JOURNAL OF VIROLOGY, May1980, p. 542-549
0022-538X/80/05-0542/08$02.00/0 Vol.34, No.2
Physical Map
of
Caulobacter
crescentus
Bacteriophage 4Cdl
DNA
BILHARABOY, LUCILLE SHAPIRO,* AND KEI AMEMIYA
Department ofMolecularBiology, Division ofBiological Sciences,Albert Einstein College of Medicine,
Bronx,
New York10461A restrictionmapof the Caulobactercrescentusbacteriophage 4Cdlgenome wasconstructedby usingthe restriction endonucleasesHindIII and HpaI.Atotal of12fragments, ranging in molecular weight from7.7 x 106to0.25 x 106,were
produced byHindIII, and 7 fragments, ranging in molecular weight from 9.0 x
106 to0.24 x
106,
weregeneratedby HpaI. The molecular weight of thegenome was estimated to be approximately 28.8 x10'
on the basis of the relative electrophoretic mobilities of the restriction fragments. The relative order of the cleavage fragments was determined by specific cleavage of isolated restriction fragments, terminal labeling of both the whole genomeand isolated fragments, and hybridization of isolated fragments to restriction fragments generated by other restriction enzymes. The genome of 4Cdl was found to be terminally repetitive, and analysis of previously determined in vivo and in vitro RNA transcripts showed that the restrictionmapcouldbe oriented such that transcrip-tionbeganonthe left andproceededtothe right end ofthegenome.Caulobactercrescentusphage pCdlis asmall, icosohedron-shaped bacteriophage containinga linearduplexDNAmolecule of molecular weight 29 x 106 (23). Its host, C. crescentus, has a defined cellcycle (14, 19), which includes mor-phologically distinct swarmer cells and stalked cells. Unlike many otherCaulobacter bacterio-phages, which infect cells onlyat acertain de-velopmental stage (2, 19), both stalked and swarmer cells are sensitive to 4Cdl infection (23). We have studied the in vitrotranscription ofphage pCdlDNAbythe C. crescentusRNA polymerase (3) and the RNA processing ofthe phage RNA transcripts (6) by C. crescentus RNase III. It has been demonstrated that the host RNA
polymerase
isresponsible
for the in vivo transcription of a portion of4Cdl
DNAimmediately
afterinfectionandthata rifampi-cin-resistant RNApolymerase,
dependentonde novoproteinsynthesis,appearsearlyafter infec-tion (Amemiya, Raboy, and Shapiro, Virology, inpress). To map theregionofearly pCdlDNA transcriptionandtolocatetheearly
promoters,wepreparedacleavagemap of thephageDNA byanalyzingthe sizes andpositionsof
fragments
produced by two restriction
endonucleases,
HpaI (7, 20) andHindIII (18,21),and
by
cleav-age of4Cdl
DNApreviouslyendlabeledbyT4 DNA kinase. Wealso determined that the ends of4Cdl
DNA contain terminallyrepetitious
DNA sequences since partial degradation by exonuclease III and
annealing
results in thecon-version oflinear DNAtocircularmolecules(17).
MATERIALS AND METHODS
Bacteria andbacteriophage. C.crescentusCB13
was used in thisstudy. C. crescentus was grown in peptone yeast extractmedium (16) at30°C. Bacterio-phage4Cdl(23) was agift from N. Agabian.4Cdlwas
propagated in C. crescentusCB13, and concentrated
phage suspensions wereobtained by the method of
Yamamoto et al. (24). The phagewere precipitated with 6% (wt/vol)polyethylene glycolcontaining 0.5 M
NaCl andsubsequently purified in CsCl gradients(3).
32P-labeled<>Cdl DNAwas prepared by growing the host in the presenceof 10ytCi of32P, (ICN, Irvine, Calif.)perml foronegeneration before the additionof
thephage. Labeled phagewereharvested and concen-trated asdescribed above.
Bacteriophage DNA preparation. Bacterio-phage4Cdl obtained from CsCl gradientswasdialyzed overnightagainst bufferD(0.01MTris-hydrochloride, pH8.0, 0.02 MNaCl,0.1 mMEDTA, disodium salt) andthen extractedthree times with distilled phenol saturated with 2x SSC (lx SSC is0.15 M sodium chlorideplus0.015Msodiumcitrate). The DNAwas then dialyzed extensively against large volumes of
bufferDand storedat4°Coverchloroform. Restriction endonuclease digestion of DNA. 4CdlDNA(1to2
jig)
wasdigestedin 0.05-mlreaction volumes containing 0.01 M Tris-hydrochloride (pH7.6),7mMMgCl2,1mMdithiothreitol,0.02MNaCl,
100,igofbovine serumalbuminperml,and1to3U of restriction endonuclease HpaI or HindIII (New England Biolabs, Inc., Beverly, Mass., or Bethesda ResearchLaboratories, Rockville,Md.).For prepara-tivegelsthe reaction volumeswereincreased upto
10-fold.Thereaction mixtureswereincubatedat37°Cfor
1to 3h,digestionwasstopped bytheaddition of0.2
volumes ofa solutioncontaining 25% (vol/vol)
glyc-542
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 34, 1980
erol, 5%(wt/vol) sodium dodecyl sulfate,and 0.025% (wt/vol) bromophenol blue, and incubationwas
con-tinuedat65°C for10min.
5'-Terminal labeling of fCdl DNA or 4Cdl
restrictionfragments. Intact <>CdlDNA orisolated HpaI fragmentsweretreated with bacterial alkaline
phosphatase to remove the5'-phosphorylgroup.The terminiwerethenlabeledbyusing [y-32P]ATP and T4 polynucleotide kinase (10).
Gel electrophoresis. 4Cdl restriction fragments
wereseparated bytwodifferent slabgel systems. In
one system the restrictionfragmentswereseparated
by electrophoresis on a 1% agarose slab gel as de-scribed elsewhere (Amemiya et al., in press). The restriction fragments were also analyzed on a 1.4% polyacrylamide-0.5% agarose composite slab gel (13 by 33 cm) (13). The electrophoresis buffer (1)
con-tained40mMTris-acetate (pH 7.8), 20mMsodium
acetate,and2mM EDTA(buffer E).Electrophoresis
wascarriedout at a constant voltage and period, as noted in thefigurelegends. The restrictionfragments
werelocatedeitherbyautoradiographyusing Kodak NS-5T X-ray filmorbysoaking the gel in ethidium bromide (1.0
[Lg/ml)
for 30min andvisualizing the fragments with a UV transilluminator (Ultraviolet Products,Inc., SanGabriel, Calif.).Isolationof DNA restriction fragments. 32P-la-beledorunlabeled4CdlDNAwasdigested withHpaI andseparatedbyelectrophoresison apreparative 1%
agarose slab gel. The restriction fragments were lo-catedbystaining the right and left edges of the slab gel with ethidium bromide.Regionsofthegel
contain-ing each fragment were cut out, and the restriction
fragmentswerethen recovered byelectrophoretic elu-tion (11). Residualagarose inthe DNAsolutionwas removed by chromatography on 0.5-ml columns of acid-washed DEAE-cellulose(DE52) (Whatman, Inc., Clifton, N.J.),asdescribed byHirsh and Schleif(9).
