Copyright©1977 American Society forMicrobiology Printed in U.S.A.
Rabies Virus
Protein
Synthesis
in
Infected
BHK-21
Cells
H. PAUL MADORE* AND JAMES M. ENGLAND
The Wistar Institute of Anatomy and Biology, Philadelphia, Pennsylvania 19104 Received for publication 22 November 1976
Rabies virus-specific polypeptide synthesis was examined under hypertonic
conditions,whichselectivelyinhibitcellular proteinsynthesis. The rabies virus
proteins(L,
G,
N, M,, M2)weresynthesized throughout thecourseof infection, withlittlechangeintheir relativeratesof synthesis. The ratesofsynthesis of the G andM,
polypeptides were more sensitive to increasingosmolarity thanthose of the L,N,and M2 polypeptides. Extrapolationtoisotonicityof the results obtained under hypertonic conditions indicated that the molar ratios of the
polypeptides synthesizedundernormalconditionswere 0.4 (L), 64 (G), 100 (N),
75 (M,), and 35 (M2). A high-molecular-weight polypeptide (-190,000),
desig-nated polypeptide L, was repeatedly detected both in infected cells and in
extracellular virus. Theestimated number ofLpolypeptide moleculesper virion
was33. Thesynthesis ofa viral glycoprotein precursor, designated gp78,
pre-cededtheappearance of thematureviral glycoprotein ininfectedcells labeled
with [3H]glucosamine under isotonic conditions. In cells labeled under
hyper-tonicconditions, littleorno matureviral glycoproteinwasdetected,buta
virus-specificglycoproteinwithanelectrophoreticmobility similartothat of gp78was
observed.Thisglycoproteincould be chased intomatureviralglycoproteinwhen
the hypertonic conditions were made isotonic. These results suggest that a
reversible block of viral glycoprotein synthesisoccurs under hypertonic condi-tions.
Rabies virus, a rhabdovirus, is a negative-strand virus that containssingle-strandedRNA withamolecularweight of
approximately
4.6 x106(40, 43).The RNAisassociatedinthevirion
with phosphorylated nucleoprotein (N),
form-ing a nucleocapsid core (40, 41). Aviral
enve-lope, which contains two non-glycosylated membrane proteins
(M,
and M2) and a glyco-protein (G), surrounds the nucleocapsid (40).The viral glycoprotein is responsible for elicit-ing the production of neutralizing antibody against the virus (53).
Rabies virus hasaneclipse periodof6 to 12h
inBHK-21 cells andproducesvirusforseveral days withoutappreciable inhibition of cellular DNA, RNA, or protein synthesis (12, 27, 47, 52). Evidence of viral polypeptide synthesis in
rabies virus-infected cells has been limited to
the detection of virus-specific polypeptides in
the cytoplasm of infected cells by
immunoflu-orescenttechniques(47, 52)andby the isolation
ofnucleocapsidcoresafter cell fractionation (8, 39). Recently,wereported thatthesynthesis of
themajorrabies structural proteins (G, N,
M1,
M2) could be detected in infected cells labeled
withradioactiveaminoacids under hypertonic conditions (26).
The RNA of this negative-strand virus would
appear torequireavirion-associated RNA
tran-scriptase for transcription (2, 21). The L
pro-tein, which ispresent insmall amounts in
in-tactVSV virions(16, 50,51),has beenshownto
benecessaryfor RNAtranscriptaseactivity (5-7, 10, 11, 31). In contrast, the presence of L protein in rabies virions has not been clearly established (40, 42), and RNA transcriptase ac-tivityhas notbeen detected inpurifiedrabies
virions (41, 48), except possibly at extremely low levels (D. Bishop, personal communica-tion). Although virus-specific RNA
transcrip-tase activity has been detected in rabies-in-fected cells (48; H. P. Madore and F. Sokol, unpublished observations), the virus-specific polypeptide (L), which may be necessary for viral RNAtranscriptase activity, had notbeen detected in our previous analysis of infected cells labeled under hypertonic conditions (26). Inthisreport,hypertoniclabeling conditions
were employed to characterize rabies-specific polypeptide synthesis during the course of
BHK-21cell infection. Inadditiontothe major
viral structuralpolypeptides,a high-molecular-weightpolypeptide (L) has been clearly identi-fied both in infected cells and in extracellular 102
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 22, 1977
virus. The effect of increased osmolarity on
rates of synthesis ofindividual viral polypep-tides andonglycosylation of viral glycoprotein
wasalsoexamined.
MATERIALS AND METHODS
Cellsand virus. BHK-21/C13 cells (25) were seri-ally passaged in Blake bottles and maintained in minimal essential Eagle medium (MEM), supple-mented with 10% fetal calf serum(growth medium) at370C(18).Rabies virus (ERA strain) and vesicular stomatitis virus (VSV) (Indiana strain) were ob-tained from Tadeusz Wiktor of the Wistar Institute. Stocks ofrabiesvirusand VSV (108 to 109 PFU/ml) werepropagatedinBHK-21cells in a manner simi-lar to thatdescribed for the preparation of labeled virus. Both cells and virus stock were shown to be mycoplasma-free by the assay of Sedwick and Wik-tor (38).
Infection and mock infection of cells. The growth medium was removed from confluent monolayers of BHK-21cells in 60- or 100-mm tissue culture dishes, and the cells were infected with 50 to 100 PFU of rabies virus (ERA strain) per cell in 1 ml or
mock-infected with an equal volume of MEM supple-mented with 0.2% bovine serum albumin (0.2% BSA-MEM). Virus was adsorbed for 30 min at370C beforeunadsorbed virus and mock infection medium were removed. Both cell culture types were
incu-batedingrowth medium at370Cin a 5%C02 atmos-phere.
Labeling of virus-infected and mock-infected cells. Growth medium wasremoved, and each cul-ture wasincubatedinEarlesaltsolution plus10%
fetal calfserum at3700 for 15 min todepleteamino
acid pools. Cells labeled under isotonic conditions
were incubatedfor various times at 370C in Earle
saltsolutionplus10%fetalcalf serum with 40,uCiof 3H-labeled aminoacidmixture perml(NET-250, 15 aminoacids, New England NuclearCorp.[NEN])or 10
,ACi
of[3H]leucineper ml (TRK 170, 50Ci/mmol, Amersham/Searle). Cells labeled under hypertonicconditions were incubated for 15 min at 370C in various hypertonic media containing different amountsof excess NaCl (toallowrunoff ofcellular
mRNA from polysomes [35]) in Earle salt solution plus 10% fetal calf serum and thenlabeled for var-ious timeswith one of the above isotopes at370Cin hypertonic media. Identical results were obtained
whenlabelingwasdone under hypertonic conditions
with [3H]leucine or 3H-amino acid mixture (after
correctingfordifferencesintherelative leucine con-tentof the individual viralpolypeptides). Cellswere
labeled with 50 ,uCi of [3H]glucosamine per ml
(NET-190, 12Ci/mmol, NEN) inisotonic or
hyper-tonic 0.2% BSA-MEM according to the procedure
previously described for amino acid labeling. The tonicity (milliosmolarity) of the various mediawas determined with an automatic osmometer (Osmette-A, PrecisionSystems, Inc., Sudbury, Mass.).
