JOURNALOFVIROLOGY, Nov. 1977, p.662-672 Copyright©1977 AmericanSocietyforMicrobiology
Vol.24, No. 2 Printed in U.S.A.
Comparisons
of the
Peptide
Maps
of
Kunjin
Virus Proteins
Smaller than the
Envelope
Protein
PETER J. WRIGHT* ANDE. G. WESTAWAY
Departmentof Microbiology, Monash University MedicalSchool, Prahran,. Victoria, Australia 3181 Received forpublication10May1977
Weanalyzedthemaps of[35S]methionine-labeled tryptic peptidesof theKunjin
virus-specified proteins NV2Y2, NV2, V2, NV1½/2, NV1, andV1. Thepeptidesof
NV1½2 are identical to those ofV2, except for one peptide contained onlyin the
latter. The maps of each of the other proteins are unique, and, consequently,
there isnoevidence ofanyoneprotein beingderived from anotherbyproteolytic
cleavage.Thetryptic peptidemaps of the abovepolypeptideswerealsocompared
withthose ofthelarger Kunjin proteins, NV5, NV4, and V3. The resolution of
the peptides of NV2½2 and NV1 is adequate to exclude any relationships with
NV5 and NV4, but a possible relationship with V3 remains, althoughitseems
unlikelyinthelightof other evidence. Since thepeptidemap of V1 iscomprised
ofonlytwo methionine-containing peptidessimilar inmobilitiesto twopeptides
found in the maps ofNV5, NV4, andV3,aprecursor, if any, of V1 hasnotbeen
positivelyidentified. Thepeptidesof NV2 and V2 (NV1½/2) are notcontained in
digestsofNV5, NV4,orV3,andtherefore, like thelatter, NV2 and V2 (NV11/2)
areindependentproductsof translation from thepositive-strandgenome.
Flaviviruses specify the translation of three
structuralproteins, V3, V2,andV1, andatleast
six nonstructural proteins, NV5, NV4,
NV2Y4,
NV2,
NVlW/!,
andNV1 (7, 12, 13). V3 and NV2are both
glycoproteins
(2, 9). In the precedingpaper we presented maps of tryptic peptides
derived from proteins prepared from cells
in-fected withKunjin virus, confirmingthat
poly-peptides NV5, NV4, and V3 are unique to
in-fected cells butdemonstrating thatNV3 is
prob-ably host coded (17). No evidence that NV5,
NV4,or V3is acleavage productwas obtained.
The other nonstructural proteins, NV2Y,, NV2,
NVlW>,
andNV1,are comparatively smallpoly-peptides of molecular weights less than 22,000
(7,13).NV2 and NV1 aredelayedin appearance
in short pulse-chase experiments, and the size
of
NV1W!
is almost identicalto that of V2, thecore protein of virions (16a). The smallest
pro-tein observed in purified virions,V1, is not
de-tected in infected cells, and ithas been
postu-lated that it results from cleavage of NV2 (8).
With other positive-strand RNA viruses,
smallproteinseither arecleavedfrom a
polypro-teinprecursorduringor soonafter translation,
or are cleaved from the immediate precursors
of structuralproteins duringmaturationof
viri-ons (reviewed in reference 3). Some molecules
of precursorproteins escapeearly cleavage and
accumulate as apparently stable products. Hence, in this report we examine the
relation-ships between the smaller
virus-specified
pro-teins,
NV2Y2,
NV2, V2,NV1W2,
NV1, andV1,
aswellasthepossibility thattheyareproductsof
proteolytic cleavages of NV5, NV4, andV3. To distinguish host from viral
tryptic peptides
inpreparations ofproteins derived from infected
cells,
double-labeledtryptic peptide
maps wereanalyzed aspreviouslydescribed(17).
MATERILS
AND METHODSDetails of all procedures used are given in the precedingpaper(17).