ThepurifiedDNA was thenconcentratedby ethanol precipitation.
32P-labeled4CdlHpaIorHindIII restriction
frag-ments for hybridization experiments were obtained from 1.0%agarosehorizontal slabgels (16by28cm) by phenol extraction andethanol precipitation ofthe
electrophoreticallyeluted DNAsamples.
DNA-DNAhybridization. Stripsofnitrocellulose filterpapercontaining ¢Cdl HpaIorHindIII
restric-tionfragmentswerepreparedasdescribed (Amemiya et al., inpress).Before hybridization thefilter strips
wereplacedinplastic bags (Seal-N-Savebags;Sears, Roebuck &Co.),wetted with 3xSSC, andthen
incu-batedat65°C for3hinmodifiedDenhardt solution (5) containing 0.2% (wt/vol) bovine serum albumin,
0.2% (wt/vol) Ficoll400(Pharmacia FineChemicals, Inc., Piscataway, N.J.), and 0.2% (wt/vol)
polyvinyl-pyrrolidone (Calbiochem,LaJolla,Calif.) in 3x SSC.
32P-labeled
0Cd1
DNA in 0.1 M Tris-hydrochloride(pH7.4)-i MNaCl-0.01 MEDTA-0.5% (wt/vol)
so-dium dodecyl sulfate-50% (vol/vol) formamide was then added to thefilters and incubated for 48 h at
37°Cwithgentlerocking.Thefilterswere then rinsed with 3xSSC,washed for 30 min at 65°C with 6x SSC
containing 0.02% (wt/vol) bovine serum albumin, 0.02% (wt/vol) Ficoll 400, and 0.02% (wt/vol)
polyvi-nylpyrrolidone,and finally washed with 3xSSC. After
CLEAVAGE MAP OF
oCdl
DNA 543thefiltersweredried underaheatlamp, autoradiog-raphy wascarried out by using Kodak XR-5 X-ray film withaCronexQuanta IIIintensifyingscreen(Du
PontCo.,Wilmington, Del.).
Exonuclease III digestion and annealing of 4CdlDNA. Limiteddigestion of 4Cdl DNA from the
3' end of each strand (16) was accomplished with Escherichia coli exonuclease III (New England
Bio-labs), asdescribedby Ritchieetal. (17). The reaction mixtures(1.0ml)contained25mMTris-hydrochloride (pH8.0), 2.5 mMMgCl2,5mM2-mercaptoethanol, 7.5
,ugof32P-labeled
oCdl
DNA, and 20 U of exonuclease III.The mixture wasincubatedat37°C for 7.5 min, and the reactionwasstopped by the adding0.1mlof 20x SSC andcoolingto4°C. Under these conditions approximately5%of theDNAwasdigested (2.5% fromeach end). Annealing of the partially digestedDNA was carried out at 65°C for 60 min. Samples were
placedonice before electronmicroscopy. For controls
weused(i) DNA which was incubated in the absence ofexonuclease IIIandcarriedthrough theannealing
procedure, and (ii) DNA which was digested with
exonucleaseIII but notannealed.
Electron microscopy. DNA samples were
pre-pared and spreadby the method of Davis etal. (4).
The DNA waspickedup oncarbon-coated (Parlodion-backed) grids, stained with uranyl acetate, and shadowed withplatinum-paladium. Sampleswere
ob-served withaSiemens 1A electron microscope.
RESULTS
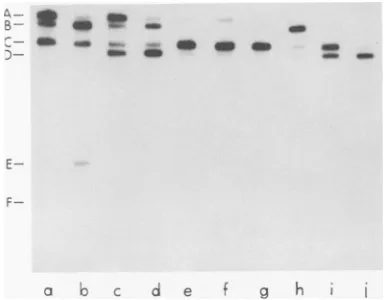
Digestion of 4Cdl DNA with restriction endonuclease. 4Cdl DNA was digested with
restriction endonuclease HindIII or HpaI, and thecleavageproductswereseparated by electro-phoresis on composite 1.4% polyacrylamide-0.5% agarose or 1.0% agarose slab gels (Fig. 1). Theautoradiograms(Fig. 1,lanesaand c) were overexposed so that the smallest HindIII and HpaI fragmentscouldbe visualized.These small fragments are difficult to visualize by staining withethidium bromide.Digestion of
oCdl
DNA withHindlll produced12fragments,which were designated AtoLaccordingto increasing elec-trophoretic mobility. Electrophoresis of ¢Cdl HpaI cleavage products yieldedsixbands, des-ignatedAtoF.Increasingthe amount of HindIIIorHpaIendonucleaseduring digestion of4Cdl DNA didnot changethe number of restriction fragments. As discussedbelow, further analysis ofHpaIfragment Crevealed that it wasactually composed of two fragments of similar electro-phoretic mobility.
Determination of molecular weights of restriction fragments. To estimate the sizes of the
oCdl
HindIII andHpaI restriction frag-ments, theirmobilitiesonagarosegels were com-pared with the relative mobilities of A DNA HindIII restrictionfragments(12) and T7 DNA HpaIrestriction fragments (11).Table 1 shows the molecular weights obtained for theoCdl
on November 10, 2019 by guest
http://jvi.asm.org/
544 RABOY, SHAPIRO, AND AMEMIYA
Hind
A
B: -A
E-G-
-;i_DC
H-1-_
E
-F
-G
-H
-J
E- F-
K-a
b
FIG. 1. Analysis of 4Cdl DNA
strictinn 1ndnnr -1'.;.QHin-dTTTtn
H/pa
I identical when the double band at HpaIfrag-mentCwas taken intoaccount.Themolecular A
weight
ofthe4Cdl
genomedeterminedby
using
-B restrictionenzymeswas
similar
tothat derivedbyWestetal.(23), who usedzonesedimentation.
c
Ordering
ofrestrictionfragments.
SeveralD methods
(see
below)
were used to order the4CdlHindIII and HpaI restrictionfragmentsin aphysicalmap.