Inpulse-chase experiments, cells in 60-mm tissue culture dishes were pulse-labeled with 100 ,Ci of
[3H]leucineinhypertonic medium (550 mOsM) for 2 h and then washed twice with 5 ml of 0.2%
BSA-RABIES PROTEIN SYNTHESIS 103 MEM(290 mOsM), followed by a chase with 2 mlof
0.2% BSA-MEM (290mOsM)for 24 h at370C.
Preparation of cellular fractions. Cells labeled underhypertonic or isotonicconditionswerewashed twice withice-cold NT buffer (0.13 MNaCl, 0.05M Tris-hydrochloride, pH 7.8). Whole-cell lysates were prepared by scraping thecells from the tissue
cul-turedish with 2 ml of ice-cold NT buffer and sonicat-ing(Accousticon) for 1 min. Nuclear and cytoplas-micfractions wereprepared after Dounce homogeni-zation, as previously described (26). Total protein contentwasdeterminedbyamodified Lowry proce-dure (30), and trichloroacetic acid-precipitable
ra-dioactivitywasdeterminedby precipitation of sam-ples in ice-cold 10%trichloroacetic acid, followed by washing with ice-cold 5% trichloroacetic acid on glass-fiber filters (WhatmanGF/C).
Preparation of labeled virus. Roller bottles of con-fluent BHK-21 cells (1.5 x 108 cells/bottle) were
infectedwith 5 PFUof rabies virus (ERA strain) per
cellor 0.01 PFU of VSV (Indiana strain) per cell. Virus was adsorbed for 30 min at330Cin 10 mlof inoculum, and then 100 ml of 0.2% BSA-MEM was
addedtoeach bottle for incubation at330C. A 14C0
labeled aminoacid mixture (0.5 ,uCi/ml, NEC-445; 15amino acids, NEN) wasadded to the rabies-in-fected cells, or [14C]leucine (0.5 ACi/ml, NEC-279,
270 mCi/mmol, NEN) was added to the
VSV-in-fectedcells. Therabies-infected cellswereharvested after 4to 5 days ofincubation at 330C; the
VSV-infected cells were harvested after 12 to 16 h of
incubation at the same temperature. The culture medium containing labeled virus was clarified by centrifugationat450xg.,for 15 minat40C,and the viruswaspurified by a modification of theprocedure described by B. Dietzschold, J. H. Cox, and L. G.
Schneider(manuscript in preparation). Inbrief,the virus waspelletedfrom theclarifiedsupernatantby centrifugationin aBeckman type 19 rotorat19,000
rpmfor 120 min at40C.The viruspelletwas
resus-pendedwith a PasteurpipetteinNTE(0.13 MNaCl,
0.05 M Tris, pH 7.8, 0.001 M EDTA) buffer and layered on a 10 to 60% sucrose gradient in NTE buffer. ThegradientwascentrifugedinaBeckman
SW27 rotor at 22,000 rpm for 90 min at40C.The vi-rusband wascollected, dilutedinNTEbuffer,and
repelleted by centrifugation in an SW27 rotor at 20,500 rpmfor 90 min at40C. The virus pelletwas
resuspendedin asmallamount of NTEbuffer over-night at40C,and thenclarifiedbycentrifugationat 800 xg.,for 10 minat40C. Labeledvirusreleased
intothe culturemediumduringthe 24-h chase(Fig.
6c) waspurified by layeringthe supernatant(2ml)
directlyonto a 10 to 50% sucrose gradientinNTE
buffer andcentrifugingasdescribedabove.
Polyacrylamidegel electrophoresis(PAGE). Cell
lysates and virus sampleswere prepared for PAGE after trichloroacetic acidprecipitation, as described by Sokol et al. (44), or precipitation in 5 volumes of absolute ethanol. The samples (in 8 M urea) were
subjectedtoelectrophoresis in continuous 7.5%
poly-acrylamidegelsin 0.1 Mphosphate buffer (pH 7.2) containing0.1%sodiumdodecyl sulfate (SDS) (44), or on discontinuous 10% SDS-polyacrylamide gels
on November 10, 2019 by guest
http://jvi.asm.org/
(23). The gels were fixed in 25% isopropanol-10% aceticacid,cut into 1-mmslices,and thenincubated
overnight at 330C in 5 ml of a 5% Protosol-liquid
scintillation fluid mixture.
RESULTS
Relative rate ofviral polypeptide synthesis in infected BHK-21 cells under hypertonic conditions (550 mOsM). The incorporation of 3H-labeled amino acids into viral polypeptides
wasmeasuredunderhypertonicconditions(550
mOsM) in infected cells during 1-h labeling periods at 6, 12, 24, 36, 48, and 72 h after infection. Analysis of the cytoplasmic and the nuclear fractions on PAGErevealedthatabout 85%of thelabeledrabies-specificproteinswere
in the cytoplasmic fraction, as previously de-scribed (26). From thesegels, the relativerate
of 3H-labeled amino acidincorporation into the viral polypeptides permilligram of total cellu-lar (nuclear + cytoplasmic) protein per hour wascalculated (Fig. 1).During the 1-hlabeling
period, 3H-labeled amino acid incorporation
into individual viral polypeptides was linear. Only 1.5 to 3.0% of the labeled polypeptides
were found in the medium. Analysis of these
released polypeptides by PAGE revealed that less than 10%of thesepolypeptidesco-migrated
withviral proteinsandthat therewas no
pref-erential release of any viral polypeptide into the medium. Inaddition, themedium used for
cells labeled for 2 h with [3H]leucine at 550
mOsM did not containdetectable labeledvirus
particles (detection limit,5%of290mOsM
con-trol), asdeterminedby sucrosegradient
analy-sis (10 to50% sucrose).
3H-labeled amino acid incorporation into G, N, and Ml polypeptides was detected by 12 h afterinfection, and incorporation into M2
poly-peptidewasnotedby24 hafterinfection/ (Fig.
