0022-538X/91/105471-06$02.00/0
Copyright © 1991,American SocietyforMicrobiology
Polymerase
Chain
Reaction Analysis
of
Defective
Human
T-Cell
Leukemia Virus
Type I
Proviral Genomes
in
Leukemic
Cells of Patients with Adult T-Cell Leukemia
BETTE
KORBER,lt
AKIHIKOOKAYAMA,"2RACHELDONNELLY,1 NOBUYOSHI TACHIBANA,2AND MYRON
ESSEX'*
Department of Cancer Biology, Harvard School of Public Health, 665Huntington Avenue, Boston, Massachusetts02115,'
and Second
Department
of
Medicine,
Miyazaki
MedicalSchool,
Miyazaki
889-16,
Japan2
Received 18March1991/Accepted21June 1991Human T-cell leukemia virus type I(HTLV-I) is the etiologic agent of adult T-cell leukemia, and the clonally derived leukemic cells all contain proviral genomes. Polymerase chain reaction with a variety of primers which span the HTLV-Igenome was used to determine that a significant fraction of patients (at least 32%) carry
deletedviral genomes in their leukemic cells. The pX region of the HTLV-I genome encoding the regulatory
genes tax and rex was preferentially retained. The fact that the tax coding region was retained provides supportingevidence that the tax protein contributes to leukemogenesis in vivo. The reasonablyhigh fractionof patients with adult T-cell leukemia carrying deleted genomes in their tumor cells suggests that the deletions have a role inleukemogenesis.
The human T-cell leukemiavirus type I (HTLV-I) is the causative agent of the human neoplasm adult T-cell leukemia (ATL) (13, 24, 37). The infection of a target cell with the virus is a prerequisite for transformation and the develop-mentof ATL. TheHTLV-Igenomecontains viral structural genes as well as the regulatory proteins Tax and Rex,
encodedin the pXregion ofHTLV-I(12, 19, 27, 32). The pX
region isexpressed as adoubly spliced mRNA with tax and rexutilizing two overlapping open reading frames (28). Tax acts in conjunction with cellular proteins (15, 26, 34) to
transactivate transcription from the HTLV long terminal
repeat (LTR) (7, 29, 33), as well as from cellular promoters
(4, 20,21, 25, 36, 38). Rex is aposttranscriptionalregulatory
protein that shifts viral mRNA expression from the doubly
spliced form that encodes Tax and Rex to the single and
nonspliced forms that encode the viral structural genes (5,
14).
The HTLV-I genome contains no homolog of a cellular proto-oncogene (27), and because HTLV-I does not inte-grate atpreferential sites inthe human genome,insertional
mutagenesis resultinginproviral activation ofacellulargene
is notalikelytrigger for developingATL(39). Because there
isa long latency from timeofinfectiontodisease
manifes-tationandalowlifetimeriskofgettingATLifinfectedwith
the virus, a multistep process is probably required for
oncogenesis (22). The Tax protein has been strongly
impli-catedintumorigenesis, although its precise role remains to
be elucidated. Taxexpressedfromanexpressionvectorwith the rest ofthe HTLV genome deleted is capable of
trans-forming cells in vitro (10, 35). Itcanstimulate expression of
the
lymphokine
interleukin-2 (IL-2) and the IL-2 receptoralphachain,aswellasadditional lymphokinesand thec-fos
gene (4, 14, 20, 21, 25). Taxmayalso influence
transforma-tionthrough itscapacity to
down-regulate
beta-polymerase,a cellular enzyme involved in host cell DNA
repair,
thuspossibly increasingthe mutation rate ininfected cells (16).
*Corresponding author.
tPresentaddress: TheoryDivision, T-10, LosAlamosNational Laboratory, LosAlamos, NM87545.
Defective HTLV-I genomes with largedeletions have been
detected in leukemic cells from patients with ATL (31, 35, 39), and inseveralcasesitwas shown that the pX regionof
the HTLV-I genome was retained (35, 39). Despite the
evidence associating tax with oncogenesis, only very low levelsofthetaxmRNA aredetected in vivo in ATL cells. In most cases, it was not detected by Northern- (RNA) blot
analysis (8) and was found in extremely low levels by
enzymaticamplificationbypolymerase chain reaction(PCR)
(17). Therefore, it has beensuggestedthattaxexpressionis
acritical component in the initiationof transformationbut is
notimportantin maintainingtumorgrowth (8).
PCRcan be usedsemiquantitativelytoestimate the
num-berofprovirusespresentinvivo(1). Weusedthistechnique
toshow that deleted HTLV genomespreferentiallyretaining
the pX region and 3' LTR are frequently found (in at least
32% ofthepatients withATLtested)in theleukemic cells of
patients with ATL.
MATERIALSANDMETHODS
Blood samplesand DNAextractions. Blood samples were
collected from 23patients withATLor
lymphoma
from theMiyazaki Medical School, Miyazaki, Japan.
Peripheral
blood mononuclearcells
(PBMCs)
wereisolated from freshwhole-blood samples by Ficoll-Hypaque separation and
washed twice in phosphate-buffered saline, and the pellet wasstoredat
-70°C
untilthe DNAwasextracted. Threetofive HTLV-negative blood samples were obtained from a
Boston blood donorcenterandwere includedin each setof
DNA preparations to serve as negative controls for PCR.