Cells and virus. Kunjin virus-infected Vero cells
werelabeled with[3S]methionine for7h, commencing
23hafter infection. Cells that had been mock infected
were labeled with [3H]methionine under the same
conditions. Kunjin virions and SHA (hemagglutinat-ingparticle of65 to80S) labeled with[3S]methionine
werepurified from infected-cell culture fluids.
Separationofpolypeptides.Radioactive cellular
and viralsamplesweredisruptedwith sodiumdodecyl sulfate-dithiothreitol and heated before
electropho-resis inpolyacrylamide gelswithacontinuous(16) or
discontinuous buffersystem (5). Polypeptides of 35S-labeled infected cellsweresubjectedto coelectropho-resis with those of 3H-labeled uninfected cells; the ratio of 5Scountsperminuteto3Hcountsperminute loaded on gels was 2:1. Proteins ofpurified virions
andof SHAweresubjectedtoelectrophoresiswithout
the additionof[3H]methionine-labeled material. Tryptic peptides. Proteinswere eluted from the
preparative gels and treated with trypsin (18), and
the peptides were separated by electrophoresis and 662
on November 10, 2019 by guest
http://jvi.asm.org/
PEPTIDES OF KUNJIN PROTEINS. II. 663 thin-layer chromatography on cellulose-coated plates.
Peptides derived from cell-associated viral proteins were eluted from the cellulose, and the ratio of 'S counts perminute to3Hcounts per minute was mea-sured. The criterion for determining whether a peptide wasviral or hostcoded was the same as that described in the preceding paper (17); i.e., peptides were desig-nated as viral if the above ratio was greater than twice the initial ratio in the digest before separation. Throughout the remainder of this paper, when refer-ringto analyzed digests of cell-associated viral pro-teins,we usetheterm"viral peptide" for the peptides satisfying the above ratio requirement. We shall not qualify "peptide" when discussing all of the peptides found in apreparation.
Peptide maps contained DL-alanine and L-valine as markers. To better compare two proteins, a mixture containing the tryptic digests of both polypeptides wasanalyzed. In some of the figures, the viral peptides of the smaller protein in the mixture (the peptides that could beidentified) are marked with arrows or
areshownseparatelyinlinediagrams.
c
V3
_l
6.4
V2
M
d 0 f
-NV5
-NV4
-V3
-NVt2i
i0
-NV 20
-NVII
-NVI
VI ft
RESULTS
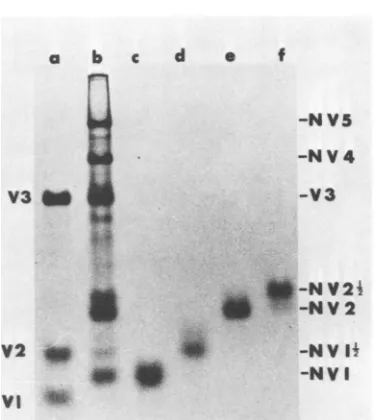
Thepeptidemapsof [35S]methionine-labeled, virus-specified proteins thatarefoundinKunjin
virus-infected cells and are smaller than V3
(namely, NV2Yz, NV2,
NV1W2,
and NV1; Fig. 1) are presented in Fig. 2 and 3. Total and netviral peptide maps are shown for NV2Y2 (Fig.
2A and B), NV2
(Fig.
2C and D),NV1W2
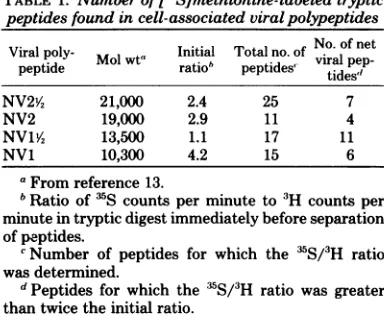
(Fig. 3C and D), and NV1 (Fig. 3E and F). The molecular weight of each polypeptide and the number of viralpeptides
resolved are listed in Table 1. The number of[3S]methionine-con-taining viral
peptides
in the two smallestpro-teins, NV1% and NV1,suggeststhat these
poly-peptides
contain ahigher proportion
(two- tothreefold) ofmethionineresidues than doNV2
and
NV2Y2 (Table
1), and V3, NV4, and NV5(17).Shapiroetal. (10) have also reported that
NV1isrelatively richer in methionine than other virus-specified proteins of Japanese encephalitis virus.