1).The rate of3H-labeled amino acid incorpora-tion into the major viral polypeptides (G, N,
M1, M2) increased gradually for at least 72 h
afterinfection. If aminoacidsfor viral polypep-tidesynthesis areavailablefromasingleamino
acidpool andthe individualpolypeptideshave
comparable stabilities, the relative rate of
amino acid incorporation into these polypep-tidesismost likely a measure of their relative rate ofsynthesis. In Fig. 2, the relative rate of amino acid incorporation into viral polypep-tidesunder hypertonic conditions (550mOsM) obtainedfrom severalexperimentswasusedto
determine the relative number of viral protein molecules synthesized per hour at 550 mOsM. These data showthatindividual viral polypep-tides are notsynthesizedinequimolaramounts
under hypertonic conditions. The L
polypep-
100-0
a w
I. 4 0
0. a.) 0.
Q
z
a) z
w
0 0. E
80
60-40
20-I0 20 30 40 50 60 70 80 TIME AFTER INFECTION (h) FIG. 1. Rateof3H-labeled amino acid incorpora-tion intorabies viruspolypeptides under hypertonic conditions (550 mOsM). Confluent monolayers of BHK-21 cells (5 X106cells perdish) infected with 50 PFUof rabies virusper cell were labeled for 1 h at 37°C with a 3H-labeled amino acid mixture (40 puCi/ ml) in 550 mOsM medium at 6, 12, 24, 36, 48, and 72 h after infection. The cells were fractionated into nuclear and cytoplasmic fractions, and 400
pg
of proteinfrom each fraction was run on continuous 7.5%SDS-polyacrylamide gels.Radioactivity incor-porated into the viralpolypeptides in each cellular fraction was obtained by subtracting radioactivitydue to residual hostpolypeptidesynthesis from the
peaks representing the viralpolypeptides. Finally,
the sum ofradioactivity incorporated into the viral
polypeptidesinthenuclear pluscytoplasmicfractions wasdividedbythe total protein in the nuclear plus
cytoplasmicfractions to give the counts per minute incorporated per milligram of totalcellular protein.
Co-electrophoresis with "4C-amino acid-labeled ra-bies virusproteinsidentifiedtheviral-specificpeaks.
tide,especially,issynthesizedat a rate approx-imately170-foldlessthan thatof the N polypep-tide. The presence of this polypeptide in in-fected cells is attimesobscuredby other high-molecular-weight polypeptides at the top of SDS-polyacrylamide gels.
Effect ofincreasing tonicity on viral poly-peptide synthesis. The rates ofNandM2 poly-peptide synthesis were enhanced relative to the ratios ofL, G, and
M,
polypeptide synthesis withincreasing tonicity (Fig. 3). The incorpora-tion ofradioactive leucine into all viral polypep-tides was inhibited astonicity increased (e.g., incorporation into viral polypeptides at 550 mOsM is 25% of incorporation at 290 mOsM [26]). Therefore, the enhancement in rate of syn-thesis ofNandM2(as compared with the otheron November 10, 2019 by guest
http://jvi.asm.org/
[image:3.508.271.460.69.274.2]VOL.22,1977~~~~~RABIESPROTEIN SYNTHESIS 105
U.-i
100-0 5 ~at
'a -i
z 25
AAe
0~~~~~~~~~~~~~
10 2I0 3I0 40 50 60 70 80
[image:4.508.55.249.65.192.2]TIME AFTER INFECTION (h)
FIG. 2. Relative numberofviralpolypeptide mole-cules synthesized under hypertonic conditions (550
m~sM)atdifferenttimesafter infection.
Radioactiv-ity (31-labeled amino acid or[3H]leucine)
incorpo-rated into individual viralpolypeptidesperhourwas determined in several experiments by 7.5%
SDS-polyacrylamide gel analysis ofcell extracts as de-scribed in Methods. Correctionsweremadefor
differ-ences in the relative leucinecontentof
steady-state-labeled viral polypeptides (the ratio of 3H-labeled aminoacidmixturef[3H]leucineis:L, 0.78;G,1.03; N, 1.21;
Ml,
0.80;M,,0.72).Thepercentageofradio-activity incorporated into individual viral
polypep-tidepeaks(countsperminuteincorporatedinto indi-vidual viralpolypeptidepeaks/totalcountsper min-ute incorporated into viralpolypeptides x 100) was convertedtomoleculesofprotein fromtheirrespective
molecular weightsandnormalized to100molecules
ofNpolypeptide.The molecularweightsusedforthe calculationwere:L, 190,000(seetext);G, 80,000; N, 62,000; M,, 40,000; M2, 25,000 (40). Except forthe 12-h andthe 36-h timepoints,thevalues obtainedare derivedfrom two tofourseparateexperiments. viral
polypeptides)
reflects a relative, ratherthananabsolute,increaseintherateof
synthe-sis of these
proteins.
Over therangeof tonicities examined (450to
650mOsM), the
plots
of the relativerateofsyn-thesis versus
tonicity
of the individual viralpolypeptides
were linear. If we assume that thislinearrelationship
extendstoisotoniccon-ditions, ashasbeen shown for VSV
polypeptide
synthesis
(32),the datainFig.
3,when extrapo-latedtoisotonicity,
give
anestimateof therela-tive ratesof
synthesis
ofindividual rabiesviruspolypeptides
under isotonic conditions(Fig.
3, dotted lines). The percentage of totalvirus-specific radioactivity
incorporated
into indi-vidual viralpolypeptides
atisotonicity
(290 mOsM) was estimated to be: L = 0.5%, N =34%,G = 33%,
Ml
=25%, andM,
= 8%. Thesevalueswereusedtocalculate the relative num-ber of viral
protein
moleculessynthesized
un-derisotonicconditions (Table1) and showthat, underisotonicconditions,theL, G, N,M1,
andM,
polypeptides
were notsynthesized
in equi-molar amounts. Furthermore, the relativenumber of
polypeptide
moleculessynthesized
wasnot
comparable
to therelative number ofpolypeptide
molecules present in extracellular virus (Table 1).Virus-specific high-molecular-weight
poiy-peptide (L) in infected cells and in purified
virus.
Synthesis
of ahigh-molecular-weight
polypeptide
wasrepeatedly
detectedby
PAGEin infected
(Fig.
4a) but notin uninfected cells(Fig.
4b). Thispolypeptide, designated
the Lpolypeptide,
wasalsopresentinpurified
rabies virions(Fig.
5a) andco-migrated
with Lpoly-peptide
of VSV.Analysis
ofpurified
rabies yiin-ons labeled with [3Hlglucosamine
and 14 C-la-beled amino acids indicated that theLpolypep-tide of rabies virus was not
glycosylated
andco
Z
50-0 C.
C.)
-.
40-a. Cl)
CC
-J o
20-0
I-0
zW.)
a.