(i) Digestion ofonerestriction fragment with the other restriction endonuclease. Thesimultaneous digestion of 4Cdl DNA with restriction endonucleases HindIII and HpaI yieldedeightnewfragments (designated by the prefix n) (Fig. 2, lane g) whichwerenot present
when
oCdl
DNAwasdigested with eachenzyme separately (Fig. 2, lanes a and b). These new fragments could be accounted for when each large HpaI fragment was redigested with HindIII(Fig. 2, lanescthrough f, and Table 2). IndividualHpaI fragmentswereanalyzed in or-dertodetermine whichHindIII fragmentswere located within the large HpaI fragments andtoc d helpidentify overlapping fragments.
Digestion of
¢pCdl
HpaI fragment A with,digefsted
Awthren
HindlIl yieldedtwo fragments (Fig. 2, lane c).mately1.0Pgof32P-labeled4CdlDNAwasdigested with HindIII orHpaI. Restriction fragments were
analyzedbyelectrophoresis, usinga1.4%
polyacryl-amide-0.5%agaroseslab gelrunataconstant50V
for16h, andfragments werevisualizedby
autora-diography (lanes aand c).Alternatively, restriction
fragmentswereanalyzedby electrophoresis, usinga
1%agarose slabgel, andfragments werevisualized
byethidium bromide staining (lanes band d). The
smallest HindIII fragments (K and L) and HpaI
fragments (E and F) are difficult to visualize by ethidiumbromidestaining. To visualizethe smallest
fragments,theautoradiogramswereoverexposed.
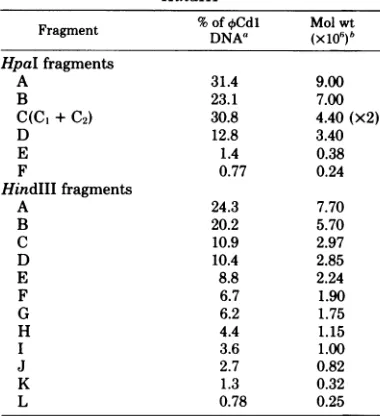
restrictionfragments. The molecular weights of
4CdlHpaI fragments ranged from9.0 x 106 to
0.24x106,and thesumof themolecularweights of theHpaI fragmentswas24.4x 106.The 4Cdl HindIII fragments ranged in molecular weight from 7.7x 106to0.25 x 106. However,thesum of the molecular weights of the HindIlI
frag-mentswas28.6x 106.Thediscrepancybetween thesumsof the molecular weightsof theHpaI and HindIII fragments was resolved when the
amountofradioactivityineachrestriction
frag-mentwasmeasured(Table 1).Itwasfound that HpaI fragment C contained twice as much
ra-dioactivelabel aswould be expectedfora
frag-mentof its size. These results suggest thatHpaI fragment C actuallyconsisted oftwo fragments of similar size (designated HpaI-C, and HpaI-C2). Thesums ofthe molecular weights of the HpaI and HindIII fragments were essentially
TABLE 1. Molecularweights of 4 CdI restriction fragments produced by digestion with HpaI and
HindIII
FragmentFragment % of
~DNA"
oCdl
Mol
(x1io)bwtHpaI fragments
A 31.4 9.00
B 23.1 7.00
C(C +C2) 30.8 4.40(x2)
D 12.8 3.40
E 1.4 0.38
F 0.77 0.24
HindIII fragments
A 24.3 7.70
B 20.2 5.70
C 10.9 2.97
D 10.4 2.85
E 8.8 2.24
F 6.7 1.90
G 6.2 1.75
H 4.4 1.15
I 3.6 1.00
J 2.7 0.82
K 1.3 0.32
L 0.78 0.25
a Determined from the distribution of32P
radioac-tivity. Gelswerestained with ethidiumbromide, indi-vidual DNA bands were cutout, and radioactivities weremeasured.
bMolecularweightswere estimated from the rela-tive mobility of each fragment compared with the mobilities ofAHindIII(12)and T7HpaI (11)digestion
products. Gelcompositionandelectrophoresis condi-tions wereasdescribed in thelegendtoFig.1.
J. VIROL.
I A8- - c-
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.503.268.458.353.561.2]CLEAVAGE MAP OF pCdl DNA 545
A
-C
_
*
XS
n2
C
-
-D
W-d 60MA4_
D:WE- *fl -n3
7
H-
_-- -PS..&*
a.m
-n4
nsl
-E
K-L- -F
[image:4.503.44.237.70.339.2]ob
cd e f gFIG. 2.Analysis of redigestion ofindividualHpaI fragments by HindIII. 32P-labeled¢Cdl DNA was digestedwithHindIII (lane a), HpaI (lane b),orboth HindIII andHpaI (lane g).ThefollowingHpaI frag-mentswereredigestedwithHindIII:HpaI-A (lane c), HpaI-B (lane d),HpaI-C,and-C2 (lane e),and HpaI-D(lanet).After redigestion byHindIII, thesamples (eachwithapproximately5x103 cpm)wereanalyzed by electrophoresis on a 1.4% polyacrylamide-0.5%
agarose slabgelwhichwas run ata constant 85 V for12 h. Thenewfragments produced bythedigestion of XCdlDNA bybothHpaIandHindIIIarelabeled (prefixed bythe lettern)ontheright.
One fragment was HindIII-C, and the other fragment was a new fragment, designated nl.
Thesumofthe molecularweights of thesetwo
fragments was 8.77 x 10', which was in close
agreementwiththemolecularweight of HpaI-A
(9.00X 106).
Digestion of pCdl HpaI fragment B with
HindIIIgaverisetoHindIII fragments Hand K andalsototwo newfragments, n2andn6 (Fig. 2,lane d). Thesumof themolecularweights of
these four fragments (6.83 x 106) agreed well with themolecularweight of HpaI fragment B
(7.0 x 106).
When HpaI fragments C1 and C2 were
di-gestedwithHindIII, sevenfragmentswere
pro-duced(Fig.2,lanee).Among thesewereHindIII
fragments E, F, G, and L. The new fragments
generated by HindIIIdigestionweren4, n5,and
n7. Unlike the HindIll digestion products of
HpaI fragments A and B, thesumof the molec-ularweights of the HpaI-C1 and -C2 digestion products (8.14x 106)wasslightlylowerthanthe
molecular weightof
HpaI-C,
plus-C2 (8.8x106).Nevertheless, these results again indicate that
HpaI fragment C is composed of two fragments having similarmolecularweights.
Digestion ofHpaI fragment D with HindIII yielded HindIII fragment J and two new
frag-ments,n3andn8(Fig. 2,lanefl.Inthiscase,the
sumof themolecularweightsof theproductsof
redigestion withHindIII (3.30 x 106) was very close to the molecular weight of the original fragment (3.04 x 106).
