0022-538X/81/020654-07$02.00/0
Viral Protein
Expression
in Producer and
Nonproducer
Clones
of Friend
Erythroleukemia
Cell Lines
RITA ANAND,1 FRANK LILLY,* AND SANDRA RUSCETTI2
DepartmentofGenetics,Albert EinsteinCollege of Medicine,Bronx,New York10461,1andLaboratory of
TumorVirusGenetics, National CancerInstitute, Bethesda, Maryland 202052
Erythroleukemia cell lines HFL/d andHFL/b, derived from tumors induced in vivo in BALB/c
(H-2d)
and congenic BALB.B(H-2b)
mice, respectively, by a polycythemia-inducing strain of Friend virus, produced bothspleenfocus-forming virus(SFFV) and its nativeNB-tropic helper virus (Friend murine leukemia virus [FMuLV]) duringearly-passage generations
in culture.Eventually
each line ceased production of both infectious viruses but retained itstumorigenic potential
in syngeneic hosts.
Virus-producer
and-nonproducer
clones of these cell lines were examined for expression of proteins encodedby
the SFFV or FMuLV genomes.Lysatesoflabeledcellsweretreated with various antiviralsera,and the precipitateswereexaminedby gel
electrophoresis. Expression
of the FMuLVenv gene-encodedprecursorprotein,
gPr85env,
wasobserved inallproducer
andmost nonproducer clones, but the FMuLV gag andpol gene products, Pr659'9 andPr2009a"'PI,
wereuniformly undetectable innonproducer clones.All HFL/d and HFL/b clones expressedappreciable
amounts ofthe SFFV-encodedenvelope
protein, gp52, includingone
exceptional
clonewhich had ceasedto express any FMuLV-encodedproteins. The molecularweight of this SFFV-encodedenvelope
protein was consistently smaller in all HFL/b clones than in
HFL/d clones,
regardless of theirproducerornonproducerstatus. Thevirus-nonproducer phe-notype thus appears to be due to shutdown ofexpressionof the 5'portion of the FMuLV genome in twoindependent cell lines.
The Friend
erythroleukemia-inducing
virus complex (FV) consists ofthespleen
focus-form-ingvirus(SFFV)
andahelper
retroviruscom-ponent,Friendmurineleukemiavirus
(FMuLV).
SFFVhas been showntobea
replication-defec-tive virus associated withspleen focus induction and therapid
development
ofsplenomegaly
and erythroleukemia in susceptible mice (29). Al-though co-infectionwithahelpervirusis essen-tial forproduction of infectious SFFVparticles, helper virus is not needed for infection and expression of the SFFV genome in fibroblast cultures (3, 31) orpossiblyevenfor transforma-tionof erythroidprecursorcells (6).Erythroleukemia cells from tumors induced byFVcan beadapted to growth inculture (15). Mice of theBALB/c
(H-2d)
andBALB.B(H-2b)
strains, congenic with respect to the H-2 locus, have been used inourlaboratorytoestablish a series of H-2-congenic erythroleukemia cell lines, designated HFL/d and HFL/b, respec-tively(12). In thesecell lines,
theexpression of infectious virus and of some viralantigenseven-tually
ceased as eachcellline wascontinuouslypassed in culture (13). Although virus-nonpro-ducercelllineswere
phenotypically
virus nega-tive, they still maintained the entire viral ge-nome, sincerenewedproduction of infectious FVcould be induced chemically. Recent studies fromourlaboratory (2)involving molecular hy-bridization ofcellular RNA with viral comple-mentary DNAprobessuggested that shutdown of Friend virus expressionin these erythroleu-kemiacell lineswasdueto amechanism involv-ing regulation oftranscription of proviral DNA. However, due to the unavailability of comple-mentary DNAprobestoindividual viralgenes, gag,pol,orenv,thepreciseandsequential role of thesegenes inthe shutdown of virus produc-tioncouldnotbeascertained.
The products of the FMuLV genome are rel-atively well known (Fig. 1). The env gene, lo-cated in the3'portionofthe viralgenome, codes for a glycosylated precursor protein,
gPr85env,
which isprocessed togp7O,themajorviral sur-facemolecule, andp15(E) (9, 10, 20). At least threeprecursorproteinsappear to be generated fromthe 5'portion of the viral genome compris-ingthegag andpol genes:
Pr2009'91"'
isprobably a precursor to reverse transcriptase, whereas gPr80gagand Pr659ag have beenreported to be two independent primary translation products of the MuLVgag gene (7,9, 18, 21, 28).gPr80gg appears to be further glycosylated and trans-portedtothecellmembrane, whereas Pr659ag is processed to form the internal structuralpro-654
on November 10, 2019 by guest
http://jvi.asm.org/
Pr65P 9 virionstructuralproteins(p15,p12p3O,plO)
gPr800"oucellsurfaceantigens \ Pr200eDPo-0/.reversetronscriptose
.+ ~ *85""-wvralenvelopeglycoprotein (gp7O) 5, gog Po/ ,nv C 3' FMuLV genome
5 ---.- 3' SFFV genome
4
[image:2.485.44.235.71.173.2]p45 gp52
FIG. 1. Diagrammatic summary of the relation-ship between the genetic maps of ecotropic MuLV and SFFV and the major protein precursors and products translatedfromthesegenomes. The dashed lines inSFFV represent the portion of this genome which is not shared withFMuLV, at least part of whichoriginates fromthe genome of anendogenous
xenotropic virus.