250 350 450 550 650
mOSM
FIG. 3. Effect of increasing tonicity on relative
ratesofrabies viruspolypeptide synthesis. Confluent monolayers (5 X 106 cells/60-mm dish) of rabies-infected (50 PFU/cell) and mock-infected BHK-21 cellswerelabeledat24hafter infection for1 h with
[3H]leucine (10 Ci/ml) under isotonic(290 mOsM)
orhypertonic(450, 500,550, 600,and 650 mOsM)
conditions. Whole-cell lysates (400 Mgof protein) of infected andmock-infected cells were run with '4C-amino acid-labeledprotein markers on continuous 7.5%SDS-polyacrylamide gels. The[3H]leucine in-corporationinto the viralpolypeptide peaks
attnibut-able to residual cellularprotein synthesis was sub-tractedafter normalizing thepolypeptidepattern of themock-infectedtotherabies-infectedcells. Thenet
[3H]leucineincorporationintovirus-specific polypep-tides (aftercorrection fordifferencesin relative
leu-cinecontentofthe viralpolypeptides) wasconverted
tothepercentage oftotalvirus-specificcounts inthe individualviralpolypeptides.
IN
-- G
IA
r - -- - - A -6 N1
CT----I- I I
VOL. 22, 1977
T
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.508.260.452.248.467.2]TABLE 1. Relative number of viral polypeptide molecules synthesized
Conditions L G N MI M2
Hypertonic (550 mOsM)a 0.60 32 100 31 79 Isotonic(290mOsM)b 0.40 64 100 75 35 Relative no. of viral poly- 1.9 101 100 52 86
peptide molecules per virionc
aPercentageof3H-labeledaminoacidincorporatedinto virus-specificpolypeptides at 550 mOsM (Fig. 3)converted torelative number of viral polypeptidemolecules synthe-sized per hour, asdescribed in Fig. 2. Thenumber of N polypeptideswas setequalto 100.
bExtrapolatedpercentage of3H-labeledamino acid in-corporated into virus-specific polypeptides at 290 mOsM (Fig. 3), converted to relativenumberof viral polypeptide molecules synthesized per hour. The number of N polypep-tide molecules was set equal to 100.
cDerived from gel analyses of three separate
prepara-tions ofpurified rabiesvirus (ERA strain), uniformly la-beled with 14C-amino acids (see text). The value for N polypeptidewassetequal to 100.
7
0-0~x
I
thus did not appear to be a dimer of the viral
glycoprotein (Fig. 5b).
Number ofL protein molecules per virion.
An estimate of the number of L protein mole-cules per virion was made from data derived from PAGEanalysisofthreedifferent
prepara-tions ofpurified rabiesvirusuniformly labeled
with"4C-labeledamino acids(Table 2). Approx-imately33 Lproteinmolecules wereestimated
tobe present perrabies virion. The numberof each ofthe othermajor structural protein mole-culespervirion estimated from these data
cor-respondedtothe valuesreportedbySokol et al.
(40).
Analysisof viralglycoprotein biosynthesis.
Avirus-specifiedglycopolypeptide labeled for2 h with[3H]leucineinrabies-infected cellsunder
hypertonic conditions consistently migrated in
PAGE faster than themarker virion
glycopro-tein (Fig. 6a). Since this component was not detected inuninfectedcellslabeled under
simi-40 50 60 70 80 90 100,
50
25
0c o
5 0
2 5
FRACTION NUMBER
START
FIG. 4. Virus-specific high-molecular-weight polypeptide (L) in infected cells. (a) Rabies-infected (50
PFU/cell) or (b) mock-infected BHK-21 cell monolayerswere labeledfor1 h with [3H]leucine(10 uCi/ml)
underhypertonic conditions (550 mOsM)24 hafterinfectionat379C, and400-pgportionsofproteinofthe whole-celllysates wereanalyzedondiscontinuous10%SDS-polyacrylamide gels.['4C]leucine-labeledVSV wassubjectedtoco-electrophoresis with the celllysates. Thearrowsrefertothe VSV markerproteins.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.508.142.376.302.614.2]RABIES PROTEIN SYNTHESIS
5 0
X
0 4
4 M12 3 Q
A
2J
i~%~ ~
I~~~~~~
3 1S 2
4
I-_ 10 20 30 40 50 60 70 80 90 100 11o +
STiRT FRACTION NUMBER
FIG. 5. Virus-specific high-molecular-weight polypeptide (L) inpurified rabies virus. Roller bottles of BHK-21 cells (1.5 x 108cellslbottle) were infected with rabies virusat5PFU/cell; the infectedcellswere
grown in 0.2% BSA-MEM for 4 days at 33°C in the presence of[3H]leucine (2 pmCi/ml) alone, or [3H]glucosamine (10 pXilml) and ["4C]leucine (2.5 pCi/ml). The virus was harvested and purified as
described inMaterials and Methods.(a) Analysis of[3H]leucine-labeledrabies virus with[14C]leucine-labeled
[image:6.508.141.375.64.344.2]VSV(12.5-cm gel). (b)Analysis of [3H]glucosamine-and[14C]leucine-labeled rabies virus(11-cm gel). TABLE 2. Number ofpolypeptide molecules per
rabies viriona
Poly- ~~~~~~~Daltons
Mole-peptid
%Total radio- Ml (oyp- cules species activity tide/virion)i/vir
ion) L 1.85 + 0.94 190,000 6 x 106 33 G 42.67 + 1.30 80,000 145 x 106 1,815 N 32.94 ± 0.43 62,000 112 x 106 1,806 M, 11.17± 0.19 40,000 38 x 106 951 M2 11.36 ±0.73 25,000 39 x 106 1,547
aPercentage of totalvirion-associated 14C-labeled amino
acidincorporatedintotheindividual viral polypeptides was determined. Thenumber ofdaltonsof each viral polypep-tideper virion wasdetermined by multiplying the percent-ageof eachviralpolypeptide in the virion times the molecu-lar weightof the total polypeptide (340.4 x 106 daltons) in thevirion(calculated from the ratio of polypeptide to RNA [74:1]inthe virion, assumingthe RNA molecular weight to be 4.6 x106daltons[40]).The number of each polypeptide molecule per virion wascalculated by dividing the number ofdaltonsof eachpolypeptide per virion by the molecular weight of that polypeptide. The molecular-weight values for the G, N, M,, and M2 are from Sokol et al. (40). The molecular weight of the L protein was estimated at 190,000 onthe basis ofco-electrophoresis with the VSV L protein (Fig. 5a) (49).
lar conditions (notshown) and could be quanti-tatively precipitated with monospecific antise-rum directed against purified virion glycopro-tein (B. Dietzschold, H. P. Madore, and J. M.