Thepeptidemapsofthe three structural
pro-teins V3 (51,300
daltons),
V2 (13,500daltons),
and V1
(8,500
daltons)
aredisplayed
inFig. 2F,
3A, and2E,respectively. The virion proteins for
peptide mapping
were prepared from purifiedKunjin virions. V3 and V1 are also found in
SHA (7,
12).
In theprevious
paper (17), wedemonstrated theidentity of thepeptide maps
of V3 from virions and of V3 from
SHA;
thepeptidemap ofV1 ofSHA (notshown) is also
identicaltothat ofV1of virions (Fig. 2E).
The lack of similarities in the amino acid
sequencesofNV5,NV4, andV3wasclearfrom
examinationoftheir
peptide
maps(17).
Weshallnow first compare the
tryptic digests
of the [image:2.500.255.443.66.276.2]smaller proteins,
NV2A2,
NV2, V2,NV1W2, NV1,
FIG. 1. Analysesoff5Slmethionine-labeled
poly-peptidesin 8%polyacrylamidegels with continuous
buffersystem. (a)Kunjin virus. (b) Infectedcells. (c)
NVI, (d) NVJV,, (e)NV2, and (/) NV2Y4 aresamples of thepreparationsusedfor trypticpeptide mapping.
and V1, as indicated
by
the peptide maps of Fig.2 and 3,and,
second, compare thesepoly-peptides
with thelarger proteins,
V3, NV4, andNV5.
Comparison
ofproteins
smallerthan V3.Thenetviralmapof
NV2Y2
(Fig. 2B) is different from that of NV2 (Fig. 2D). The differencewasconfirmed
by analyzing
amixture ofNV2VY
and NV2 (Fig. 4A).NV1W2
possesses at least five peptidesnotfound inNV2¼2,
asshownbycom-parison of their maps (Fig. 3D and2B,
respec-tively) and from
analysis
of theprotein
mixture (Fig. 4C and D). Thepeptides
ofNV1 (Fig. 3F) andV1 (Fig. 2E) were notfound inNV2A2 (Fig.
2B). Thus, V1, NV1,
NVlW2,
and NV2 are notproducts ofproteolytic cleavage of
NV242.
Thepeptides of V1 (Fig. 2E), NV1 (Fig. 3F),
and
NV1Y2
(Fig. 3D) are distinct from those ofNV2 (Fig. 2D). The
peptides
of V1(Fig. 2E)
andNV1 (Fig. 3F) are notfound in the mapof
NV1W2
(Fig. 3D), and V1(Fig. 2E)
isnotderivedfrom NV1 (Fig. 3F). Taken together, these
re-sults show that viral
proteins NV2Y/!,
NV2,NV1W2,
NV1, and V1 each haveauniqueaminoacid sequence and noneis derived
by
cleavage
of alarger
polypeptide
inthegroup.Comparison of NV1% with V2. The maps of
NV1W2
(Fig.30)
and V2(Fig. 3A)
arealmostidentical. The
only
obvious difference betweenthe twois the presenceofanadditional
peptide
in V2 (arrow,
Fig. 3A).