teins p30,p15,p12,and plO.
Several translational products of the SFFV genome have also been identified (Fig. 1). A relatively well characterized one is gp52 (also designatedgp55) (5, 24, 26,33).The gene encod-ing this protein isapparentlythe result of recom-binationbetween the env genes ofFMuLVand axenotropicMuLV (26, 30).The gp52 molecule shows marked immunological cross-reactivity with thegp70productof anumber of mink cell focus-inducingstrains ofMuLV,which also ap-pear to derive fromenvregionrecombinational eventsbetween
ecotropic
andxenotropic
strains of MuLV (8, 11, 16). Certain strains of SFFV have also beenshowntocode foranother prod-uct,p45,which is encoded in the 5'regionof the virus and isantigenically
relatedtocertain prod-ucts oftheFMuLVgag gene(27).Thepresent
study
explores
theexpression
of theproteinsencodedbytheFMuLVand SFFV genomes in relation to the shutdown of infec-tious virusproductioninthe variouserythroleu-kemia cell clones.
Metabolically
labeledtrans-lationalproductsof FMuLVwere identifiedby immunoprecipitation and gel electrophoresis, using antisera to gag and env gene proteins. SFFVgene productswereidentified with anti-sera raised
by
immunizing
rats withsyngeneic
nonproducercellsinfected
only
with SFFV. MATERIALS ANDMETHODS
Cell lines. Frienderythroleukemiacelllinesused inthese studies have beenpreviously described
(12-14). Theircharacteristicsareoutlined in Table1.
Controlcell linesincluded normalratkidney cells (NRK)orFisherratembryocells(FRE) infected with SFFValone,FMuLValone,both FMuLV andSFFV,
orneither. These lineswereprovided bythegenerosity
ofD. H. Troxler and E.M.Scolnick of theNational Cancer Institute.
Antisera. Rat anti-SFFVserum waspreparedby immunizing Fisher rats with syngeneic SFFV-FRE
cells.The rats were injected subcutaneously at multi-ple sites with a total of 107 cells each and developed solid tumors in 2 to 3 weeks after injection. Antisera from two rats were pooled and inactivated at 56°C for 30 min. Goat antisera toRauscher MuLV (RMuLV) gp7O and Pr65O" were provided by the Division of Cancer Cause and Prevention, National Cancer Insti-tute. Goat anti-Pr65O'' serum was prepared against the purified RMuLV gag precursor and reacts with all gagproteins.
Metaboliclabeling of cells.Subconfluent mono-layers offibroblast lines in 100-mm petri dishes or 6 x106 cells/ml from each mouse cell suspension culture were labeled with 100 MCi of [3H]leucine per ml in leucine-free medium containing 1% dialyzed fetal calf serum.Pulsingwascarried out for 30 min at 37°C.
Fortunicamycin inhibition experiments, cells were treated with 0.5,ugoftunicamycin per ml (E. Lilly & Co.,Indianapolis,Ind.) for1h and thenlabeled with 100uCi of[3H]leucineperml for 30 min.
Immunoprecipitation of pulse-labeled
ex-tracts.Labeledcellswerewashed in cold phosphate-buffered saline and then lysed in extraction buffer (0.01 Msodium phosphate buffer, pH 7.6, 0.1 M NaCl, 1%TritonX-100, 0.5%deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]). Extracts were then spun at 2,000 x g for 20 min and pretreated overnight by incubation withnormal ratserum(5
id/ml)
and 50ulofStaphylococcus aureus per ml (a 10% Formalin-fixedsuspension of Cowan strain I [17]). Extracts were clarified by centrifugation at2,000 x gfor 20min. Immunoprecipitationswere setupat4°Cby incubat-ing1 ml of eachextractwith10 1d ofratanti-SFFV serum per mlorwith5,ulof various goat anti-RMuLV
seraperml.After16hof incubationat4°C,S.aureus
wasadded (50pl/ml) and incubationwascontinued for2hmore.Reaction tubeswerethencentrifugedat
2,000 x gfor 15 min, and the pelletswere washed three times with extraction buffer andsuspendedin
samplebuffer (50 mMTris-hydrochloride,pH 6.8, 1%
SDS,1%,B-mercaptoethanol,10%glycerol,and 0.001%
bromophenolblue)inpreparationforpolyacrylamide gelelectrophoresis.