England, unpublished observations), it was
considered to be virus specific. When rabies-infected cells were labeled for 2 h with
[3H]leucine under hypertonic conditions and chased for 24 h in isotonic medium, a large
proportion of the radioactivity that was
previ-ously associated with the faster-migrating in-tracellular glycopolypeptide shifted to the slowermigrationrateofthe virionglycoprotein
(Fig. 6b). A small proportion of the
radioactiv-ity associatedwith the intracellular glycopoly-peptide still migrated more rapidly than the virionglycoprotein (Fig. 6b). The [3H]leucine-labeled glycopolypeptide that was synthesized under hypertonic conditions was incorporated into virusreleased into the mediumduringthe 24-hchase in isotonicmedium, asshownby co-migration with the rabies 14C-labeled virion
glycoprotein that was labeled in isotonic
me-VOL. 22, 1977 107
I
0
2
I
I
0
a.)
U J
z
J
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.508.59.253.423.533.2]N0201 A000
u
IS
25M-c. I c-)
L
W 10- 50
Z
~~~~~~~~0
1020~ ~3 4 0 60 7
---r
---F
35- 30-
25-20 6 P10u0
ti5 (05
i0 (0PK
-
2.5I
10 20 30 40 50 60 70°
SAT FRACTION NUMBER
FIG. 6. Pulse-chaseanalysis ofintracellular viral
glycoprotein synthesized under hypertonic
condi-tions. Cultures(5 X106cells/60-mmdish)of rabies-infected (50PFU/cell) BHK-21 cells grown at370C for24 h were eitherpulse-labeled with [3H]leucine (100 gCi/ml) for2 h at 550 mOsMat 370C (a) or
pulse-labeledfor2hat550mOsMandthenchased for24hat37°Cin290mOsM medium (b). Whole-cell lysates containing 50,000 trichloroacetic acid-precipitablecpm (a) and(b) andextracellular virus from (b) above containing 30,000 trichloroacetic acid-precipitablecpm(c)wereanalyzedon discontin-uous10%SDS-polyacrylamidegels. Thearrows
in-dicate 14C-amino acid-labeled rabies virus marker proteinssubjectedtoco-electrophoresiswiththe
sam-ples.
dium (Fig. 6c). These results indicatethat hy-pertonicconditions cause the accumulation of a morerapidly migratingform of the viral
glyco-protein. During an isotonic chase, however,
this more rapidly migrating form of
glycopro-tein is converted to a form that co-migrates
with virion glycoprotein and becomes associ-ated withextracellularvirus.
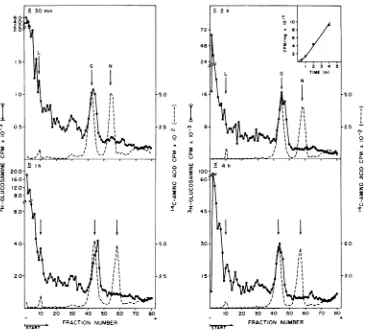
In effortsto detectthe synthesis ofa faster-migrating formof viral glycoprotein under iso-tonicconditions(290 mOsM),infected and unin-fectedcellswerelabeled with [3H]glucosamine. As showninFig.7aandb, aprominent hetero-geneouspeakof[3H]glucosaminelabel was de-tected intheinfected butnotin the uninfected
celllysates.Thisviral-specificglycopolypeptide showed two components on the gel, a major component, whichco-migratedwith thevirion
glycoprotein, and a minor component, which
migrated morerapidlyas ashoulder. Both
com-ponents could be quantitatively precipitated
withantibody directed against virion
glycopro-tein (B. Dietzschold, H. P. Madore, and J. M.
England, unpublished observations). We have
designatedthe morerapidly migrating
compo-nent gp78 to distinguish itfrom the
glycopro-tein present inextracellular virus (gp8O). The faster-migrating component exhibited approxi-mately thesameelectrophoreticmobilityasthe virus-specific peak labeled with [3H]glucosa-mineunderhypertonic conditions (Fig. 7cand d).
To examine thepossibilitythat gp78 was an
intermediateinthebiosynthesis of rabies virus glycoprotein (gp8O), infected cells were pulse-labeled with [3H]glucosamine under isotonic conditions forintervals from 30min to4 h (Fig. 8). After 30 min oflabeling, the majority of
[3H]glucosamine, whichwasassociated with
vi-rus-specificglycopolypeptide, was inthe faster-migrating component, gp78 (Fig. 8a).After1h
of labeling, a portion of the [3H]glucosamine
label was in a shoulder of the gp78 peak that migrated with the gp8O marker (Fig. 8b). After
2h (Fig. 8c) and 4 h (Fig. 8d) oflabeling, the majorityofthe[3H]glucosaminelabelwas
asso-ciatedwithgp8O. This shift of[3H]glucosamine label fromgp78 togp80 withincreasedlabeling
time suggests that gp78 is a precursor ofthe
mature viralglycoprotein (gp8O).
DISCUSSION
Using hypertonic conditions to selectively
suppress cellular protein synthesis, we have
characterized several aspects of rabies virus protein synthesis in BHK-21 cells. The rabies virus structural polypeptides L, G, N,
M,,
M2are synthesized continually throughout the
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.508.65.247.63.501.2]RABIES PROTEIN SYNTHESIS 109
1Y
0
N
0.
w
z
4 0
C-)
IS
en
I20
I.0
-2
0
0
4 0
-30 -15
10 20 30 40 50 60 70 80 90 160 10 20 30 40 50 60 70 80 90 100
[image:8.508.56.455.59.382.2]FRACTION NUMBER
FIG. 7. Labelingofrabies-infectedoruninfected cells with[3H]glucosamineunder isotonic (290mOsM)or
hypertonic(550mOsM)conditions. Confluent monolayers(5 x 106cells/60-mm dish)ofBHK-21 cellswere eitherinfectedwithrabies virus (100PFU/cell) (a and c)ormock-infected(bandd)andincubatedat37'C
for40 h.Cells were then labeled under isotonic conditions(290mOsM) for2hwith[3H]glucosamine(50 XciI
ml) (a and b). After pretreatment for 15 min withhypertonicmedium (550mOsM),other cells werelabeled
for2h inthe same hypertonic medium containing[3H]glucosamine (50 puCi/ml)(c andd). The whole-cell
lysates containing27,000 trichloroaceticacid-precipitable cpm wereanalyzedon discontinuous10%
SDS-polyacrylamidegels. Arrows indicate 14C-amino acid-labeled rabies virus proteins.
courseof infection. Incontrasttothesynthesis of the NS polypeptide ofVSV, which has been showntobepreferentially
synthesized
early
in infection (17), none of the rabies-specificpoly-peptides appearedtobesynthesizedat agreater
relativerateearlyininfection.