ChromosomalDNA wasisolatedby sodium
dodecyl
sulfate(SDS)-proteinase
K digestions of the cells at56°C
and thenby
phenol-chloroform
extractions and ethanolprecipitation
ofDNA
(31).
DNA wasresuspended
in lx TE(10
mMTris[pH 7.8], 1 mM EDTA) by rocking for 24to 48 h at room
temperature, and DNA concentrations were determined
by
measuring the absorbance ratioof 260/280nm. HUT102cells (24) were used as anHTLV-I-positive
control,
and HUT78cells(9, 23)wereusedas an
HTLV-I-negative
control.DNA5471
on November 10, 2019 by guest
http://jvi.asm.org/
was extracted in a tissue culture facility in an area separate from that where the PCR products were analyzed.
Amplification of DNA. A 2-,ug portion of PBMC DNA was typically used peramplification reaction (there are approxi-mately 140,000 human genomeequivalents in 1
,ig
of DNA). Each PCR experiment included a set of negative control DNAs that had been coextracted with the ATL DNA, a negative control with DNA from the uninfected cell line HUT78, and a reaction with cocktail only and no added DNA. In addition, a 10-fold dilution series was included in each set with serial dilutions of HTLV-I-infected HUT102 cell DNA diluted in a background of 2 ,ug of uninfected HUT78 DNA to determine the sensitivity of each experi-ment. (The number of HTLV-I genomes per cell in ourHUT102culture was estimated bycomparing theintensityof
the PCRamplification product signals from serial dilutions of
HUT102 cells with dilutions of a known number of
mole-cules of the HTLV-bearing plasmid pMT2, cut with the restriction enzyme PstI. Using this method, we estimated that there were seven HTLV-I genomes perHUT102 cell.) The MgCl2 and primerconcentrations were optimized for eachprimer pair.Amplification reactions containedtemplate DNA, 1.25 mM of each deoxynucleoside triphosphate, 50 mM KCl, 10 mM TrisHCI (pH 8.3), 1.5 to 4 (usually 2.5) mM
MgCl2,and 0.2 to 1 ,uMeach primer in a
100-plI
totalvolume.Reaction mixtures were heated to 95°C for 3 min and then cycled 39 times for 1 min at 95°C, 45 s at 55°C, and 30 s at 720C, using an MJResearchthermocycler. The primer pairs had a range ofsensitivities even when optimized. All PCR experiments were assayed by viewing the product on an ethidium bromide-stained gel to ascertain that the appropri-ate-sized PCR product gave the predominant band, coupled with a dot-blot assay (3).
PCR primers and probes. Allprimers gave a strong distinct band of the appropriate molecular weight on an ethidium bromide-stained gel that reacted specifically with the appro-priate oligonucleotide probe when analyzed by Southern blotting with HUT102 cell DNA as a template (with the exception of the primers that go across the splice site). The following list of primers either gives a reference or, if the primers are described here for the first time, the 5'-3' sequence of the oligonucleotides used and the bases which define theouterboundaries of the amplified region. The base numbers correspond to the GenBank sequence, accession number J02029. Primers SK43 and SK44 amplify from the tax region and were probed with SK45 (18). SG231 and SG238 amplify from the pol region and were probed with SG232 (11). SG452 and SG453 amplify from the env region and were probed with SG228 (11). (SG452 and SG453 are essentially equivalent to SG221 and SG227, respectively, in Greenberg et al. [11], except SG452 and SG453 are both lacking the first three 5' bases ofSG221 and SG227.) Primers RM3 andRM4 amplify from the LTRs (bases 420 to 720) and were probed with RM3/4: RM3, GCCATCCACGCCGGT
TGAGTCGCGTTCTGC; RM4, CCAACGGAGTCGCCGG
TACTTGGCCGTGGG; RM3/4, GTGCCTCCTGAACTGC
TGCTGCCGCCGTCTAGG. Primers BKgagl and BKgag2
amplify across the border of the 5' LTR in the gag region (bases 660 to 910) and were probed with RM4: BKgagl,
CCCTTTCATTCACGACTGACTGCCGGCTTG;
BKgag2,CTGGAGGAAGTTAAGCCAGTGATGAGCGGC.
BKprol and BKpro2 amplify from theprotease region (bases 2180 to 2380) and were probed with BKprol/2: BKprol, CATTAGATCCCGCCCGTCGGCCCG; BKpro2, GAGGTGAGCT
TAAAGTGATCTTGG; BKprol/2, GTTCTCAAGTAATA
CTCCCTCAAAAATACATCCGTATTAGG.
BKtaxl andBKtax2
amplify
across theborder of the 3' LTR within thetax gene
(bases
8101 to8320)
and wereprobed
with BKtaxl/2:
BKtaxl,
GGCCTAAAGATGGCCAGCCATCTTTAG; BKtax2,
GGGGCTCATGGTCATTGTCATCTGCC;
BKtaxl/2,
TACTCTCACACGGCCTCATACAGTACTCTTCCTTTC.