SDS-polyacrylamide gel electrophoresis. Seven percentpolyacrylamidegelswith3.5%
stacking
gelswere prepared, and electrophoresis wascarried
out as previously described (19).
125I-labeled
bovineserumalbumin(68,000)and RMuLVp30(30,000)were
used as molecular weight markers. Gels were then
processedforfluorography(4)and
exposed
toKodak RoyalBlueX-Omat filmat-70°C.RESULTS
HFL/d and HFL/b cell lines: virus pro-duction and
tumorigenicity
in vivo. The HFL/d and HFL/b cell lines and their clonal derivatives originated from thespleens
of BALB/c and BALB.B mice,respectively,
in-fected with FV of NB-tropic host range (14). Each cell line produced both infectious SFFV andFMuLV (titers,>102 infectiousunits/mlofculturesupernatant)inearly passagesin culture. TheHFL/dline ceased
producing
both viruses very early, and no frozen stocks of this lineon November 10, 2019 by guest
http://jvi.asm.org/
656
TABLE 1. Tumorigenicityandexpressionof infectiousvirusbyclonesofratandmousecelllines
Cell line Symbol Tumora
SFFV
FMuLV
yin mor
~~~~~~(FFU/m-l)b
(PFU/rnl)
Rat
SFFV-FRE/FMuLV 8.5x102 2.5x105
FMuLV/NRK 0 6.25x102
SFFV/NRK 0 0
SFFV/FRE 0 0
NRK NT NT
Mouse
HFL/d-NB cl. A V+gp70+ + 15.8x 104 6.25x106
HFL/d cl.4 V-gp70+ + 0 0
HFL/d cl. A V-gp70- + 0 0
HFL/b cl. B2 V+gp70+ + 7.1x 102 2.5x103
HFL/b cl. C4 V-gp70+ + 0 0
a5x
l06 cells
were inoculatedsubcutaneously into young syngeneic recipients, and micewereobserved fortumorgrowth.
bCulture
supernatants
weretestedbythespleenfocus assay in vivo.FFU, Focus-formingunits.'Culture
supernatants
weretestedbythe XCplaqueassay.before shutdown exist. TheHFL/bline ceased virusproduction much later in itspassage his-tory,andthis shutdown has occurredrepeatedly atapproximately thesame cumulativepassage level when stocks frozen before shutdownwere reestablishedinculture (22).
Characteristicsof the various clones of the cell linesused inthesestudies,representing different phenotypeswith respect tovirus
expression,
are summarizedinTable 1.Clonesrepresentingthe HFL/d cell line include: (i) a typical nonpro-ducerclone, HFL/d clone4, which expressesthe viralenvelope antigen (VEA) ofFV(12) andits majorcomponent,gp7O (14), calledV-gp70+; (ii) a unique nonproducer clone, HFL/d clone A, derived fromaV-gp70+population
butlacking
expression ofVEA orgp7O, calledV-gp7o-; and (iii)twovirus-producer clones obtained fromthe V-gp70- line by either treatment with 5-bro-modeoxyuridine (HFL/d-Bu) orsuperinfection withSFFV-freeNB-tropic FMuLV (HFL/d-NB clone A), bothcalledV+gp70+. Clones represent-ingtheHFL/b cell lineinclude: cloneB2,which producesbothSFFVandFMuLV and expresses VEAantigens(V+gp70+); and clone C4, which is avirusnonproducer butexpresses VEAantigen (V-gp70+).NoHFL/b clones which have ceased to express VEA antigen have as yet been ob-served.
Each of thesecloneswasstudied forthe pro-duction of infectious virus detectable in either the XC plaque assay for FMuLV (25) or the spleenfocus assay for SFFV (1).All lines,virus producers aswell asnonproducers, gave rise ta tumors in syngeneic mice 1 to 3 weeks after subcutaneous injectionof4 x 106 cells (Table 1).
Demonstration ofspecificity of antiviral sera.Various antisera to be used in
characteri-zation ofthe
erythroleukemia
cell clones were analyzed for their specificity, using rat fibro-blastsinfected withFMuLValone,SFFValone, or both FMuLV and SFFV. Anti-serum to Pr659' precipitatednotonly FMuLVgaggene products(Pr200"ag'P,
Pr65V",
andp40[21];Fig. 2, lanes 11 and 12), but also the homologous SFFV gag protein (p45) (lane 13). Anti-gp7O serum precipitated the FMuLV env gene pre-cursor,gPr85env
(lanes9 and10), aswellasthe analogous protein encoded by SFFV,gp52(lanes 7, 8, and 10). Rat antiserum prepared against SFFV-nonproducer cells precipitated FMuLV gag gene products (lanes 3 and 4) but not FMuLVenvgeneproducts.The rat serumalso precipitated the SFFV-encoded gp52 (lanes 2 and4).The above antiserawere thenusedbelow to characterize the FMuLV and SFFV proteins expressedinthe variousmouseerythroleukemia cellclones.