(ii)5'-TerminallabelingofqCdl DNA and
isolated fragments.Todeterminewhich 4Cdl
HindIII and HpaI restriction fragments were
located at the termini of the genome, the 5'
termini were labeled with [_y-32P]ATP before digestion with HindIlI or HpaI. In addition,
pCdl HpaI fragments A, B, C, and D were labeled at their termini before being digested
withHindIII in order todetermine which
frag-mentswerelocatedatthe ends of theindividual HpaIfragments (datanotshown).
Digestion ofthe 5' terminally labeled pCdl
DNA with HindIIIrevealed thatHindIlI
frag-ments C and F were located at the ends, and
TABLE 2. RedigestionofthemajorcpCdlHpaI fragments with HindIIIa
Fragmentsgenerated IsolatedHpaI fragment by redigestionwith
HindIII
Molwt Molwt
Fragment
(x106)
FragmentA 9.00 nl 5.80
C 2.97
B 7.00 n2 4.60
H 1.15
n6 0.76
K 0.32
C(C,+C2) 4.40(x2) E 2.24
F 1.90
G 1.75 n4 0.86
n5 0.80
n7 0.34
L 0.25
D 3.40 n3 2.15
J 0.82
n8 0.33
aThe
largestHpaI
fragments were isolated and redigested withHindIII as described in the text. The molecular weights of the pCdl HpaI, HindIII, andnewfragments (designatedby theletter nfollowedby
anumber) produced by digestionswith bothHpaIand HindIII were determined as described in Table 1, footnote b.
VOL. 34,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.503.251.442.374.588.2]546 RABOY, SHAPIRO, AND AMEMIYA
cleavage of the labeled genome with HpaI showed that HpaI fragments A and
C,
were at theends. Theassignment ofHpaIfragment C1 to the end of the genome, rather than HpaI fragment C2, is discussed below. Both fragments fromtheHindIIIdigestion ofterminallylabeled HpaIfragmentAwerelabeledsince there were only twofragmentsproduced. Fragments n2 and n6 were found to be labeled when terminally labeled HpaI fragment B was digested with HindIlI. Digestion of terminally labeled HpaI fragmentsC,
and C2 yielded fourlabeled frag-ments, namely fragments HindIII-F and n5 be-longing toHpaI-C,
and fragments n4 and n7 belongingtoHpaI-C2. Finally,whenHpaI frag-ment Dwas terminallylabeledand digestedwith HindIII, fragments n3and n8 were found to be labeled.(iii) Reciprocal DNA hybridization. To help identify overlapping fragments and their order in a physical map, individual
32P-labeled
fragments cut by one restriction enzyme were hybridizedto Southernblots (22) of restriction fragments generated by a second restriction en-zyme. The twosmallestHpaIand HindIII frag-mentswere not used as hybridization probes in thisstudy because of thedifficultiesinobtaining adequate quantities. In several instances they couldnotbeobserved after autoradiography of thehybridization filter, possiblybecause of the poor retention ofsuchsmallfragments to nitro-cellulosefilters (22).
Figure3shows thehybridization patternwhen individual
32P-labeled
oCdl
HpaI fragments werehybridized
to theoCdl
HindIIIcleavage
fragments. WhenHpaIfragmentA wasused as theprobe,it
hybridized
toHindIIIfragments A,
C,and F (Fig.3,lanea).Hybridizationwasalso observedtoHindIII
fragments
BandH,butwe believe thistobethe resultofsome contamina-tion ofHpaI fragmentAwithHpaIfragment
B. This appearslikely
because whenHpaI-B
wasused as the
probe,
ithybridized
strongly
to HindIIIfragmentsB and Hbesideshybridizing
to HindIII fragments D andK
(Fig.
3, lane b). WhenHpaIfragments
C,
andC2
wereusedas aprobe,theyhybridizedtoHindIII fragments A, B, C, E,F,G,and I(Fig.3, lanec).When HpaI-D wastheprobe,it
hybridized
to HindIIIfrag-mentsD,I,and J(Fig.3, lane d).
Figure 4 shows the
reciprocal
hybridizations
when individualHindIII
fragments
wereusedas probesagainstHpaIcleavage
fragments.
Cross-contaminationcanbeseenbythe relative inten-sities ofhybridization
tospecific
HpaI
frag-ments. Nevertheless, the results from this
hy-bridization study and the
reciprocal study
de-scribed abovehelpeddefine theoverlapping
re-gions. In addition, homology between the twoJ. VIROL.
e
A- _
B
C- D-E-P
F-
G-!
-Oso4
4-
K-(
FIG. 3. Hybridization of individual 32P-labeled HpaI fragmentstothe4Cdl HindIII restriction pat-tern. Approximately5 x 10 cpmof each HpaI
frag-ment was usedin thehybridization assay and
ana-lyzed as described in the text. The autoradiogram
showsthehybridization patterns withHpaI-A (lane a), HpaI-B (lane b),HpaI-C1 and-C2 (lane c), and HpaI-D (lane d). The HindIII restriction pattern of
4oCdl
DNAisshownschematicallyattheleft.endsof the pCdl genome wasrevealed bythis study, since HindIII fragment F hybridized to HpaI fragments C1andC2 andtoHpaIfragment A (Fig. 4, lane fl and HindIII-C hybridized to
the samefragments (Fig.4, lane c). In the recip-rocal experiments, HpaI-A hybridized to both HindIIIfragmentsC and F (Fig.3, lanea),and HpaI fragments C1 and C2 (or whatwe desig-nated
HpaI-C,
as being at one end of the ge-nome) hybridized toboth HindIII fragments C andF (Fig.3,lanec).With the information obtainedbythe proce-dures described above, the restriction map of
4Cdl DNA was constructed (Fig. 5). Based on
the analysis of in vivo and in vitro4Cdl tran-scripts (Amemiya et al.,in press), thephysical map of 4Cdl DNA has been oriented suchthat transcription of the genome begins at the left
end andproceedstoward theright. HindIII frag-ment F andHpaIfragmentC1,therefore,should contain the earlypromotersrecognized by the
host RNApolymerase (Amemiyaetal.,inpress). We know thatHpaIfragmentC
actually
consistson November 10, 2019 by guest
http://jvi.asm.org/
[image:5.503.293.433.65.330.2]
B-B= * __-_-- - _
- -_
E-
F-ci cc d e g h
FIG. 4. Hybridization of individual 32P-labeled HindIIIfragmentstothe 4CdlHpaIrestriction pat-tern. Approximately 2 x 104 cpm ofeach HindIII fragment was used in the hybridization assay and
analyzedasdescribedin thetext.Theautoradiogram shows thehybridizationpatternwith HindIII-A(lane a), HindIII-B(laneb), HindIII-C (lane c),HindIII-D
(lane d), HindIII-E (lane e), HindIII-F (lane
t),
HindIII-G (lane g), HindIII-H (lane h), HindIII-I (lane i), and HindIII-J (lanej). TheHpaIrestriction pattern of pCdlDNA isshownschematicallyatthe
left.