BKpoll
andBKpol2 amplify
from thepol
gene(bases 3385 to 3505) and were
probed
with BKpoll/2: BKpoll, TACAAAGGCATACTGATCCC; BKpol2, CAGGGT
TTGGACTAGTCTA;
BKpoll/2,
AGTTCAATCATTAGTGCAGCTGCGGCAGGCCCTGTCACAGAACTGCC. The
primerpairRM3and
spl
wouldamplify
acrossthefirstsplice
junction
if cDNA made from aproperly spliced
mRNAwaspresent(base421tothe
splice donor, splice
acceptortobase5163) and were
probed
withBKspll,
which crosses thesplice junction:
spl,
CCGACGGGTCTTGGGCATGCAGCTCGC; BKspll, CGTCCGCCGTCTAGCTTCCTGGTC.
Southernblot and dotblot.
Hybridizations
wereperformed
by standard
techniques
(3). Reinforced nitrocellulose(Opti-bind; Schleicher&
Schuell)
wasused forblotting,
and DNAwas bound to the nitrocellulose by using a Stratagene UV cross-linker. For dotblots, 30
p.l
of the 100-p.1 PCRproduct
was used. Dot blots were probed with T4 polynucleotide
kinase
32P-end-labeled
oligonucleotideprobes. Stringencyof the wash was determinedempirically
foreach primerpair-probe combination to ensure that the negative control
reac-tions did not give an inappropriate background.
Generally,
three20-minwashes in2x SSC(1x SSC is 0.15 MNaCl
plus
0.015 M sodium
citrate)-0.2%
SDS at55°C wereused. Afterautoradiography, spotswere cutfromthe nitrocellulose, and
theirradioactivity was counted forquantitation. For
South-ernblotsofPCRproduct,20 p.1of thePCRproductwasused per lane and probed with oligonucleotide probes. Genomic Southern blots used 20 p.g of DNApurifiedfrom PBMCs and cut with PstI and were probed with the HTLV-I-bearing
plasmid pMT2radiolabeled with 32P by nick translation (3).
Quantitative PCR. For the more stringent
quantitation,
three PCR reactions were done on each patient sample,
using 10,000, 1,000,and 100cellequivalents ofinputDNAto
serve as the initial PCR template. The intensities of the
signals were compared with those of PCR reactions with
2,
20, 200, 2,000, 20,000, and 200,000 molecules of input
HTLV-I DNAina background of 10,000cell equivalents of DNA from HUT78 cells, each reaction done in triplicate.
Duplicatedot blots were performedfor each reaction. After
autoradiography, the spots were cut out and their
radioac-tivitywascounted, and the number of HTLV molecules ina
patient samplewasestimatedby comparisonwith the HTLV
dilutions.
RESULTS
DNA samples derived from PBMCs of nine patients with ATL (patients 1 to9, Table 1) were screened with a set of
eightPCRprimers that span the HTLV-I genome(Fig. 1). In
a semiquantitative assay in which the PCR signal amplified from 1 p.g of PBMC DNA was compared with 10-fold serial dilutions of HUT102 cell DNA, a trend became obvious: some samples gave strong PCR signals only from primers chosen from the 3' (pX or LTR) region of the genome and faintsignals from the gag-pol region (samples 2, 7, 8, and 9). Other samples (1, 3, 4, 5, and 6) gave strong signals from all parts of the HTLV-I genome. Strong signals would be expected because at least 10%of the PBMCs in patients with ATL are leukemic cells that carry the virus. This indicated that in fourof nine samples, the 5' gag andpol regions of the genome were deleted in the clonally derived
on November 10, 2019 by guest
http://jvi.asm.org/
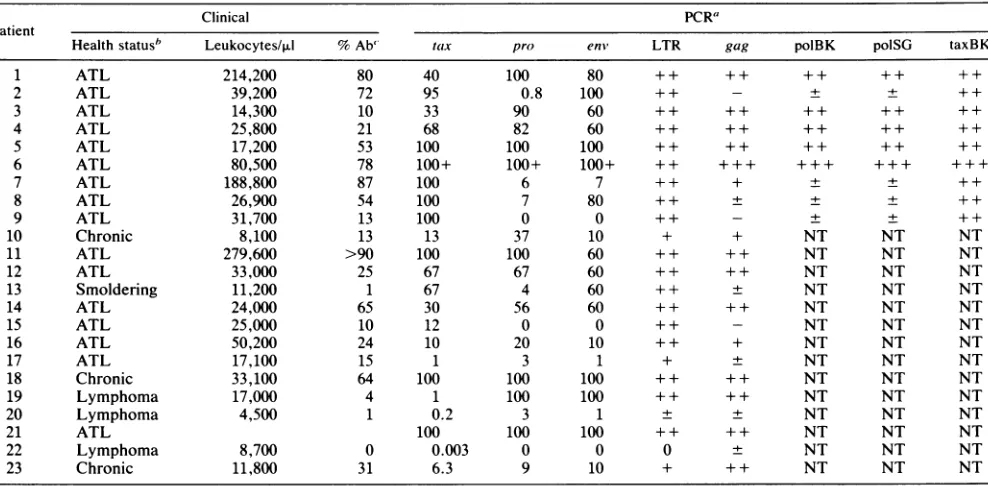
TABLE 1. ClinicalstatusandPCR results from 23HTLV-1antibody-positive patients
Clinical PCR"
Patient
Healthstatusb Leukocytes/,ul %Ab' tax pro enm, LTR gag polBK polSG taxBK
1 ATL 214,200 80 40 100 80 ++ ++ ++ ++ ++
2 ATL 39,200 72 95 0.8 100 + + - + + + +
3 ATL 14,300 10 33 90 60 ++ ++ ++ ++ ++
4 ATL 25,800 21 68 82 60 ++ ++ ++ ++ ++
5 ATL 17,200 53 100 100 100 ++ ++ ++ ++ ++
6 ATL 80,500 78 100+ 100+ 100+ ++ +++ +++ +++ +++
7 ATL 188,800 87 100 6 7 + + + + ++
