4.510.275.465.73.210.2]2 34 3 4
FIG. 5. SacI and BglII restriction patterns of MuMTVproviralsequencesin GRtissue,
GR-Mtv-2-tissue, andGR 3A cell DNAs. (A) Each tissue DNA
(9 ug)andGR 3A cell DNA (2 ,ug)wasdigested with
either SacI (A) or BglII (B) and analyzed by the
Southern blotting procedure and autoradiography. Slot 1, GR mammarytumorDNA; slot 2, GR liver DNA; slot 3, GR-Mtv-2- liverDNA; slot 4, GR 3A cell DNA. The 1.6x 106- and 4.0x 106-dalton SacI and 2.6x106-dalton BglII fragmentsareindicatedby the arrows.
contrast to the large degree of amplification
observedin mammary tumors fromothermouse
strains (13, 15). Thissuggestedthat the degree
of amplification of theGR-MTV-2 provirusmay
be limited. Comparison ofSacl digests of GR
liver and GR 3A tumorcell line DNA,however,
demonstrated extensiveamplificationofthe 1.6
x 106-and 4.0 x 106-dalton SacIfragments(Fig.
5A). The 2.6 x 106-dalton BglII fragment was
also (and exclusively) amplified in GR 3A cell
DNA (Fig. 5B). Since theGR 3A cell linewas
derived from a GR mammary tumor, we
con-clude thatamplification of theGR-MTV-2
pro-virus can be extensive under some
circum-stances.
DISCUSSION
Amplification ofproviral geneshas been
ob-served in severaltumorsystems (5, 10).
Ampli-fication ofMuMTVproviralsequences in mouse
mammarytumorshas beendocumented in
sev-eral mouse strains whichtransmitamilk-borne
virus (e.g., BALB/cfC3H and RIII) (13, 15).
However,noamplificationofMuMTVcould be
detected in the mammary tumors of GR mice
(13, 15). This, togetherwith thefact that
milk-borne viral infection is notnecessary for tumor
development (4),hasledtothespeculationthat mammarytumorigenesis in GR micemay
rep-resent aspecialcase(BentvelzenandHilgers,in
press).
Our results indicate that MuMTV proviral
amplification does occur in the mammary
tu-J. VIROL.
-ioLJ6
AW ..
*Noi"
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.510.106.231.305.598.2]mors of GR mice. Thus, mammary tumors of GR mice resemblethose of otherstrains exhibit-ingamilk-borne virus. The relatively small de-greeofamplificationweobserved in GR
mam-mary tumorsundoubtedly accounts for its
fail-ure to be detected byliquid hybridization (13,
15).
Theproviralamplificationweobservedin GR mammary tumors appears to involve only one of the endogenousGR proviruses. Thisprovirus
isnotpresentinthegenomeofGR-Mtv-27mice
(Fig. 5). TheGR-Mtv-2- line originatedby cross-ing GR mice withastrain (C57BL) exhibitinga
lowmammary tumorincidence (25). GR-Mtv-2
mice have a very low incidence of mammary
tumors,donottransmitamilk-bornevirus, and
have fewer endogenous MuMTV proviral
se-quencesthanGR mice(12,25). Thus,ourresults
suggest that the genetic locus responsible for
mammarytumorsinGR mice is the GRprovirus (GR-MTV-2) thatwe found amplified inthese
tumors.Since all GRmammary tumorsthatwe
haveexamined exhibitamplification of this pro-virus,weinfer that GR-MTV-2amplification is critical in GRmammarytumorigenesis.
The majority of the restriction enzymes we
haveusedcleave the MuMTV provirusesone or two times and, thus, should give rise to
high-molecular-weight DNA fragments containing
bothproviraland theflankingmousesequences.
WefindthatMuMTV-specific,
high-molecular-weight fragments can be detected in tumor DNAs after restriction enzyme-Southern blot analysis (Fig. 2). The sizes of these fragments
varyfromtumor totumor,suggesting thatthey
arenotinternalfragmentsof the MuMTV pro-virus, but rather are the provirus-mouse
se-quences. The number of proviral integration
sites in the mouse genome is apparently large,
possiblyin thehundreds,andnositeappearsto
be preferred (6). Thus, our finding of specific
proviral-mousesequences suggeststhat thevast
majority of cellsinthetumor masscontain the
amplified provirusinthesame integrationsite.