The relative rates ofsynthesis of the rabies
virus polypeptides do not correspond to their relative proportions in the intact virion. This observation indicates that theprotein
composi-tion ofvirions is not solely determined by the relativerate atwhich the viralpolypeptidesare
synthesized andsuggests thatadditional mech-anismsunderlieviral assembly.In the absence ofdata that compare the absolute rate of
syn-thesis of a given polypeptide to the rate at
which thatpolypeptide isincorporated into
vi-rus, noconclusioncanbe made concerningthe
rate of synthesis of a given viral polypeptide and arate-limitingstep inviral assembly.
Lodish(24)hasproposedamodel for the regu-lation ofa- and f3-globin synthesis which
pre-dicts that a reduction in the overall rate of
peptide chaininitiation willcausethe preferen-tial translation of mRNA species with higher
rate constants of peptide initiation. Since hy-pertonicconditions have been shownto specifi-cally suppress peptide chain initiation, this model can account forthe differential inhibi-tioncausedby hypertonicconditionsof the
syn-thesis ofvariouscellular (33)andviral (rabies, VSV [32], and MMTV [36]) polypeptides.
Ifthis modelis indeed correct, the different
sensitivities amongthesynthesesof L, N, and M2, aswellasG andMl,polypeptidesto
hyper-tonic conditions suggests that at least three
VOL. 22, 1977
on November 10, 2019 by guest
http://jvi.asm.org/
I
0
a-0)
30min
8.0-5
1.0-
ft7
II
II I I II
z
f 200
<n 16.0
0
°
12.0-80
ID
6.0-4.0
2.0
-5.0
TI
-2.5 N,
o
X X
(
E
2
J
0
9 A,
-50
-25
I-0
z
0 20 30 40 50 60 70 80 10 20 30 40 50 60 70 80
FRACTION NUMBER FRACTION NUMBER
START lSTART
FIG. 8. Kinetic analysisof intracellularviral glycoproteinsynthesis.Confluent monolayers(5 x106 cells!
60-mmdish) wereinfected with rabies virus (100PFUlcell),and thenincubated for40hat37TC. Cellswere
exposedto[3H]glucosamine (50
gCi/ml)
inisotonicmediumfor 0.5 h (a),1h(b), 2 h (c), and4h (d). Whole-cell lysates (450 pgofprotein) were analyzed ondiscontinuous 10%SDS-polyacrylamidegels. 14C-amino acid-labeled rabies virus proteinsweresubjectedtoco-electrophoresis with the cell lysates. Totalincorporation of[3H]glucosamineintoacid-precipitablematerialas afunction of time is shown in the insert of (c).ribosome-binding sites (with different initia-tion efficiencies) exist in rabies virus mRNA. Since eukaryotic mRNA apparently contains only one functional initiation site (9, 13, 22), thesedata suggest that there isaminimum of threediscrete rabies virus mRNA species. Con-sistent with this argument are observations that the synthesis ofindividual poliovirus pro-teins, whichare translated fromasingle
poly-cistronic mRNA molecule with one initiation site (13, 35), does not show differential inhibi-tionbyhypertonic conditions (34), whereas the synthesis of individual VSV proteins, which are translated from several monocistronic mRNAmolecules (4, 20, 28, 29, 46), is differen-tially inhibited by hypertonic conditions (32,
34).
A majordifferencebetween rabies virus and VSVhas been the inabilitytodetect RNA
tran-scriptase activity in purified rabies virions (41, 48). RNA transcriptase activity could not be
detected in purified rabies virions by in vitro
assays that were sensitive enough to detect RNAtranscriptasewithaspecific activity 4,000 times less than that present in VSVvirions (H. P. Madore and F. Sokol, unpublished observa-tions).
In VSV, the high-molecular-weight L poly-peptide has been shown to be necessary for RNA transcriptase activity (5-7, 10, 11, 31). Since a high-molecular-weight polypeptide analogous to the VSV L polypeptide had not
beenclearly demonstratedinrabiesvirions(40, 42), itwaspossibletoattribute thedeficiencyin RNAtranscriptase activity inrabies virionsto
the absenceorextremely lownumber of L poly-peptide molecules per virion. In the present work, however, wehave detecteda high-molec-ular-weight polypeptide (with an electropho-retic mobility similar to that of the VSV L
polypeptide) in rabies virions and in infected
cells, and we have estimated that each rabies b Ih
I
1
I
I I
III
I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.508.80.448.59.389.2]RABIES PROTEIN SYNTHESIS 111 virion containsapproximately 33 Lpolypeptide
molecules (Table 2). In comparison, aVSV vir-ion contains about 60 L protein molecules (3). Therefore, if we assume that the Lpolypeptides of rabies virus and VSV arefunctionally simi-lar, the extremely low level of RNA transcrip-tase activity in rabies virions does not seem to result from an absence or extremely low num-ber of L polypeptide molecules in the rabies virion.
In rabies-infected cells labeled for 2 h with [3H]glucosamine under isotonic conditions, two forms of virus-specific glycoprotein are ob-served: the major form, designated gp80,
co-migrates with the viral glycoprotein; a minor
form, designated gp78, is seen as the
faster-migrating shoulder of the major viral glycopro-teinpeak (Fig. 7a). Kineticanalysis of the
bio-synthesis of these two forms has revealed that
the initial appearance of the faster-migrating gp78 and the subsequent accumulation of the
gp8Oiscoupled withthe slowdisappearance of
the gp78(Fig.8a-d).These resultssuggest that the faster-migrating gp78 isanintermediatein
the synthesis of gp8O. Furthermore, gp78 may represent anincompletelyglycosylatedform of the viral glycoprotein, since neuraminidase-treated viralglycoproteinmigratesonPAGE at
a rate that approximates that of intracellular
gp78 (B. Dietzschold, unpublished
observa-tions). Glycoproteinsynthesis of another rhab-dovirus, VSV, also appears to involve incom-pletelyglycosylatedintermediates that migrate faster than the mature glycoprotein in
SDS-polyacrylamide gels (1,4, 17, 20, 28, 29,45, 46).
The rabies-specific glycoprotein in
infected
cellslabeledwith aminoacids (Fig. 6a) or with
glucosamine (Fig. 7c) underhypertonic
condi-tions migrates in SDS gels with a mobility similar to that of gp78. The observation that this faster-migrating glycoprotein can be chased under isotonicconditions into an
intra-cellular
glycopeptide
that co-migrates withgp8O and is subsequently released with
extra-cellular virions (Fig. 6b and c) suggests that hypertonic conditionsmay reversiblyinterfere withthe biosynthesis of rabiesvirus glycopro-tein. The faster migration rate of the product
synthesized under hypertonic conditions
sug-gests that interference occurs at an
intermedi-ate that requires supplementaryglycosylation
to formthematureviralglycoprotein. Whether this component is (i) identical to gp78, (ii) an-other intermediate not resolved from gp78, or
(iii)anaberrantstepinviralglycoprotein
mat-uration remains to bedetermined. A
compari-sonof the sizeandionexchange distribution of
the oligosaccharides of the viral glycoproteins
isbeing carriedout to answerthisquestion.