8 ATL 26,900 54 100 7 80 ++ ± + + ++
9 ATL 31,700 13 100 0 0 ++ - + + ++
10 Chronic 8,100 13 13 37 10 + + NT NT NT
11 ATL 279,600 >90 100 100 60 + + + + NT NT NT
12 ATL 33,000 25 67 67 60 + + + + NT NT NT
13 Smoldering 11,200 1 67 4 60 + + ± NT NT NT
14 ATL 24,000 65 30 56 60 + + + + NT NT NT
15 ATL 25,000 10 12 0 0 + + - NT NT NT
16 ATL 50,200 24 10 20 10 + + + NT NT NT
17 ATL 17,100 15 1 3 1 + ± NT NT NT
18 Chronic 33,100 64 100 100 100 + + + + NT NT NT
19 Lymphoma 17,000 4 1 100 100 ++ ++ NT NT NT
20 Lymphoma 4,500 1 0.2 3 1 + + NT NT NT
21 ATL 100 100 100 ++ ++ NT NT NT
22 Lymphoma 8,700 0 0.003 0 0 0 ± NT NT NT
23 Chronic 11,800 31 6.3 9 10 + ++ NT NT NT
a The PCR primers correspondto thefollowing: tax,SK43andSK44;pro,BKproland BKpro2;
enm,
SG452 andSG453; LTR,RM3 andRM4;gag,BKgagl and BKgag2; polBK, BKpoll andBKpol2;polSG,SG231 andSG238;and taxBK, BKtaxl and BKtax2. The numbers under thePCRprimer columnstax, pro, andenvreferto an approximateestimate:(number of viral genomes/number of human genomes)x 100%. ++, veryintense,saturatedsignal; +,strongsignal; ±, faint signal;-, nosignal; NT,nottested. Thesensitivity ofeachreaction(minimumnumber ofinput moleculesrequired forapositive PCR)wasasfollows: tax, 2; pro, 20;enm,20; LTR, 200;gag,200;polBK,200;polSG,200;taxBK,200.bHealth statusreferstoclinicalstatus at thetimeblood sample was drawn. ATL, acute type;chronic,achronic type;lymphoma,T-celllymphomawithor
withoutleukemic change; smoldering, smoldering type. ' Percentageof abnormal cells in peripheralblood.
ing leukemic cells, with the 3' region preferentially pre-served. Previous instances ofdeleted genomes in samples
frompatients withATLhave been described (2, 31,39).We
obtainedanadditional 14samples frompatientstoattempt to
determine how frequently the deleted genome occurred in
ATLpatients and theapproximate boundariesof the deleted
region, usingthe PCR mapping technique described above.
A subsetof five PCRprimers that wererepresentative of
sd
CAP
2074 2784
proI
thesetthat had been used on the
original
ninesamples
wasused on the entire set of 23
samples (Table 1).
In threesamples,
thetumorcells were not presentin theperipheral
blood inadequate
numbers to be differentiated fromback-ground
HTLV-infected nontumorperipheral
blood cells(samples 17, 20,
and22),
andforanadditionalcase(sample
21),
hematological
information was not available. Of theremaining
19samples,
6(32%)
had the 5' endoftheir genome5202 6668 7324 8381
env tax
sasd
I I
aaa
I l t- ll
23 777 824 21132243
pol
sa
LTR 51
5209 8300 9054
SG231/ RM3/4 BKgagl/2 BKPro1/2 238 BKPol1/2
_I
[E
421-720 660-881 2180-2380 2800- 3385-3505
3038
SG4521
RM3/spl 453
421-sd; 5796-6106 sa-5163
SK43144
-U-7358-7516
BKtaxl/2
RM3/4 8101-8320
FIG. 1. Map oftheHTLV-Iproviralgenomeindicatinglocationsofamplification products.Theboundaries and locations of the LTRs and the gag,pol,env, tax, andprotease(pro)genesaregivenin the upperdrawing.Thelocations ofthesplicedonor(sd)andspliceacceptor (sa) sitesaremarked.Directly belowarethelocations andouterboundaries ofthePCR
amplification
products
for eachofthenineprimer
pairs
used. Theblack boxes areprimersthatreactedstrongly with all samplesfrompatientswith ATL. Boxes filled in with dots indicate PCR
primersthatlie in the deletedregion,whichreactedweaklyin 6 of 19ATLpatients. SG452/453wasretained in one-half of thepatientswith
deletedHTLV genomes and wasdeleted in the other half. Theblankbox forRM3/splindicates the primersthatamplifyacross thesplice
junctionwhichgavenoPCRproductforanypatientsampletested.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.157.470.526.664.2]1 2 3 4 5 6 7 8 9 10 11 12
RM3/4 * *. **.:.*.*@ ***
gag // * *... ...#4..