Expression ofFMuLV-coded proteins in mouse tumorcellclones.
Suspension
cultures of clones ofthe HFL/d and HFL/b cell lines werepulse-labeled with[3H]leucine
for 30min,and
lysates
of the cellswere treatedwithanti-viralsera.Electrophoresisoftheproteins precip-itated with anti-RMuLV gp7O serum revealed thatgPr85envwas detectable inall clones, irre-spectiveof theirvirus-producer or -nonproducer status, with the exception of HFL/d clone A (V-gp70-) (Fig. 3, lane 4). Thegp7Oproduct of gPr85env was detected only at very low levels aftera30-min pulse, but a 2-h chase after the
labeling
period ledto amarkedenhancementofgp7O
detection (datanotshown) andpermitted confirmationthat its presenceorabsence corre-latedwiththatofgPr85enU.Similarly prepared cell lysates were also
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.485.49.449.80.217.2]1
2 3
4
m
Pr
20gogpoL
56 7
8 910
11121314
Pr209
68K-
a.
am
p40-
*N30K-r
Anti SF.V
Anti
SFFV
4I041
a..
-_ _p
I I
Anti
gp70
-gPr 85
env
-Pr
659°9
pS2
p45
I I
Anti
Pr659°9
FIG. 2. Identification of viral proteins in clones of rat cells infected with SFFV, FMuLV, orboth. Cells
were metabolically labeled with [3H]leucine for 30 min, 'and lysates were treated with various antisera. Antigen-antibody complexes were extracted with a 10% Formalin-fixed suspension of S. aureus and analyzed by electrophoresis on a 7% polyacrylamide gel containing SDS. Rat anti-SFFV antiserum (lanes 1-4), goat
anti-RMuLVgp7Oserum(lanes6-10),orgoatanti-RMuLVPr659' serum (lanes 11-14)was used to precipitate uninfected NRK (lanes 1, 6, and 14), SFFV-FRE (lane 7),SFFV-NRK (lanes 2, 8, and 13), FMuLV/NRK (lanes 3, 9, and 12),orSFFV-FRE/FMuLV(lnes 4, 10, and 11) cells. Lane 5 represents'"I-labeledmolecular weight markers bovine serum albumin (68,000) and RMuLVp3O (30,000).
1
2 3
4
5
6
_ W 0
\_-Pr8~Pr
5@n
[image:4.485.45.441.55.228.2]ff:n_
~gp
48
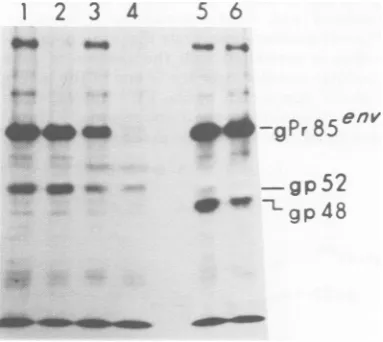
FIG. 3. Immunoprecipitationofenvgeneproducts from mouse erythroleukemia cell lines. Clones of HFL/d and HFL/b cell lines were labeled with
[3Hlleucinefor30min.Celllysateswereprecipitated bygoatanti-RMuLVgp70serum.Different
virus-pro-ducer and-nonproducercelllinesusedwere HFL/d-Bu, V+gp70 (lane1); HFL/d-NBclone A Vgp70+ (ane2); HFL/d clone 4, V-gp70+ (lane 3); HFL/d
clone A, V-gp70-(lane4); HFL/b cloneB2,V+gp70'
(lane5);andHFL/b cloneC4, V-gp70 (lane 6).
treated indifferentexperiments withgoat
anti-serum prepared against the RMuLVgag gene precursor,Pr659ag(Fig. 4). Incontrasttotheenv gene product of FMuLV, the expression of
Pr65"" and gPr80gag correlated with the
virus-producer phenotypein thevariousclones(lanes
1 and 5). Virus-nonproducer clones showed no detectablelevels of this protein (lanes 2-4). This samecorrelationwith virus production was seen withrespect toexpression of the high-molecular-weight precursor,
Pr200"9`"',
a finding con-firmed in separate studies using goat anti-RMuLV reverse transcriptase antiserum (data notshown).Theseexperimentsdemonstrated that the vi-rus-nonproducer phenotype among the eryth-roleukemiaclonesexamined correlated withthe absence ofgene productsof the 5' portion but notwith thoseof the 3'portion of the FMuLV genome.