The reversible block of rabies glycoprotein
biosynthesis caused by hypertonic conditions
appears tobeanalogoustothe reversible block in Semliki forest virus glycoprotein synthesis, which occurs wheninfected cells are grown in
sugar-free medium (15). In contrast to hyper-tonic and sugar-free conditions, the impaired glycosylation of influenza (19, 37) and Semliki forest virus (14)glycoprotein intermediates
in-duced by 2-deoxyglucose or excess glucosamine cannotbereversedafterremovalof these inhib-itors. Thisirreversibleblockageinglycoprotein biosynthesis isattributedto incorrect glycosyl-ationinduced bythe inhibitors (15).
ACKNOWLEDGMENTS
The skilled assistance of Len Milewski and Len Kaplan isgratefullyacknowledged.
This work wassupported by Public Health Service grant AI-09706 from theNational Institute for Allergy and Infec-tiousDiseases, a fellowship (CA-01463) from the National CancerInstitute,andgrant RR-05540 from the Division of Research Resources.
LITERATURE CITED
1. Atkinson, P. H., S. A. Moyer, and D. F. Summers. 1976.Assemblyofvesicular stomatitis virus glycopro-teinand matrix protein into HeLa cell plasma mem-branes.J.Mol. Biol. 102:613-631.
2. Baltimore, D., A. S. Huang, and M. Stampfer. 1970. Ribonucleic acid synthesis of vesicular stomatitis vi-rus.II. AnRNA polymerase in the virion. Proc. Natl. Acad.Sci. U.S.A. 66:572-576.
3. Bishop, D. H. L., and P. Roy. 1972. Dissociation of vesicular stomatitis virus and relation of the virion proteins to theviral transcriptase. J. Virol. 10:234-243.
4. Both, G. W., S. A. Moyer, and A. K. Banerjee. 1975. Translation andidentification of the mRNA species synthesized in vitro by the virion-associated RNA polymerase of vesicularstomatitis virus. Proc.Natl. Acad. Sci. U.S.A. 72:274-278.
5. Emerson, S. U., andR. R.Wagner. 1972. Dissociation andreconstitution ofthetranscriptaseand template activitiesofvesicularstomatitis BandT virions. J. Virol.10:297-309.
6. Emerson, S. U., and R. R. Wagner. 1973. L protein requirementforinvitroRNAsynthesisby vesicular stomatitis virus. J.Virol. 12:1325-1335.
7. Emerson, S. U., and Y-H. Yu. 1975. Both NS and L proteins arerequiredfor in vitro RNA synthesis by vesicularstomatitis virus. J.Virol. 15:1348-1356. 8. Hummeler, K., N. Tomassini, F. Sokol, E. Kuwert, and
H.Koprowski.1968.Morphologyof thenucleoprotein componentofrabiesvirus.J.Virol. 2:1191-1199. 9. Glanville, N.,M.Ranki,J.Morser, L.Kariainen,and
A.Smith. 1976.Initiation oftranslation directed by 42Sand 26S RNAsfromSemlikiForest Virus in vitro. Proc. Natl.Acad. Sci. U.S.A. 73:3059-3063. 10. Hunt, D.M., S. U. Emerson, and R. R. Wagner. 1976.
RNA- temperature-sensitive mutants of vesicular stomatitis virus: L-protein thermosensitivity ac-counts for transcriptase restriction of group I mu-tants.J.Virol. 18:596-603.
11. Hunt, D. M., and R. R. Wagner. 1974. Location of the transcriptiondefect in group I temperature-sensitive mutants ofvesicular stomatitis virus..J. Virol. 13:28-35.
VOL. 22, 1977
on November 10, 2019 by guest
http://jvi.asm.org/
12. Iwasaki, Y., T. J. Wiktor, and H. Koprowski. 1973. Early events of rabies virusreplicationin tissue cul-tures. An electron microscopic study. Lab. Invest. 28:142-148.
13. Jacobson,M.F.,and D. Baltimore. 1968. Polypeptide cleavages in the formation ofpoliovirus proteins. Proc.Natl.Acad. Sci.U.S.A. 61:77-84.
14. Kaluza,G.1975.Effect ofimpaired glycosylationonthe biosynthesisof Semliki forest virusglycoproteins.J. Virol. 16:602-612.
15. Kaluza,G. 1976. Earlysynthesisof Semlikiforest vi-rus-specific proteins in infected chicken cells. J. Vi-rol. 19:1-12.
16. Kang,C. Y.,and L. Prevec. 1969. Proteins of vesicular stomatitis virus. I. Polyacrylamide gel analysis of viral antigens. J. Virol. 3:404-413.
17. Kang,C.Y., andL.Prevec. 1971.Proteins of vesicular stomatitis virus.III. Intracellularsynthesis and
ex-tracellularappearanceofvirus-specificproteins. Vi-rology 46:678-690.
18. Kaplan, M. M., T. J.Wiktor,R. F.Maes, J.B. Camp-bell,and H. Koprowski. 1967. Effect ofpolyionson theinfectivityofrabiesvirus in tissueculture: con-struction ofa single-cycle growth curve. J. Virol. 1:145-151.
19. Klenk,H-D., C.Scholtissek,and R. Rott. 1972. Inhibi-tionofglycoproteinbiosynthesisofinfluenzavirusby D-glucosamine and 2-deoxy-D-glucose. Virology 49:723-734.
20. Knipe,D., J.K.Rose, andH. F.Lodish.1975. Transla-tionof individualspecies of vesicular stomatitis viral mRNA. J.Virol. 15:1004-1011.
21. Knudson, D. L. 1973. Rhabdoviruses. J. Gen. Virol. 20:105-130.
22. Lachmi, B.-E., and L. Kaariainen. 1976. Sequential translation ofnonstructural proteins in cellsinfected with a Semliki Forest virus mutant. Proc. Natl. Acad.Sci. U.S.A. 73:1936-1940.
23. Laemmli,U. K. 1970. Cleavage ofstructural proteins during the assembly oftheheadofbacteriophageT4. Nature (London)227:680-681.
24. Lodish, H. F. 1974. Modelfor the regulation of mRNA translationappliedtohaemoglobin synthesis. Nature
(London)251:385-388.
25. MacPherson,I., andStoker, M. 1962.Polyoma trans-formation ofhamster cell clones-an investigation ofgenetic factorsaffecting cellcompetence.Virology
16:147-151.