POSITION 0 20 40 60 80 100
T
Hndm f G 0 HAK 0 JI l E A C
T T T 7
[image:6.503.45.238.66.216.2]Hpo I Cl E 0 0 C2 F A
FIG. 5. Restriction mapofpCdlDNA. The cleav-age mapofpCdlDNAconstructed withHindIIIand
HpaIrestriction endonucleases isshown.Restriction
fragments were labeled andorderedasdescribed in
thetext.Only the positionsof the smallestfragments (HindIII-L andHpaI-F) are uncertain. Early
tran-scriptionofthe genomebeginsattheleftandproceeds
totheright.
of two fragments (Cl and C2); one of these is
located at the leftterminus,and that fragment
wasdesignatedHpaI-Cl. HpaI-C2wasassigned
to the region further to the right. The exact
placement of the two smallest fragments,
HindIII-L andHpaI-F,remains inquestion. Terminally repetitious sequences. Two
different experiments indicated that the 4Cdl
genome might be terminally repetitious. The
results ofreciprocal hybridization studies (Fig.
3 and 4) showed that HindIll fragment F
hy-bridizedto bothHpaI-Aand-Cl (Fig. 4,lanef) and thatHindIII fragment C hybridizedto the
same fragments as HindIII-F (Fig. 4, lane c).
The second observation which indicated that thegenomemayhaveterminally repetitious se-quences camefrom invitro transcription
map-pingstudies (Amemiyaetal.,inpress).Asingle
RNAtranscript synthesizedinvitrobyC.
cres-centusRNApolymerase with 4Cdl DNAasthe template was found to hybridize to restriction fragments located at both termini. To determine whether terminally repetitious sequences were
present in 4Cdl DNA, the intact genome was
partially degraded (approximately 5%) with E. coli exonuclease III (16) and then allowed to
annealat65°C for 60min,asdescribedby Rit-chie et al. (17). Electron micrographs of the digested and annealed molecules are shown in Fig. 6. Only linear molecules were observed in controlsamples of
oCdl
DNA that hadnotbeen treated with exonuclease III but had otherwise beensubjected totheincubationandannealing conditions (Fig. 6A). Treatment of 4Cdl DNA with exonuclease III but without the annealing procedure also yielded only linear molecules (data not shown). However, when 4Cdl DNA was partially degraded by exonuclease III and allowedtoanneal,asignificant proportionof the molecules were circular (Fig. 6B). Approxi-mately 10% of the exonuclease III-treated and annealedmoleculeswerecircular,whereas in thetwo controlsamplesno circularmoleculeswere
seen. These results, together with those dis-cussedabove,indicate that
oCdl
DNA contains terminallyrepetitiousnucleotidesequences.DISCUSSION
A physical map of the Caulobacter phage pCdl genome wasconstructed by using restric-tionendonucleases HindIII and HpaI. A total of
12fragments were produced byHindIll, and 7 fragments were generated by HpaI. Although
somecross-contamination between the large re-strictionfragmentsoccurred during the isolation procedure, the relative order of the restriction fragmentscould be deduced. Only the positions of the smallestfragments(HpaI-F and HindIII-L) are still uncertain. The molecular weight of ¢Cdl DNA wasestimated to be approximately
28.8 x 106, asdetermined by the relative electro-phoreticmobilities of the restriction fragments. This is in close agreement with the results of Westetal. (23), who determinedthemolecular weighttobe29.0 x
10k
by zone sedimentation.Hybridization studies with restriction frag-mentsand in vitro transcription mapping (Ame-miya et al., in press) indicated that common sequences were located at the termini of the DNA. The presence of terminally repetitious sequences was confirmed by the observation that circularmolecules were formed after limited exonuclease III digestion and annealing of the exposed single strands. Terminally repetitious sequenceshave been shown to be present in T3, T7 (17), and T5 (15) DNA. It has become ap-parentthat there are many similarities between
oCdl
andthe E. coli bacteriophage T7 (8, 25).VOL. 34,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.503.46.239.341.380.2]-"'..- ...-,-.1 .;...i..:.- ., .,... ...-..--.,,,-..,-,t .,....,:.:,..-..'t.%I....7.-.i .,- -,-.... ,... ..
.... ...:..,.,,...:-:-.. -.-" .. .; .,.:....I,...
,.... -...--I1. z,,...:.,.-.,,..,..
-, .-.. .,.:".,-..."-,:,..-'. ,:..-...-,,'i..,",...;....
..::..,,,.. .:, t". -,---,-,- ..-. ,,---,-. -,..,-.I..
::.! ,.---,--....",.-...`.`--'...-.'-- ..'...1 -.- .,,...,!.I..1.'!.-...,-,I...I...,,-..1,.,Ili...:,.";.'.-..-..'--,'.1,::;:,:---,-,..:,...!...
."..,:,:% ..',-;.1-.-.,7.', -.'..
,.,,..:,,-.,,-...l.. i...,!-.---...:--..,," -,,. -.r,tl...,j...:i,.,:"'....,..-...,I-,...-q'i...-;-...-..:-.-O...-:-,...-..: ..-:! :-....,..,'...-I.,...
,..; 1; "...--,..
.---.-..
''... ..%....----....I..,-.-...,:..d...:..."--.. ., --,.,jj-,;..-...,.--!-.-..-...".a. ..;I... .-,1...I..:...,".
.---....!,. ...
,....,...-..-I..
..Fl,"'....,..-,.-.,--,.,,-"p..'It.... '-.:1'.,-.,--:7...1...-...,.-,,-I..:...-..','-':!...1..-,.-.'-.,..--..-..:-... ,-.. ...".--.,.,. ...-,>-.-,.-.-.. .- ...:..-.,, ..,.! ...-,-:.,...,--:---..-.,, ...'...,""... .. ..,-II..-:-...,.,....-....-:...,.-..-I,-I--.--... .:li
1,...
i;,..;,.,....,.,.I.) ... ,"'--...:. -t.,-,.,;..f!.-,.,:.--..
..-'.-,..,..'!.:.I
j...,11:,:.,...----;,-..-I...,,....I%-I..., ...
.-,,,:.:....-.:.I.I..--:-.!....:.-....--...-,-;. -,".,..,...:.-1; ..: ..-...:.-....::.:..-..:.,!.--...-I-.. .., ....-:--.-
-.--,-,-.".;-:.::....-:-....;.,-...