Expression of SFFV-coded proteins in mouse tumor cell clones. The SFFV gene product, gp52, was precipitated by antibodies presentinratanti-SFFVandgoat anti-RMuLV gp70 antisera. When these sera were used to precipitate radiolabeled
lysates
ofthepanel
oferythroleukemia
cell lines(Fig.
3 and5),
gp52couldbedetectedineach of theHFL/d clones whetheror nottheyproduced virusorexpressed FMuLV env gene products (Fig. 3, lanes 1-4; Fig.5,lanes1,2,5, and6).Thiswasalsotrueof the virus-producer and -nonproducer HFL/b clones,but the sizeof themajorprotein precip-itated with anti-gp70 and anti-SFFV sera was always slightly smaller,
approximately
48,000 daltons (Fig. 3, lanes5and6; Fig. 5,lanes3, 4, 7, and8).Thesmallerproteinwasalsoaglyco-protein, since it, like gp52,
incorporated
[3H]-mannose
during
a 1-hpulse
(data
notshown).
Smaller amounts of a
52,000-dalton
protein
couldalso be detected in these cells.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.485.46.238.313.484.2]658 ANAND, LILLY, AND RUSCETTI
1
2
3
4
Pr6
:- f SQ
qPr80990
00W0;0<:4iWt
irom
FIG-.4f-.
Imuopei:tto
of
ga gen-oe-nonproducerclonesoftheHFL/d and HFL/b cell lineswerepulse-labeledwith[3H]leucine for30min. Celllysateswereprecipitatedwithgoatanti-RMuL V
Pr65'g'serumand-separatedon a7%polyacrylamide
gel containingSDS. Clones analyzedwere HFL/d-NB clone A, V+,gp70+ (lane 1); HFL/d clone 4,
V-gp70+(lane 2);HFL/dcloneA, V-gp77- (lane 3);
HFL/b cloneC4,V-gp70+(lane 4);andHFL/b clone
B2, V~gp70+(lane 5).
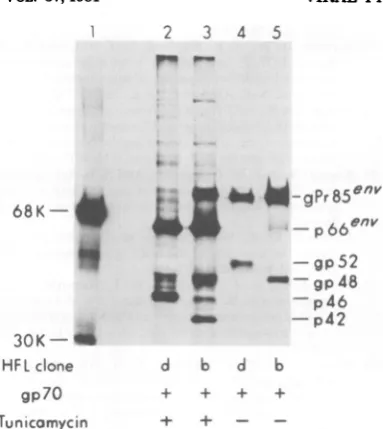
Toascertain whether the difference in
molec-ular weight of HFL/d gp52 and HFL/b gp48
resided in the protein or carbohydrate portion
of the molecule, the virus-producer clones of
both lines were treated with tunicamycin, an
inhibitor ofglycosylation, before labeling with
[3H]leucine.Smallergp52-related proteinswere
expressedinbothclones aftertunicamycin
treat-mentof thecels(Fig.6,lanes2and3),and the
difference in molecular weight between these
unglycosylated proteins appeared to be about
thesame asthat between gp52andgp48.
Con-version of FMuLV gPr85en to Pr66" in the
presence of tunicamycin (lanes 2 and 3)
con-firmned that glycosylation was indeed inhibited in this experiment. It thus appears that the
difference between the HFL/d gp52 and the
HFL/bgp48resides in theirproteinmoieties. In experiments not shown, spleen cells from bothBALB/cand BALB.B mice infected9days
earlier with FVexpressedagp52butnot agp48. Thus, the expression of gp48 in clones of the HFL/bcell lineis most
likely
anidiosyncracy of this line which isnotrelatedtoitsH-2b
type.AlthoughtheSFFV-encoded
gag-related
pro-tein,p45,could beconsistently
immunoprecipi-tated withgoat
anti-Pr65Vqg
andratanti-SFFV sera from SFFV-infected rat celllines(Fig. 2),
its presence in any ofthe mousecell lineswas
notreadily detected, suggestingthat itwas ex-pressed only atverylow
levels,
ifatall. Ithas previously been shown that certain strains of SFFV donotexpressp45(27), andthestrainof SFFVin thelines studied here may be sucha p45-negative strain. Rat anti-SFFV seradid,
however, precipitate
Pr65Vag
in all producer clones (Fig. 5, lanes 5 and 8), but not in any nonproducer clones (Fig. 5,lanes6and7), con-finning thedata obtained with goatanti-Pr65ag serum.DISCUSSION
Two
FV-inducederythroleukemia cell lines of independent origin, HFL/d and HFL/b, have given risetoclones representing both the virus-producer and virus-nonvirus-producer phenotypes. Presentstudiesdemonstrate that virus nonprod-uction is correlated with the absence of gene products encodednearthe 5'end of the helper FMuLV component of the FV complex. Gene products encodedin the 3' envregion of FMuLV continuedtobe detectableinnonproducercells,
1 2 3 4 5 6 7 8
env
gPr85
X4
gp52- Pr65909
5
-p48
Virus +-- +
gp7O + - + + + - + +
l~l
Serum anti gp7O antiSFFV
FIG. 5. Comparativeanalysis of viral proteins of
mousecell lines by immunoprecipitation with goat anti-RMuLVgp70serumandratanti-SFFVserum.