13 14 15 16 17 18 19 20 21 22 23
RiM3/4* @@** * *
-24 25 26 RM3/4
BKWGf/12
27 28 29 30 31 32 33 34 35 RY 3/4 *
Bfgog//2_
FIG. 2. Dot blot showing semiquantitative PCR results with DNAfrom patients with ATL. Spots 1to23arePCRproductsfrom the DNA from patients with ATL, correspondingto the numbers given in Table 1. Spots 19, 20, and 22arefrom DNA frompatients
with lymphoma with low levels of HTLV-infected tumor cells
amongtheir PBMCs. Spots 24to26arenegative controls with DNA
derived from healthy blood donors. Spots 27 to 33 are serial
dilutions of HUT102 cell DNA into HUT78 cell DNA. They contained, respectively, approximately 1,000,000, 200,000, 20,000, 2,000, 200, 20, and 2 copies of HTLV-I input DNA. (Longer
exposuresof this blot revealed specific PCR product withasfewas
200copies of HTLV-I [spot 31]), butsomeof thecontrastwaslost in thesamples from patients with ATLatthis exposure;therefore,
alightexposureisshown which doesnotshowsignal in all dots in whichasignalcanbe detectedwith longer exposures.) Spot 34 isa
HUT78 DNAnegative control, andspot35 isa zeroDNAnegative control. A2-rig portion of DNAwasused for each PCR.
deletedinthe leukemiccells (Table 1, samples 2, 7, 8, 9, 13,
and 15). Lowlevels ofintactgenome were present inthese
samples, presumably owing to the very low number of HTLV-infected nontumor cells circulating in the periphery
and amplified by the PCR technique. Three of six samples
retained the DNA amplified by the env primers, and the
other three were deleted in that region. All 19 gave strong
signals with LTR or taxprimers. It was not clear whether anyof the 5' LTR was present. The primer pair RM3 and
RM4amplifywithinthe LTR, and the PCR signal from these primers could be generated entirely by the 3' LTR. The BKtaxland BKtax2 primer pair amplify across the 3' LTR
border in the taxgene and gave a strong reaction with all samples tested (Table 1; Fig. 2). The BKgagl and BKgag2 primer pair amplify in the gag gene at the border of the5'
LTR;thissequence wasnotpresentin thetumorcells ofthe
patients with deletedgenomes (Table 1; Fig. 2).
An example ofa PCR experiment with the primer pairs
RM3 and RM4 and BKgagl andBKgag2isshownin Fig. 2. Amorestringent quantitativetestwasdoneonall 23 samples
(Table 1;seeMaterials andMethods) with the primers SK43
andSK44, which amplify from thetaxregionpresent in the tumorcells of allsamples; the primersBKprol and BKpro2,
which amplify from the protease region embedded in the deleted region; and the primers SG452 and SG453, which amplify in theenvregion thatwasretained in one-half ofthe
cases with partial genomes. These primers were chosen
becausethey were the most sensitive (SK43 and SK44are
abletodetectasfewas2 initialinput HTLV molecules, and BKprol and BKpro2 and SG452 and SG453 are able to detect 20 molecules) and they represented each class of result.
Clinical datawereavailable for all butonepatient, includ-ing the leukocyte count (Table 1). In some cases, this number was very high (e.g., samples 1, 7, and 11), which indicates that the PBMCs are composed almost entirely of
leukemic cells. Patient 7(Table 1)showsaparticularlyclear
example of a deleted HTLV-I genome in tumor cells
be-cause,despite theextremely high number of leukemic cells and the intense PCR signal from the taxand LTRprimers,
thePCRprimersfrom the 5' portionof the genome display
only a weak response. The other five patients who carried
deleted genomes in their tumor cells, while having a less
overwhelming number of leukemic cells in theirperipheral
blood, still showeda clearquantitative contrastwhen
com-paringPCRamplification products from differentregions of
the genome.
Aldovini etal. (2)reported an instance in whicha
defec-tive HTLV-Iwascloned withadeletion thatsuggesteditwas
an integrated cDNA that had been copied from a properly
spliced RNA. To determine whether this had occurred in
someofthedeletedgenomesdescribedhere,wetested PCR
primers that amplify across the first splice site (RM3 and spl). Noneofthesamples fromthe23patientswithdefective
orintactgenomesreacted with this
primer pair.
Although it islikelythat severalpatients havemorethan
onecopyofHTLV DNAintegrated into their leukemic cell genomes(39),patient6wasinteresting in that DNA derived from this patient, with all primers tested in
comparisons
made by serial dilutions, repeatedly gave a more intense
PCR signal than HUT102 DNA. We estimated that our
HUT102linecontainssevenHTLVcopiespercell.Patient 6 may have had 10 or more copies of HTLV
provirus
perleukemic cell.
Southern blot
analysis
of DNA frompatients
with ATLwasdone on threesamplesthatcontained defectivegenomes andonthreesamplesthatcontained intactgenomes
(Fig.
3).Genomic DNA was cut with the restriction enzyme PstI,
which yields fourconserved DNAfragmentsthat lie within the HTLV genome (31).Thesefragmentswerepreserved in thegenomesthat thePCRresults indicatedwereintact. The
Pstlfragments which represent the 5'portion of the HTLV
genome were lost in thedefective viruses.