..;-.,-... ,...,..1...
-.,:-,.-,...-,.,.-. ;, ...,...'.;-...,.j".--I.I...,..
;:,...:...II-.---,..7.:..:...
-...
.,!Z.-,j 7.,.'.. .,-:,--...1: :....-,-%.! -.--',-, -.I.,...,.. ..I. 'f..--.--I,-.-,.11'...,,..-..;.:...:....:.... ...%..:--;.,%..-,.,.r-,--,Jf---,,I.-.-'.-,-;-,:v....,.... ,...--...-j.... .-.... .- ,-!.-,-...,,..k.1:---....---..,.:.. -. 1,...:.
..- ..."...I....--,..'.
!...,,---..,.-,,,,,,..,.-. .-..,..--,-,,---;:,,.-,,F ,!....,.-....,...'... ... -... ,...,..-1:--...,-..,"..7...:,...,--,;.,...:--...; ...,.... .,..-,...,,..,...-..---....I..:.:...1,..,----.:.11I..;.
--.-.-.I.--,...'-'... .-
..-,.-. ,.-,,-.-I'.-...I...t..:...,.!,,f..",-If... --.:
-.--,--%..-,--!,--:--., ..:. --.,..:--:.:.-I!.-, '...,....,....
-...,-i..I...I-" ,-.,.-- ....,..,.'.'....-...t..,-."...-:...-.-1.... :...
.t-...
-.,-....
-ZI.--;.1 ,f:..---,---.,.::--,...-.I.,----"-..,--,...".,.: '.-AI.4---1,,; 1.--..-,,.,,.t;.--:,-%-,-.-.. 1, ,--,:::,,-!.,.'.:,;.,:...1,"...I...-!-:-.,"-'..,, ...I-,- 11,...":'. ...,.R,-..--,
...-11.,,.,-.,.: ,.... ....-L.--...,",-.
.,-..,.,..,;... .,.:..,...--,::...
Z.-1:1,...i...1.,.. -...I.."...."-;,,...--,:... ,-,:
...,-... ..--.-,o...--...I..-,.-..
....,,,..-d..-,-I....;..t...:.,.1.--. .i.- ...-..;:-,:...A --.-,I.-,",,`.,...:
..,-.-.. ...
j'... ..;..4.. ,..,,, ...
"'."-,.,..-:--- -.:...; .,.-,...,---...
,...-,7.1,---".... ...-...
,..,
.,....-,.:-..:--:...1-.-.",..:..&.
.... ,.. -...----.%,:.-'..----,,.;.... I..-.-,:-:,,:I....,.,...
.,-.'.! -,,..--,...-I....-....1-.,...
..--...,...,,-....
,,. .., ....;,...:.'-,'.t-- :-, -,-.,.. -.. .I.I.:- -.:...':.;:..-..-,-..!...:..---,I... :-.
..,%.-.:..-....,...:..,...
...,,-.,-.. .,...--...I-.--,...---,.-:1- ----;-k,.. ,...-.
...--.-..-...- ... ...-...-..
...,,,.. ,-.-..:...?.,.".-....-...,:-...I-, .-...-.-:'-.-.-..,-,-,,6,--.,.--...,..4,.",-...,
,....I....-:--.1 ::i.. -..-:,...-..,.
Z ..."-,.i.?-.... ...1-.?...,,,.-....
,...;.-1..,-...-...-...:....:-.,L,...
-. .,..-...;. ...A"
,'..:..:,..,....,...;.,..:.1..:., ...
...,v,.I...,..-...,..I,..I---..t,,.,,:4...I. -.-..,...
:.%-.,....I...-..IQ: ....-...,I
.. ...,--,-- ",,,.-..-- -..-7.,-.'--.I'.-.---,-..,-...,... .--%,--, --.... ... :;.."..,,.-,"-'...,. -. -....--.
...-.... -..:...,... .-.,!...--.-,-....I-..
,.:-...".., ,,,.,..- ...,1--,,.:P!,...
.--1.j...,.-.,,:.,-,-,...-....I..
...-.-".". I. ...; .:'.,...,...,,.
.,F.. ..,14.-:---..-r. .,.' .:-,-...--:!...,
,.. ."'-,-,...,-,-...,-....
..;-...,...,,..,;....
.,..,...;.:-...I.-.....1...
:.tI.-,.-.
.,--..,..I....,:I---.I
--...,...,-,..,..-II.1.--...
...1...,-,-.
.-.:.:
,-..1.:i...z,,,,.... .. ---....I...:-...,.-... ..11.-...,-:;,.-.......,...,..-..i-..,:,.,.I.- ...--...,..,...-..-.,.. ...t.,....--..-.I-...-..I. ..., .-:...:...-,..
..-...;,;.----.,-..:..,-...,...-..'...,.,...,.. ..:..:...-,----...2...,-...:..-,c:6..-..-..- ...-..
...,;,j.,.",--..'.- I.-,...;----.-.I..
.'.-".t,:,:... ..1...-,.
-..-...I.:,....N"..
....,....,,-
...,...:.1..-..,..-,...
.. ,,..-,...,---:." .. ,--,I..:;Z..,:,,.-
.,-r,..I,--.:I..:,-...
-1... ...,..."...'...-...r. .--....i"...-...
--.--,..."...,..,!.:,-.'r.:,...,-.,,---...:,-.I..I....-...:-I.,....I.:...,... ,..
I:.-;,:,-.-Z...-.-,.!,..;'..."'!...,,,... . -:'-...1-,... ,.::".. -,...:..-,:..,:,..l.. .,.
..II.,.,.
I,,I-.1--...:.,.-,--..,...I.1...,.'-....-....,,:. --, ,-....,...1....., ,-1..., ... ...-. .,,: .:...-...--!,.,--l.,.-'....,...,..,..,.,-,.. ... ...1-..-... ...'... .,.... ,..,... .i6,
.,,.-,..,--:,. .,...I...:..
-..,.-- I-,-..-...; ,...,--...
.-,;.- .... ,-,.-'. .:. ..---,C--: .,-...,... --.---..,,- ---.-- -....,I
,.,,-, ,...1 ....,.,--..I
., A. .: ,. -,...;,;. -...4.."..-$,...-t,I,-,--,.;....:.;,.,.----.,., 1 I-.-.I..,,.-.-',--l-.--I.. I... ...--, -_:.-.I... -.1-...I-..'... ..'... ::,:, ...
-,.1,.,.(I,\,
",...,
....,,,.,..-f.,.
."...,...1..1-....
..--.,1, I--I-.,,.,. .. p., -..-...-.."',-I-,.,..--.... ..:,:...,,
,-,-"--.,!I..-!.-...,!,,...1:1.,...-...:-
e.-.,-,.-..-.:...