HFL/dandHFL/bvirus-producerand-nonproducer cloneswerelabeled with
[3H]leucine
for30min.CeUlysatesweredividedintotwoportionsand immuno-precipitated separatelywithgoatanti-RMuLVgp70
serum(lanes1-4) andratanti-SFFVserum(lanes
5-8). Various cell linesused wereHFL/d-NB clone A,
V+gp70+ (lanes1 and5); HFL/d clone A, V-gp70-(lanes2 and6); HFL/b clone C4, V-gp70+ (lanes3 and 7); andHFL/b cloneB2, V+gp70+ (lanes4and
8).
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.485.50.240.59.343.2] [image:5.485.254.441.390.533.2]1 2 3 4 5
68K-1
30K-HFL clone
gp7O
Tunicomycin
-gPr
85&nv
-p66env-gp52 gp48
-p46
p42
d b d b
+ + + +
_-FIG. 6. Viralgeneproducts in tunicamycin-treated
cells. HFL/d-NBcloneA, V+gp70+ (lanes2and4) and HFL/b cloneB2, V+gp70+ (lanes3and 5)were
incubated either in thepresence(lanes2 and 3) or
absence(lanes4and5) oftunicamycin for1h, labeled
with[3H]leucine for30min, and then
immunoprecip-itated with goat anti-RMuLVgp7O serum. Lane 1
represents'I-labeledmolecular weight markers.
althoughoneofthenonproducer lines (HFL/d,
V-gp70+) eventuallygaverisetoadifferent
var-iant (HFL/d, V-gp70-) whichexpressedneither 5'nor3'genesofFMuLV. Incontrast,allclones,
producer and nonproducer alike, clearly
ex-pressed the gp52 product of the 3' end of the SFFVgenome.
Our present resultsare consistentwith other
recentstudies inthislaboratory which involved analysis bynucleic acidhybridization techniques of theexpression ofFMuLV-specificand SFFV-specific RNA sequences in the cytoplasm and
nucleus of thesesameproducerandnonproducer clones (2). These studies indicated that both
typesof clones expressed essentiallyall SFFV-specificRNAsequencesand all RNAsequences
shared by both SFFV and FMuLV. However, nonproducercloneswerepartiallyortotally
de-ficient in expression of FMuLV-specific RNA
sequences. The presentstudies havepermitted
ustoidentifytheportionof theFMuLVgenome
whose expression is deficientinthesecell lines.
Clones found earlier to express only 75% of
FMuLV-specific RNA sequences are those
which express gene products of the 3' but not
the 5' end of FMuLV, and the unique clone
found earliertoexpressno detectable
FMuLV-specific RNA sequences also fails to express
products of either endof the viralgenome.
Thenonproducererythroleukemiacellclones analyzed in this study are apparently different fromthe single nonproducer clone analyzed by Racevskis and Koch (23), who concluded that the nonproducer status ofcellsthat they were studying was due to a defect in the processing of
Pr6Y5g
rather than to its failure to be tran-scribed. Thus, more than one mechanism may beresponsible for the nonproducer phenotype of a givenerythroleukemia clone.The SFFV-encoded gp52 molecule was de-tected inallclones of both the HFL/d andHFL/ b cell lines. However, in HFL/b clones, itwas routinelyfainter in theautoradiographs, and a newband, absent inHFL/d clones, was always present at a position corresponding to about 48,000 daltons. This new protein showed the samepattern of reactivity withthe various an-tiseraasgp52. Treatmentof the cell lineswith tunicamycin to inhibit glycosylation produced
parallel
effects on the electrophoresis patternsofgp52 in HFL/d cells and gp48 in HFL/b cells, theapoprotein of gp52 being about 46,000 dal-tonsand thatof gp48beingabout 42,000daltons. Preliminary observations of electrophoretic pat-terns of productsofpartial enzymatic digestion of gp52 and gp48 also show them to be very similar (datanotshown). Thepresenceofgp48 inHFL/bcellsappears to be an idiosyncracy of
this
particular
line, since onlygp52andnotgp48could be detected in cells from the enlarged spleens of BALB.B mice infected in vivo with FV. Apossible explanation for the phenomenon is thatgp48originates fromavarient SFFVenv gene; since gp52isalso detected in these
cells,
it seemspossible that the HFL/b line originated fromaprogenitorcell containingmorethan one SFFV proviral genome, one of which bears a mutation in the gp52 region. Consistent with thisideaistheobservationthat somestrains of SFFV code forslightly
smaller gp52 molecules (27,32).Each of the clones examinedinthese studies has retained its
capacity
to form tumors upon reintroductionintosyngeneic
mice.Thefinding
that one ofthese
clones, HFL/d
cloneA,
failstoexpress anyFMuLVgeneproducts while
contin-uing
toexpressthe SFFVgp52 lendssupportto the ideathatSFFV and notFMuLV is respon-sible for theoncogenicpotential
of the FVcom-plex. Also, the observation that the SFFV-en-codedgp52 isexpressed ineach of the
erythro-leukemiacellclones,whereas theSFFV-encoded p45 is not, is consistent with thehypothesisthat the oncogenic
potential
of SFFV resides in its env gene. However, additionalV-gp70-
clones of FV-inducederythroleukemia
celllines,
tu-morigenic as well asnontumorigenic,
must be analyzedinordertoestablishafirm correlationon November 10, 2019 by guest
http://jvi.asm.org/
[image:6.485.46.238.59.274.2]between SFFV gp52 expression and leukemic transfornation.