DISCUSSION
The preferential retention ofthe 3' part of the HTLV-I
genome in leukemic cells ofpatients with ATL who carry
defective genomes in their leukemic cells points to the
potential importance ofthetaxregioninoncogenesisinvivo.
Itis possible thatgeneration ofa defective proviruscan in
some casescontribute to oncogenesis because the tax
pro-tein may be freed from the regulatory constraints of the intactHTLV-Igenome. Thenormal modulatinginfluence of
the rex gene product (6), which switches viral expression
fromtaxandrextotheviral structuralproteins by
posttran-scriptional regulation (5, 6, 14, 30), may be lostin someof
thedeleted genomes. Other constraints that may normally be
imposed by regulated transcriptionfrom the viral LTR may
beoverriddenby transcription ofthedeletedvirus pXregion from cellular promoters proximal to the proviral insertion
site.
Kinoshita et al. (17) used PCR to detect mRNA from
on November 10, 2019 by guest
http://jvi.asm.org/
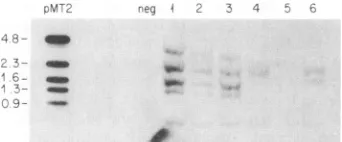
[image:4.612.68.282.79.290.2]-M_ eg 4 4 _
FIG. 3. Southern blot analysis of genomic DNA from PBMCs from patients with ATL. Lane pMT2 is plasmid pMT2cutby the restriction enzyme PstI. The 2.3-, 1.6-, and 1.3-kb fragments are
internal fragments from the HTLV genome. The 0.5-kbfragment
frequently found in HTLV DNA (26) (see lanes 1 to 6) is not conserved in pMT2. The 4.8- and 0.9-kb fragments contain both HTLV-I DNA andvectorDNA. A20-,ug portion ofDNAcutwith PstIwasused for each of thefollowing lanes. The lanemarkedneg
used DNA from a blood sample from a noninfected blood donor.
Lanes 1 to 3 were PBMC DNAs from patients who carry intact
HTLV genomes in theirleukemic cells (patients 1, 5, and 6),and lanes 4 to 6 were DNAs from patients with deleted genomes
(patients 2, 7, and 9). The internal Pstl sites correspond tobases 1392to3036 (1.6 kb), 3035to4316 (1.3 kb), 4330to6755 (2.3kb), and 6755to7294(0.5kb). The 2.3-kb band is derived fromen'sequences
andthe end of the polgene. Lanes 4 and 6 showaconserved 2.3-kb
bandbuthave lost the 1.6- and 1.3-kb bands. Lane 4 is derivedfrom DNAfrom patient 10,which doesnot reactwiththe envprimers,so
itislikely that the band in thiscaseis duetoanewPstIlsite. Lane
5 has lost alltypical Pstlbands, gaininga newbandat3kb. Alllanes retained the 0.5-kbband. Other bands of various sizesareprobably
atinsertion sites andarecomposed ofboth HTLV DNA and human
flankingsequence DNA.
patients with ATL and from healthy HTLV-I-infected indi-viduals; the PCR primer pair used crossed the second splice
site in the HTLV-I genome. Only low levels oftranscript wereobserved, and inatleastonepatient withATL, notax mRNAwasdetected.Thedefectivegenomesdescribedhere
may generate tax transcripts that may not be properly spliced and would not be detected by PCR primers that
flankedthe splice site. It would beinteresting todetermine
whether thelevel oftaxmRNAobservedinthe PBMCs from
patients with ATL may be higher in some cases with tax primersthatdo notcrossthe splice junction.
Because of the potential for the presence of several HTLV-I proviralgenomesin ATL leukemic cell DNA(39),
it ispossiblethatdefectiveprovirusispresentinsomeof the
leukemic cells that also contain intact genomes. The pres-ence of the defective genome would be masked and not detected by PCRorthe Southern blot analysis. Therefore,
the 32% of patients with ATL that we found harboring
defectivegenomeintheir leukemic cells is alower estimate. The frequency with which deleted genomes are observed suggeststhat such deletions contributetodevelopingATL in
somecases.Ifderegulationof thetaxgenethroughmutation
or deletion of cis-acting regulatory sequences can be a
contributing factor to leukemogenesis, it could help to
ex-plainthelong latencyfrom viral infectiontodisease. Appro-priate mutagenic events and deletions would be rare; thus,
infected individuals would havealow lifetimerisk of
devel-opingATL onceinfectedwith HTLV-I. ACKNOWLEDGMENTS
This work was supported by Public Health Service grants CA39805and HL33774 from the National Institutesof Health. Bette Korberwas aLeukemiaSocietyof Americafellow.
We thank Mary Fran McLane for assistance and advice and Tun-Hou Lee and Garth Ehrlich for useful scientificdiscussions. We
alsothankEdward Hamiltonfor editorialassistance andmanuscript
preparation.
REFERENCES
1. Abbott,M.A.,B.J. Poiesz,B.C.Byrne, S. Kwok, J. J.Sninsky, andG. D. Ehrlich. 1988.Enzymaticgeneamplification:
qualita-tive and quantitative methods fordetecting proviralDNA
am-plified in vitro.J. Infect.Dis. 158:1158-1168.
2. Aldovini, A., A. DeRossi, M. Feinberg, F. Wong-Staal, and G. Franchini. 1986. Molecular analysis of a deletion mutant provi-rus of type I human T-cell lymphotropic virus: evidence ofa doubly spliced X-lor mRNA. Proc. Natl. Acad. Sci. USA 83:38-42.