,-- ,--:.'..-I."..
-.-.,..,...
...1.-...,_....Z.,..II...,,.-...,.,..---....-..,.,----II
,- I-.-..--,...--I.i.1...I--.... ..-,..:, .. .:..-,,,.... ,...
...7.",.-".,....:.-:.,..,--,..---,'t,.:-..-,...
....-,-;,,.--I--: --,...-.I..-,.. !,...-., ..:-...,..,...
..,.:.,.7.,
I.,
.i..--
I..:..;...,:.. I.. --,..!...,...,...I...
.,.,...
,...,..._.I...-...-..7...,--,. ,-,-... ,...:.I...";..."...:..,-I'.4-!I....
.,I.
.-.--.-,-%...,...-.:."...I-i'.,..-,,. ,.---.-....I.. .:
.1:-I-...,
..i..:...-:-..--'...,.
.,..,.,..:-,.-...9..-,!....--...-I.
...1---- ,.;-...--...--,-...'....-,..,,-,'--.,:-'.,:.-;.-P.': ...L--...,....-,.-...,...-,,.-...
I..-,..-, -"...-.,,.
..-....:.I.
..,...:-...-1.-....,..-.-,....:..,.,.,...;,.:,. .:...I.1..1....,...,:...1.I..
I.,....e.:...
-I.-...-.-".-..II..
..
...,.-....I,.I...-:-,-i...,,....,.-I..,.,..:...-.:-%...I---l----...,7...I...-.1.,I...-..:,.1..-...-...,I
I.' i.: .-.;-...-..-;..t,.".-...,-:i.: .1...--
:...-,...",
.-....%,-.--.-, ,- ...-!. :.-...,... ..,,...I..-.1....,:1,,...II.,...-..
-....:--...,....-...IL;-..,:..,.-
--...i.-.--.,.-....-"---....I.I....
I.,..
.,-,.1I,.,.I...
,.---.,:I....-..,--.,...:..,.,...1..--.-i,...%....,....,1....-.,-,-I-,..--,.:.-:.,,. .-..-,.j-.:.,..-:,,-.:.'..,, .--;-,-1!,.:.,,."-.-...;.:...
-..;...
-.I...-,,.."I.,,j.."..,.--...-.;..:I...,....,...,...I...I...-.-...-,.'.;.,...-I-.,I.-....,..11I--...,..-... ...,.,..,..-I...,...,...,..1...:!.,,,--,- --..,....--t,--...1-....It'.I...:.. ...., ....-I--. .-,----....
..-.,,--.-
,--,-,.,..,!.
%--.,--:.:....,...,..:,...--,...,.,.--,".`-,.-...I.-...,-...-.--...--i .,:-,..,..-,.,,- .:...:-,,...,:,.-,-...-..:...,.
...,...
...,...,:...-.,:,. ..,,..,-...;.I,..,..,.,.,...I.
...I...'..:..-::.:....1;.,.,.1..-..
...I.-,-,-::...-..-..,.,,....-...,---..:.I.-...:---..!:.
....:..,.-!-..,..I.I.-..-I....I.-...--..,.-- i.,.--.1.I.--.I... -...--...-I....:.1. ...,...,-...---...11
.l..-I-,-,,-I--
I.-I--.,...
...,-..-%.,...,.,..:.,..I...iI...:...,%.. :;1;"; .:...--,.-.
..1. ..,I... .,..-.:-.. ...-.-7 .:.,,,.,-..-. --,-.-.-- ..-:.,..,
...",.-...."...-",,."..,.-,.. .,-,...,....",....;.:..,....-..-I...;,-...-,--17.,...,
-..-.- ...-.'....,-, ..---..-,.--... .'-'..,&
.-...-.., :...:.. ... --.-1- -.
- , :.,-,.--,.: ... .,.: ,.:... ,,
.:.,..-,I-I-: ..
.:.4,.,--..--,,-Z" .. ,
...:,Z. ..,,,I,,..- ,..,,--.. P' ,P- .-f..
..:...,...1,,-.,----...,.-..-..,-,z... , ...,..,.,-,:..,.,.t ,-;i,.,;...-., ..
..I-:... ,... ...,I.- -,.,...-:.,--!---'-,..-I...:.- 'o -,..,.-.. :-0 ,
.,.--.-,, ..,,.,.;.,1:-1....,I...., .:,",", -,;--- ..---,i-,,.,;."I.,.,-,-t..
.., ....,-.-.,....--,l--.: "-..:,-.,,!-j,;-".i,,.,-.,1--,:..;.,-- \-....-...I... ,-.,6-., -.". -.. -,-"?. --,--,-..,.-:.l. .,.-.--:....-":.-.,-,"', ,-"(.-.--,.--C'.,-,.I..,-,.1.:,. ..
,...! .-,,-:...-.I..1. ..",-,-..-.--....---.-1. "' ,,,
,-....,-
..;..-- ---.,:.,.--.--.:.-- -.'t..,..,4.-., ....,.,.,,.,..I.,o...r.,i..,....-.. . .,,..,
-. -, ,'. ..-:---.--,..1..,-:.---.1,
'S.;.-.--...r."-1,-.-.:.,.t:,.-..-.e-- -,...'-...-...,--',-....,,.--1,,,.,....,...,....,,'....:-,---,- ...,-."-..-I...., ".I- .,..--.C,-,...-,%.-,;,;-. ..li
.,..-.. .,,---.,...'..I..".,,----.-. -.--j/...II,.,,--.:....f--'::....1...:?, 11,,...,!;i, "I..-.--".'".'-..';"'.'. p-4dp --,-.').,"-I,,lp.-;"I-.-.-,...'t '.--..-
-,.-i,-4,....'.-".C--...,,, .- i,1,.,,-
-x--.. -,-..r..,.... ,,.;-...'.'--", .;.". ..-S..
.-,---,--.-,.,.. .--, I... f..
--,
,-...--...^,,--- ,---...--,,,-,.,#... ,..-,.,.--.i,."'!., -...,,: -.. .:.,,-1
t..-FIG. 6. Electronmicrograph ofexonucleaseIII-treatedo4CdlDNA.4Cdl DNAwaspartially digestedwith exonuclease III and then allowedtoannealasdescribed in thetext. (A) 4Cdl DNA withoutexonuclease III digestionbut incubated underannealingconditions. (B) oCdl DNA treated with exonuclease III (approxi-mately5%digestion)and allowedtoanneal. Bar= 1.0 Lm.