This impliesthat thetumor mass contains the
progeny ofa unique cell (monoclonality) or a
limited number of subsets of cells (limited pol-yclonality). Others have concluded that GR
tu-mors areclonalonthebasis ofhonnonal
respon-siveness (21). In addition, restriction mapping
data have suggested a clonal origin for C3H
milk-virus-induced BALB/c mammary tumors
(6).
The variability in the provirus-mouse
frag-ments seenfromtumor to tumor (Fig. 2) rules
outthepossibilityofsimplechromosomal
dupli-cation.Instead,the-additionalprovirus(es)must
beintegrated into other sites in themouse
ge-nome.Whether thisoccursbya processof
pro-viralexcision-reintegrationorby superinfection
ofmammarycells by virions is unknown.
Our finding of proviral amplification in GR
mammary tumorsstrengthens the correlationof
MuMTV gene amplification with mammary tumorigenesis. Geneamplificationmayresultin aselectiveadvantagefor the cell (18). Selection
ofone or a few cellshaving amplified proviral
sequences would also account for the
(postu-lated) clonal origin of thetumor. Theselective advantage conferred upon the cell may be re-lated to the integration site of the amplified provirusor to anincrease inMuMTV mRNAor geneproduct(s), orboth.In any event,proviral
gene amplification, asobserved in
MuMTV-in-ducedmurinemammary tumorsand other
on-cornavirus-related tumors (5, 10, 23), deserves seriousconsiderationas afundamental process leadingtoneoplastic transformation.
ACKNOWLEDGMENTS
Wethank L. J. T.Youngfortechnicalassistanceand D. Freer for densitometerscans.
This work wassupported by Public Health Service grant5 RO1CA21454awardedbytheNational Cancer Institute.
LITERATURE CITED
1.Andervont, H. B., and W. J. McE:leney. 1939. The influence of foster nursing upon the incidence of
spon-taneousbreast cancer in strainC3Hmice. PublicHealth Rep. 54:1597-1603.
2. Bentvelzen, P. 1968. Resistance to small amounts of Bittnermammarytumorvirus inoffspring of C57BL female mice with the virus. J.Natl. CancerInst.41: 757-765.
3. Bentvelzen, P. 1974. Host-virus interactions inmurine
mammarytumorigenesis.Biochim.Biophys.Acta355: 236-259.
4.Bentvelzen, P.,J. H. Danam,P. Hageman, and J.
Calafat. 1970. Genetic trn on ofviruses that incitemammarytumor inmice. Proc. Natl. Acad. Sci. U.S.A. 67:377-384.
5. Berns, A., and R. Jaenisch 1976. Increase of
AKR-specificsequences intumortissues of leukemic AKR mice. Proc.Natl. Acad.Sci.U.S.A.73:2448-2452. 6. Cohen,J.C.,P.R. Shank,V. LMorris,R.Cardiff,
andH.E.Varmus.1979.Integrationof the DNA of mouse mammarytumorvirus in virus-infected normal andneoplastictissue of themouse.Cell 16:333345.
7. Cohen,J.C., and H. E. Varmus. 1979. Endogenous
mammarytumorvirusDNA varies amongwild mice and segregates during inbreeding. Nature (London)
278:418-423.
8. Dion,A.S.,U.L.Heine,A.A.Pomenti,J.Korb,and G. H. Weber. 1977. Electrophoretic analysisofthe molecular weight of murine mammary tumor virus RNA.J.Virol. 22:822-825.
9.Gross-Bellard, M,P.Oudet,and P.Chambon.1973.
Isolation ofhigh-molecular weightDNA from
mam-malian cells. Eur. J. Biochem.36:32-38.
10. Jaenisch,R.1976.Germ lineintegrationandMendelian transmon of the exogenousMoloneyleukemia virus.
Proc.Natl. Acad.Sci.U.S.A.73:1260-1264.