3. Berger, S. L., and A. R. Kimmel. 1987. Guide to molecular cloning techniques. Methods Enzymol. 152:582-587.
4. Cross, S. L., M. B. Feinberg, J. B. Wolf, N. J. Holbrook, F. Wong-Staal,andW.J.Leonard. 1987.Regulation ofthe human interleukin-2 receptor alpha chain promoter: activation of a non-functional promoter by the transactivatorgene ofHTLV-I. Cell49:47-56.
5. Dokhelar, M.-C.,H.Pickford, J. Sodroski, and W. A.Haseltine. 1989. HTLV-Ip27rex regulates gag and env proteinexpression. J. Acquired Immune Defic. Syndr. 2:431-440.
6. Dokhelar, M.-C., H.Pickford, J. Sodroski,and W. A.Haseltine. 1989. The potentialforhomeostatic regulation of the Xregion proteinsofthe humanTcell leukemia virus typeI. J. Acquired Immune Defic. Syndr. 2:588-594.
7. Felber, B.K.,H. Paskalis, C. Kleinman-Ewing, F. Wong-Staal, and G. N. Pavlakis. 1985. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeat. Science 229:675-679.
8. Franchini, G., F. Wong-Staal, and R. C. Gallo. 1984. Human T-cellleukemia virus (HTLV-I) transcripts in fresh andcultured cells ofpatients withadult T-cell leukemia. Proc. Natl. Acad. Sci. USA81:6207-6211.
9. Gazdar, A. F.,D. N. Carney, P. A. Bunn, E. K. Russell, E. S. Jaffe, G. P. Schecter, and D. W. Guccion. 1980. Mitogen requirements for the in vitro propagation ofcutaneous T-cell lymphomas. Blood55:409-417.
10. Grassmann, R., C. Dengler,I. Muller-Fleckenstein, B. Flecken-stein,K. McGuire, M.-C. Dokhelar, J. Sodroski, and W. Hasel-tine. 1989. Transformation to continuous growth of primary human T lymphocytes by human T-cell leukemic virus type I X-region genes transduced by a herpesvirus saimiri vector. Proc. Nat]. Acad. Sci. USA 86:3351-3355.
11. Greenberg, S. J., G. D. Ehrlich, M. A. Abbott, B. J. Hurwitz, T. A.Waldmann, and B.J.Poiesz. 1989. Detection ofsequences homologous to human retroviral DNA in multiple sclerosis by geneamplification. Proc. Natl. Acad. Sci. USA86:2878-2882. 12. Haseltine, W. A., J. G. Sodroski, R. Patricia, D. Briggs, D.
Perkins, and F. Wong-Staal. 1984. Structure of the 3' terminal region of type ll human T lymphotropic virus: evidence fora new coding region. Science 225:419-421.
13. Hinuma, Y., K. Nagata, M. Hanaoka, M. Nakai, T.Matsumoto, K. Kinoshita, S. Shirakawa, and I. Miyoshi. 1981. Adult T-cell leukemia:antigeninanATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA 78:6476-6480.
14. Inoue, J.-I., M. Yoshida, and M. Seiki. 1987. Transcriptional (p4Ox) and posttranscriptional(p27x-III) regulators are required for the expression and replication of human T-call leukemia virus type I genes. Proc. Natl. Acad. Sci. USA 84:3653-3657. 15. Jeang, K. T., I. Boros, J. Brady, M. Radonovich, and G.
Khoury. 1988. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p4Ox-responsive 21-base-pair sequence. J. Virol. 62:4499-4504.
16. Jeang, K.-T., S. G. Widen, 0. J. Semmes, and S. H. Wilson. 1990. HTLV-I trans-activator protein, tax, is atrans-repressor of the human beta-polymerase gene. Science 247:1082-1084. 17. Kinoshita, T., M. Shimoyama, K. Tobinai, M. Ito, S.-I. Ito, S.
Ikeda, K. Tajima, K. Shimotohno, and T. Sugimura. 1989. Detection of mRNA for the taxl/rexl gene of human T-cell leukemia virus type-I in fresh peripheral blood mononuclear
on November 10, 2019 by guest
http://jvi.asm.org/
cellsofadult T-cellleukemia patients and viralcarriersbyusing the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:5620-5624.
18. Kwok, S., G. Ehrlich, B. Poiesz, R. Kalish, and J. J. Sninsky. 1988. Enzymatic amplification ofHTLV-Iviralsequences from peripheralblood mononuclearcells andinfected tissues. Blood 72:1117-1123.
19. Lee, T.-H., J. E.Coligan, J. G. Sodroski, W. A. Haseltine, S. Z. Salahuddin, F. Wong-Staal, R. C. Gallo, and M. Essex. 1984. Antigens encoded by the 3'-terminal region of human T-cell leukemia virus: evidence for a functional gene. Science 226:57-61.
20. Maruyama, M., H. Shibuya, H. Harada, M. Hatakeyama, M. Seiki, T. Fujita, J. Inoue, M. Yoshida, and T. Taniguchi. 1987. Evidence foraberrant activation oftheinterleukin-2autocrine loop by HTLV-I encoded p4Ox andT3/Ti complex triggering. Cell 48:343-350.