548
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.503.94.434.32.614.2]oCd1
These phages have a small icosahedral head
(diameter, approximately 60nm) andpossess a short tail. Their genomes are double-stranded linear moleculeswith similar molecularweights (29.0x 106for
oCdl
and26.5x 106 forT7) (11). The ends of bothoCdl
DNA and T7 DNAare terminally redundant. Furthermore,atleasttwo aspectsof theinfection processare commontothese lyticphages: (i) thehost RNApolymerase is involved in the early transcription program, and (ii) arifampicin-resistant RNA polymerase isrequired for lategeneexpression.
We have oriented the 4Cdl restriction map
such thatphage transcription beginsatthe left
end of the mapandproceedstotheright. Both in vivo and in vitro transcription studies have identified the restriction fragment where
tran-scription is initiated. Our investigation of the temporalprogramofphage transcription
(Ame-miya et al., in press) indicated that the host RNApolymerase is involvedinthetranscription of the phage genome early after infection and that a rifampicin-resistant RNA polymerase, whoseappearanceisdependentondenovo pro-teinsynthesis, is required for later transcription. With the construction of the physical map, we have identified whichregions of thegenome are involved in the early transcription program of thephage and which regions are dependent on denovoproteinsynthesis forgeneexpression. In vivo, theearly phage RNA transcripts produced by the host RNA polymerase hybridized pri-marily to HindIII fragments F, G, and B and HpaI fragmentsCi and B. RNAtranscripts
syn-thesized invitrohybridized tothe same region of thephysicalmap.Weare nowintheprocess oflocating thepromotersontherestriction
frag-mentsin theearly regionofthe 4>Cdl genome.
ACKNOWLEDGMENTS
Thisinvestigationwassupported by grantGB42545X from
the National ScienceFoundation(to L.S.), byPublic Health
Service grant GM 11301 from the National Institutes of
Health, and by PublicHealth ServicecorecancergrantNIH/ NCI P30CA113330-8from the National Institutes of Health.
L.S. isarecipient ofaHirschl TrustAward, and B.R. isa
recipient ofaMartinFellowship.
We thank N.Agabian for phage4Cdland JaneFant for
her excellent assistance with theelectronmicroscopy.
LITERATURE CITED
1. Aaij, C.,and P. Borst. 1972.Thegel electrophoresisof DNA.Biochim.Biophys.Acta269:192-200.
2. Agabian-Keshishian,N.,and L. Shapiro. 1971. Bac-terial differentiationandphageinfection.Virology44:
46-53.
3. Amemiya,K.,C. W.Wu,andL.Shapiro. 1977. Cau-lobactercrescentusRNApolymerase.Purification and characterizationofholoenzymeandcorepolymerase.J. Biol. Chem.252:4157-4165.
4. Davis, R. W., M. Simon, and N. Davidson. 1971. Elec-tron microscope heteroduplex methods for mapping regions of base sequence homology in nucleic acids. MethodsEnzymol. 21:413-428.
5. Denhardt,D. T. 1966. A membranefiltertechniquefor thedetectionofcomplementaryDNA.Biochem. Bio-phys.Res.Commun. 23:641-646.
6. Dunn, J.J.,and F.W.Studier. 1973.T7earlyRNAs are generated by site-specific cleavages. Proc. Natl. Acad.Sci.U.S.A. 70:1559-1563.
7. Gromkova, R., and S. H. Goodgal. 1972. Action of Haemophilus endodeoxyribonuclease on biologically activedeoxyribonucleic.J.Bacteriol.109:987-992. 8. Hausmann, R. 1976.Bacteriophage T7genetics. Curr.
Top.Microbiol.Immunol.75:77-110.
9. Hirsh, J., and R. Schleif.1976.High resolution electron microscopic studies of geneticregulation.J.Mol. Biol. 108:471-480.
10. Maxam, A. M.,andW.Gilbert.1977. A newmethod for sequencing DNA. Proc.Natl. Acad. Sci. U.S.A. 74:560-564.
11. McDonell,M. W., M. N.Simon, and F. W.Studier. 1977. Analysis of restrictionfragmentsof T7 DNA and determination of molecular weights by electrophoresis inneutral and alkalinegels.J.Mol. Biol.110:119-146. 12. Murray, K., and N. E. Murray. 1975. Phage lambda
receptor chromosomes for DNAfragments madewith restrictionendonucleaseIIIofHaemophilus influenzae and restriction endonucleaseI ofEscherichia coli. J. Mol. Biol.98:551-564.
13. Peacock,A.C., and C.W.Dingham. 1968.Molecular weight estimationandseparationof ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry7:668-674.
14. Poindexter,J.S. 1964.Biologicalproperties and classi-ficationof theCaulobacter group. Bacteriol. Rev. 28: 231-295.
15. Rhoades, M., andE. A.Rhoades.1972. Terminal repe-tition inthe DNA ofbacteriophage T5. J. Mol. Biol. 69:187-200.
16. Richardson, C. C., I. R. Lehman, and A. Kornberg. 1964. Adeoxyribonucleicacidphosphatase-exonuclease from Escherichia coli. J.Biol. Chem.239:251-258. 17. Ritchie,D.A., C.A.Thomas,Jr., L.A. MacHattie,
andP.C. Wensink.1967.Terminal repetitionin non-permuted T3 and T7bacteriophageDNA molecules. J. Mol. Biol.23:365-376.
18. Roy, P.H., and H.0. Smith. 1973. DNA methylases of Haemophilus influenzae Rd. I.Purification and prop-erties. J.Mol. Biol.81:427-444.
19. Shapiro, L.1976.Differentiationin the Caulobacter cell cycle. Annu. Rev. Microbiol.30:377-407.
20. Sharp, P. A., B. Sugden, and J. Sambrook. 1973. Detectionof two restriction endonuclease activities in Haemophilus parainfluenzaeusing analytical agarose-ethidium bromide electrophoresis. Biochemistry 12: 3055-3063.
21. Smith, H. O., and K. W. Wilcox. 1970. A restriction enzyme from Haemophilusinfluenzae. I. Purification andgeneral properties.J. Mol. Biol. 51:379-391. 22. Southern, E. M. 1975. Detection of specific sequences
amongDNAfragments separatedby gel electrophore-sis. J.Mol. Biol.98:503-517.
23. West,D.,C.Lagenaur,andN.Agabian. 1976. Isolation andcharacterizationofCaulobacter crescentus bacte-riophageoCd1.J.Virol. 17:568-575.
24. Yamamoto, K. R.,B. M. Alberts,R. Benzinger, L. Lawthorne, and G.Treiber. 1970. Rapid bacterio-phage sedimentation in the presence of polyethylene glycoland itsapplication to large-scale virus purifica-tion.Virology40:743-744.
VOL. 34,1980