ACKNOWLEDGMENTS
The progressof these studies has been abettedby discus-sionswith Alan S.Berkower, Ruy Soeiro,and Richard A. Steeves. We thank IdaShapirofor expert secretarial assist-anceand KennethBlumbergforhelpwith virus assays.
Supportfortheworkwasfrom the National Cancer Insti-tuteunder Public Health Service grant09173.R.A.was sup-ported by Public Health Servicetraininggrant 09060from the National Cancer Institute.
LITERATURE CITED
1.Axelrad, A. A., and R. A. Steeves. 1964. Assay for Friendleukemia virus:rapidquantitative method based on enumeration ofmacroscopic spleenfoci in mice. Virology 24:513-518.
2. Berkower, A.,F.Lilly,and R.Soeiro.1980.Expression of viral RNA in Friend virus-inducederythroleukemia cells. Cell 19:637-642.
3. Bernstein, A., T.Mak, and J. R.Stephenson. 1977. The Friend virus genome: evidence for the stable asso-ciation of MuLV sequencesandsequences involvedin erythroleukemic transformation. Cell 12:287-294. 4. Bonner, W.M., and R. A.Laskey.1974.Afilmdetection
method for tritium-labeledproteinsandnucleic acids in acrylamidegels. Eur. J. Biochem. 46:83-88.
5. Dresler,S.,R.Martin,J. J.Murray, and D. Kabat. 1979.Glycoprotein encoded by the Friendspleen focus-forming virus. J. Virol. 30:564-575.
6. Eckner, R. J., and K. L. Hettrick.1979.Persistence and pathogenicity of defective spleenfocus-formingvirus. Decreasedtransplantability of hemopoietic cells as a marker forpreleukemicchange. J. Exp. Med. 149:340-357.
7. Edwards, S.A., and H. Fan.1979.gag-related polypro-teins ofMoloneymurineleukemia virus: evidence for independent synthesis of glycosylated and unglycosy-lated forms. J. Virol. 30:551-563.
8. Elder, J. H., J. W. Gautsch, F. C. Jensen, R. A. Lerner, J. W. Hartley, and W. P. Rowe. 1977. Bio-chemical evidencethatMCF murine leukemia viruses areenvelope (env) gene recombinants. Proc. Natl. Acad. Sci.U.S.A. 74:4676-4680.
9. Evans,L.H.,S.Dresler,and D.Kabat.1977.Synthesis andglycosylation of polyprotein precursors to the in-ternalcoreproteins of Friend murineleukemia virus. J. Virol. 24:865-874.
10. Famulari,N.G.,D.L.Buchagen, H. D. Klenk, andE. Fleissner. 1976. Presence of murine leukemia virus envelope proteinsgp7Oandp15(E)inacommon poly-protein of infected cells. J. Virol. 20:501-508. 11.Fischinger, P.J., N. P.Dunlop, and C. S.Blevins.
1978.Identificationof virusfound in mouse lymphoma inducedby HIX murineoncornavirus. J.Virol. 25:532-535.
12. Freedman, H. A., and F. Lilly. 1975. Properties ofcell lines derived from tumors induced by Friend virus in BALB/c andBALB/c-H-2b mice. J. Exp. Med. 142: 212-223.
13. Freedman,H.A.,F.Lilly,andR.A. Steeves. 1975. Antigenicproperties of cultured tumorcelllines derived from spleens of Friend virus-infected BALB/c and
BALB/c-H-2'mice. J. Exp. Med.142:1365-1376. 14.Freedman,H. A., F.Lilly,M.Strand, and J. T.