26. Madore, H. P., and J. M. England. 1975. Selective suppression of cellularprotein synthesisin BHK-21 cells infected with rabies virus. J. Virol. 16:1351-1354.
27. Matsumoto, S. 1974. Morphology of rabies virion and cytopathologyof virus infectedcells.Symp. Ser. Im-munobiol. Handb. 21:25-34.
28. Morrison,T.G.,and H. F.Lodish.1975.Siteof synthe-sisofmembrane and nonmembrane proteins of vesic-ularstomatitis virus. J.Biol. Chem.250:6955-6962. 29. Morrison, T., M. Stampfer, D. Baltimore, and H. F.
Lodish. 1974.Translation of vesicular stomatitis mes-sengerRNAbyextractsfrommammalian andplant cells. J.Virol. 13:62-72.
30. Oyama, V. I., and H. Eagle. 1956. Measurement of cell growthintissueculture with aphenol reagent (Folin-Ciocalteau). Proc.Soc. Exp. Biol. Med. 91:305-307. 31. Naito,S., and A. Ishihama. 1976. Function and
struc-ture of RNApolymerase from vesicular stomatitis virus. J.Biol. Chem. 251:4307-4314.
32. Nuss, D. L., and G. Koch. 1976. Differential inhibition ofvesicular stomatitis viruspolypeptidesynthesisby hypertonic initiationblock.J.Virol. 17:283-286. 33. Nuss, D. L., and G. Koch. 1976. Variation in the
rela-tivesynthesisofimmunoglobulin G and non-immu-noglobulin G proteins in cultured MPC-11 cells with
changes in the overall rate of polypeptide chain initi-ationandelongation. J. Mol. Biol. 102:601-612. 34. Nuss, D. L., H. Oppermann, and G. Koch. 1975.
Selec-tiveblockage of initiation of host protein synthesis in RNA-virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 72:1258-1262.
35. Saborio,J.L., S-S.Pong,and G. Koch. 1974. Selective andreversible inhibition of initiation of protein syn-thesis in mammalian cells. J. Mol.Biol. 85:195-211. 36. Schochetman, G., and J. Schlom. 1976. Independent polypeptide chain initiation sites for the synthesis of different classes ofproteins for an RNA tumor virus: mouse mammarytumor virus.Virology 73:431-441. 37. Schwarz, R. T., and H-D. Klenk. 1974. Inhibition of
glycosylation of the influenza virus hemagglutinin. J. Virol. 14:1023-1034.
38. Sedwick, W. D., and T. J. Wiktor. 1967. Reproducible plaquing system for rabies, lymphocytic choriomen-ingitis,and otherribonucleic acid viruses in BHK-21/ 13S agarose suspensions. J. Virol. 1:1224-1226. 39. Sokol,F. 1973.Purification ofrabies virusandisolation
of itscomponents, p. 165-178. In M. M. Kaplan and H.Koprowski(ed.), Laboratory techniques in rabies. World Health Organization, Geneva.
40. Sokol, F. 1975.Chemicalcompositionand structure of rabies virus, p. 79-102. In G. M. Baer (ed.), The natural history of rabies. Academic Press, Inc., New York.
41. Sokol, F., and H. F. Clark. 1973. Phosphoproteins, structural components of rhabdoviruses. Virology 52:246-263.
42. Sokol, F., H. F.Clark,T. J. Wiktor, M. L. McFalls, D. H. I. Bishop, andJ. F. Obijeski. 1974. Structural phosphoproteins associatedwith ten rhabdoviruses. J.Gen. Virol. 24:433445.
43. Sokol, F., H.D. Schlumberger,T. J. Wiktor, and H. Koprowski. 1969.Biochemical and biophysical stud-ies on the nucleocapsid and on the RNA of rabies virus.Virology 38:651-665.
44. Sokol, F.,D.Stancek,and H.Koprowski.1971. Struc-tural proteinsofrabiesvirus. J.Virol. 7:241-249. 45. Toneguzzo, F., andH. P. Ghosh. 1975. Cell-free
syn-thesis ofvesicular stomatitis proteins: translation of membrane-boundpolyribosomal mRNAs. FEBS Lett. 50:369-373.
46. Toneguzza, F., and H. P. Ghosh. 1976. Characteriza-tionandtranslation of methylatedand unmethylated vesicular stomatitis virus mRNA synthesized in vitro by ribonucleoproteinparticles from vesicular stoma-titisvirus-infectedLcells. J. Virol. 17:477491. 47. Tsiang,H., and P. Atanasiu. 1971. Replication du virus
rabiquefixe en suspension cellulare. C. R. Acad. Sci. 272:897-900.
48. Villarreal,L.P., and J. J.Holland.1974.Transcribing complexes in cells infected by vesicular stomatitis virusand rabiesvirus. J.Virol. 14:441-450. 49. Wagner,R. R.,L.Prevec, F. Brown, D. F.Summers,
F. Sokol, and R. MacLeod. 1972. Classification of rhabdovirus proteins: a proposal. J. Virol. 10:1228-1230.
50. Wagner,R. R., T. A.Schnaitman, and R. M. Snyder. 1969. Structural proteins of vesicular stomatitis vi-rus.J.Virol. 3:395403.
51. Wagner, R. R., T.C. Schnaitman, R. M. Snyder, and C. A. Schnaitman. 1969. Protein composition of the structural components of vesicular stomatitis virus. J.Virol. 3:611-618.
52. Wiktor, T. J. 1973. Tissue culture methods, p. 101-123. InM. M.Kaplan and H. Koprowski (ed.), Laboratory techniques in rabies. World Health Organization, Geneva.
53. Wiktor, T. J., E.Gyorgy,H.D.Schlumberger, F.So. kol,and H.Koprowski. 1973. Antigenic properties of rabies viruscomponents. J. Immunol. 110:269-276.
on November 10, 2019 by guest
http://jvi.asm.org/



![FIG. 5.BHK-21grown[3H]glucosaminedescribedVSV Virus-specific high-molecular-weight polypeptide (L) in purified rabies virus](https://thumb-us.123doks.com/thumbv2/123dok_us/1546534.107188/6.508.59.253.423.533/glucosaminedescribedvsv-virus-specific-molecular-weight-polypeptide-purified-rabies.webp)

![FIG. 7.polyacrylamideforforeitherlysateshypertonicml) Labeling ofrabies-infected or uninfected cells with [3H]glucosamine under isotonic (290 mOsM) or (550 mOsM) conditions](https://thumb-us.123doks.com/thumbv2/123dok_us/1546534.107188/8.508.56.455.59.382/polyacrylamideforforeitherlysateshypertonicml-labeling-ofrabies-infected-uninfected-glucosamine-isotonic-conditions.webp)