21. Nagata, K., K. Ohtani, M. Nakamura, and K. Sugamura. 1989. Activation ofendogenous c-fos proto-oncogene expression by human T-cellleukemiavirustypeI-encodedp40" protein in the human T-cell line, Jurkat.J. Virol. 63:3220-3226.
22. Okamoto, T., Y. Ohno, S. Tsugane, S. Watanabe, M. Shi-moyama, K. Tajima, M. Miwa, and K. Shimotohno. 1989. Multi-step carcinogenesismodel for adult T-cellleukemia.Jpn. J. CancerRes. 80:191-195.
23. Poeisz, B. J., F.Ruscetti, J. Mier, A. Woods, and R. C. Gallo. 1980.T-celllines established fromhuman T-lymphocyte neopla-sias by direct response to T-cell growth factor(mycosis fun-goides, Sdzary syndrome and acute lymphocytic leukemia). Proc. Natl. Acad. Sci. USA 77:6815-6819.
24. Poiesz, B. J., F. W. Ruscetti, A. F. Gazadar,0. A. Bunn, J. D. Minna, and R. C.Gallo.1980.Detectionandisolationof type C retrovirus particles from fresh and cultured lymphocytes of a patient withcutaneousT-celllymphoma.Proc. Natl.Acad.Sci. USA77:7415-7419.
25. Ruben, S., H.Poteat, T. H. Tan, K. Kawakami, R. Roeder, W. Haseltine, and C. Rosen. 1988. Identification of cellular tran-scription factors required for regulationofinterleukin2receptor gene expression by the human Tcell leukemiavirus tat gene product. Science 241:89-92.
26. Ruben, S. M., A. Perkins, and C. A. Rosen. 1989. Activation of NF-KB bytheHTLV-I trans-activator protein tax requires an additional factorpresent in lymphoid cells. New Biol. 1:275-284.
27. Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide se-quenceof theprovirusgenomeintegratedinleukemiacell DNA. Proc. Natl.Acad.Sci. USA80:3618-3622.
28. Seiki, M., A. Hikikoshi, T. Taniguchi, and M. Yoshida. 1985. Expression ofthe pXgeneof HTLV-I: general splicing mech-anism in theHTLVfamily. Science 228:1532-1534.
29. Seiki, M., J. Inoue,T. Takeda, and M. Yoshida. 1986. Direct evidence thatp4Ox of human T-cell leukemia virustype I isa trans-acting transcriptional activator. EMBOJ. 5:561-565. 30. Seiki, M., J.-I. Inoue, M. Hidaka, and M. Yoshida. 1988. Two
cis-acting elements responsible for posttranscriptional trans-regulation ofgene expression ofhuman T-cell leukemiavirus type I.Proc.Natl. Acad. Sci. USA85:7124-7128.
31. Siomi, H., T. Nosaka, T. Saida, H. Miwa, Y. Hinuma, S. Shirakawa, N.Miyamoto, T. Konko, K.Araki,M. Ichimaru,A. Miura, and M. Hatanaka.1988.Twomajorsubgroups ofhuman T-cellleukemia virus type-IinJapan. Virus Genes 1:377-383. 32. Slamon, D. J., K.Shimotohno, M.J. Cline, D. W. Golde, and
I. S. Y. Chen. 1984. Identificationofthe putativetransforming protein of the human T-cell leukemia viruses HTLV-I and HTLV-II. Science 226:61-65.
33. Sodroski, J.G.,C. A. Rosen, W. C.Goh,and W. A. Haseltine. 1985. A transcriptional activator protein encoded by the x-lor region ofthe human T-cell leukemia virus. Science 228:1430-1434.
34. Tan, T. H., M.Horikoshi, and R. Roeder.1989.Purification and characterization of multiple nuclear factors that bind to the TAX-inducible enhancer withinthe humanT-cellleukemiavirus type I longterminalrepeat. Mol.Cell. Biol.9:1733-1745. 35. Tanaka, A., C. Takahashi, S. Yamaoka, T. Nosaka, M.
Masa-toshi, and M. Hatanaka. 1990.Oncogenic transformationby the tax geneofhuman T-cell leukemiavirus type I invitro. Proc. Natl. Acad.Sci. USA 87:1071-1075.
36. Wano, Y., M.Feinberg, J. Hosking, H. Bogerd, and W. Greene. 1988.Stable expression of thetaxgene oftypeIhumanT-cell leukemia virus inhumanT-cellsactivatesspecific cellulargenes involvedingrowth.Proc. Natl. Acad. Sci. USA 85:9733-9737. 37. Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemiaand itsimplication in thedisease. Proc. Natl. Acad. Sci. USA 79:2031-2035.
38. Yoshida, M., and M. Seiki. 1987. Recent advances in the molecular biology of HTLV-I: trans-activation of viral and cellulargenes. Annu. Rev. Immunol. 5:541-559.
39. Yoshida, M., M. Seiki, S. Hattori, and T. Watanabe. 1984. Genome structure of human T-cell leukemia virus and its involvement inthe development ofadultT-cell leukemia/lym-phoma,p.141-148.In R.Gallo, M. Essex, and L.Gross(ed.), Human T-cell leukemia/lymphoma virus. Cold Spring Harbor Laboratory, Cold Spring Harbor,N.Y.