Au-gust. 1978.Variations in viral gene expression in Friend virus-transformedcell lines congenic with respect to the H-2locus. Cell 13:33-40.
15. Friend,C.,W.Scher, J. G. Holland, and T. Sato. 1971. Hemoglobin synthesis in murine virus-induced leu-kemic cells in vitro. Stimulation of erythroid differen-tiationby dimethyl sulfoxide. Proc. Natl. Acad. Sci.
U.S.A. 68:378-382.
16.Hartley, J. W., N. K.Wolford, L. J. Old, and W. P. Rowe. 1977. A new class of murine leukemia virus associatedwithdevelopment of spontaneous lympho-mas. Proc.Natl. Acad. Sci. U.S.A. 74:789-792. 17. Kessler, S. W. 1975. Rapid isolation ofantigens from cells
withstaphylococcal protein A-antibody absorbant: pa-rameters of theinteraction ofantibody-antigen com-plexes with protein A. J. Immunol.115:1617-1624. 18. Kopchick, J. J., W. L. Karshin, and R. B.Arlinghaus.
1979.Tryptic peptide analysis ofgagandgag-polgene products of Rauscher murine leukemia virus. J. Virol. 30:610-623.
19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head ofbacteriophage T4. Nature (London) 227:437-439.
20. Naso, R. B.,L. J. Arcement, W. L.Karshin, G. A. Jamjoon, and R. B.Arlinghaus. 1976. A fucose-de-ficient glycoproteinprecursor toRauscher murine leu-kemia virus gp69/71. Proc. Natl. Acad. Sci. U.S.A. 73: 2326-2330.
21. Naso, R. B., W. L. Karshin, Y. H. Wu, and R. B. Arlinghaus. 1979. Characterization of 40,000 and 25,000daltonintermediateprecursors to Rauscher mu-rineleukemia virusgaggeneproducts. J. Virol. 32:187-198.
22. Plata, F., M. M.Goodenow, and F.Lilly.1980.Studies ofcloned Friend erythroleukemiatumorcells: modula-tion of the tumor-specific Tlymphocyte response by infectious Friend virus production in vitro.J.Exp. Med. 151:726-742.
23. Racevskis, J.,and G. Koch. 1977. Viralprotein synthesis inFriend erythroleukemia cell lines. J. Virol. 21:328-337.
24. Racevskis,J., andG.Koch. 1978.Synthesisand proc-essing of viral proteins in Friend erythroleukemia cell lines.Virology 87:354-365.
25. Rowe, W.P.,W. E.Pugh, and J. W.Hartley. 1979. Plaque assaytechniques for murine leukemia viruses. Virology 42:1136-1139.
26. Ruscetti, S.,D.Linemeyer,J.Feild,D.Troxler,and E. M. Scolnick. 1979. Characterizationofa protein found in cells infected with the spleenfocus-forming virus that sharesimmunological cross-reactivity with thegp7O found in mink cellfocus-inducingvirus parti-cles. J. Virol. 30:787-798.
27. Ruscetti, S., D. Troxler, D.Linemeyer, and E. M. Scolnick. 1980. Threelaboratory strains of spleen fo-cus-forming virus: comparison of their genomes and translationalproducts.J. Virol.33:140-151.
28. Schultz, A.M.,E.H.Rabin, and S. Oroszlan. 1979. Post-translational modification of Rauscher leukemia virus precursorpolyproteins encoded by the gag gene. J.Virol. 30:255-266.
29. Steeves,R. A.1975.Spleenfocus-forming virus in Friend and Rauscher leukemia virus preparations. J. Natl. CancerInst.54:289-297.
30. Troxler,D.H.,D.Lowy,R.Howk,H.Young, and E. M.Scolnick.1977.Friend strainofspleen focus-form-ing virus is a recombinant between ecotropic murine type-C virus and env gene regionofxenotropic type-C virus. Proc. Natl. Acad. Sci. U.S.A.74:4671-4675. 31. Troxler,D.H., W. P. Parks, W. C. Vass, and E. M.
Scolnick. 1977.Isolation ofa fibroblast nonproducer cell linecontaining the Friend strain of the spleen focus-formingvirus.Virology 76:602-615.
32. Van Griensven, L. J. L. D., and M. Vogt. 1980. Rauscher "mink cell focus-inducing" (MCF) virus causeserythroleukemia in mice: its isolation and prop-erties.Virology101:376-388.
33. Yoshida,M., and H. Yoshikura. 1980. Analysis of spleen focus-formingvirus-specific RNA sequences coding for spleen focus-forming virus-specific glycoprotein with a molecularweightof55,000 (gp55). J. Virol. 33:587-596.