11. Michalides,R., and J. Schlom. 1975.Relationshipin nucleic acid sequencesbetweenmousemammarytumor
on November 10, 2019 by guest
http://jvi.asm.org/
114 FANNING, PUMA, AND CARDIFF
virus variants. Proc. Natl.Acad. Sci. U.S.A. 72:4635-4639.
12. Michalides, R., L. Van Deemter, R. Nusse, and R. VanNie. 1978. Identification of the Mtv-2 gene respon-sible for theearly appearance ofmammarytumors in the GR mouseby nucleic acid hybridization. Proc. Natl. Acad. Sci. U.S.A.75:2368-2372.
13. Michalides, R.,G.Vlahakis,and J.Schlom. 1976.A
biochemical approachtothestudyof thetransmission of mouse MTV inmousestrainsRIII and C3H. Int. J.
Cancer 18:105-115.
14. Moore, D.H.,C. A.Long,A. B.Vaidya,J.B.Sheffield,
A. S. Dion,and E. Y.Lasfargues. 1979.Mammary
tumorviruses. Adv. Cancer Res. 29:347418. 15.Morris, V. L., E. Mederios, G. M. Ringold,J. M.
Bishop, and H. E. Varmus. 1977. Comparison of mouse mammarytumorvirus-specific DNA in inbred, wildand asian mice, andintumorsand normal organs from inbred mice.J. Mol. Biol. 114:73-91.
16. Murray,K., and N. E. Murray. 1975.Phage lambda receptor chromosomes for DNA fragments made with restrictionendonucleaseIIIofHaemophilusinfluenzae andrestriction endonuclease Iof Escherichia coli. J. Mol. Biol.98:551-564.
17. Myers,J.C.,S.Spiegelman,and D. L. Kacian.1977.
Synthesis of full-length DNA copies of avian myelo-blastosis virus RNA inhigh yields. Proc. Natl. Acad. Sci. U.S.A.74:2840-2843.
18.Nowell, P. C. 1976.The clonal evolution oftumor cell populations. Science 194:23-28.
19. Rigby,P.W.,M.Dieckmann,C.Rhodes,and P.Berg.
J. VIROL.
1977.Labellingdeoxyribonucleic acid tohigh specific activity in vitroby nick translation with DNA polym-eraseI.J.Mol. Biol. 113:237-251.
20. Ringold,G.M.,E.Y.Lasfargues,J.M.Bishop,and H. E. Varmus. 1975.Production ofmousemammary
tumor virusby cultured cells in the absence and pres-ence ofhormones: assay by molecular hybridization. Virology 65:135-147.
21. Sluyser,M.,T.Nouwen,J. Hilgers,and J.Calafat.
1977. Levels of mammary tumorvirus in
hormone-de-pendent and -independent mouse mammary tumor
cells.Cancer Res.37:1986-1990.
22. Southern, E. M. 1975.Detection ofspecific sequences amongDNAfragments separated by gel electrophore-sis. J.Mol. Biol. 38:503-517.
23. Steffen,D., S. Bird, W. P. Rowe, and R. A. Weinberg. 1979. Identification of DNA fragments carrying eco-tropic proviruses of AKR mice. Proc. Natl. Acad. Sci. U.S.A.76:4554-4558.
24. Taylor,J. M.,R. lilmensee, and J.Summers. 1976.
Efficient transcription of RNA into DNA by avian sarcomaviruspolymerase.Biochim.Biophys. Acta 442: 324-330.
25. VanNie, R., andJ.deMoes. 1977. Development of a congeneicline of the GRmouse strain without early mammary tumors. Int. J. Cancer20:588-594. 26. Varmus, H. E., J. M. Bishop, R. C.Nowinski, and N.
H. Sarkar. 1972.Mammarytumorvirusspecific nu-cleotide sequences inDNA ofhigh and low incidence
mouse strains. Nature (London) New Biol.
238:189-190.
on November 10, 2019 by guest
http://jvi.asm.org/