Thedesignation
of theviral
polypeptide
ofapproximately
13,500
dal-VOL. 24,1977
on November 10, 2019 by guest
http://jvi.asm.org/
664 WRIGHT AND WESTAWAY
A
NV2;2
4w
Qtep
C
NV2
0s
0>
E
1vIrion
Ob
0D
0
_**
A
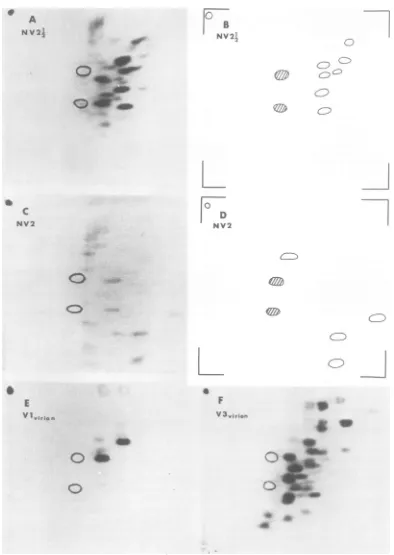
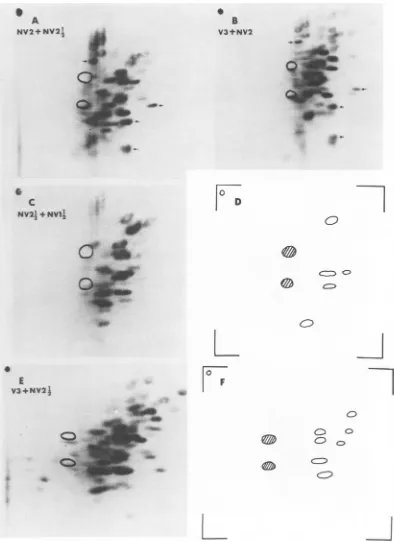
FIG. 2. Tryptic peptidemapsof ?S~methionine-labeled proteins. Electrophoresiswasfrom leftto right; chromatographywasfromtoptobottom. Theopencircles mark thepositions ofalanine and valinemarkers.
(A)NV2i4 prepared from infectedcells.(B)Netviralmapof NV2I4. (C)NV2prepared from infectedcells.(D)
Net viralmapofNV2.(E) V1and(F)V3prepared from purifiedvirions.
e
CB
NV2,
L
D
NV2
C/A.9.Xz
F
V3,VIj
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.500.65.459.68.623.2]B
V3eelI
o t
I
DO
_ V
e
a
w }
o .
S
t [image:4.500.55.446.36.637.2]*4 at
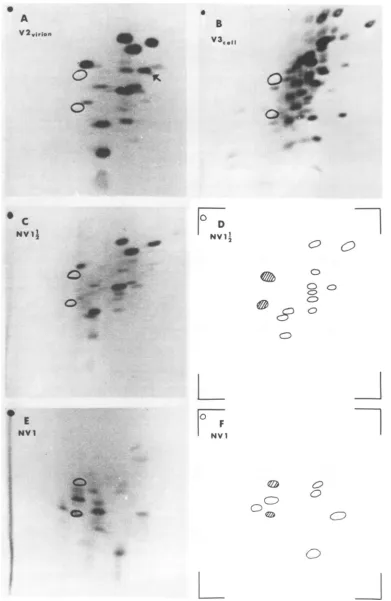
FIG. 3. Tryptic peptidemapsof
rSlmethionine-labeled
proteins. (A) V2prepared frompurifiedvirions. Arrow indicatesdifference between V2 and NV1½. (B) V3from infectedcells.Fordesignation ofhost and viralpeptides,seereference17. (C)NVJVpreparedfrom infectedcells. (D)NetviralmapofNV1½2. (E) NVI preparedfrominfectedcells. (F)Net viralmapof NVI.665
A
V2yirion
"a
& #P ov
e
40
c
NV
I'
NV120
0
Co
C) 0
0
0
5F9
0E
NV1
0 F
NV1
0
0
CD
C
0
0
C
on November 10, 2019 by guest
http://jvi.asm.org/
666 WRIGHT AND WESTAWAY
TABLE 1. Numberof
P5S~methionine-labeled
tryptic peptides found in cell-associated viralpolypeptidesViral poly- Initial Totalno.of No of net
peptide wt ratio td viral pep-pttpeptdes' tides"
NV2V, 21,000 2.4 25 7
NV2 19,000 2.9 11 4
NV1A4 13,500 1.1 17 11
NV1 10,300 4.2 15 6
aFromreference13.
bRatio of'S counts per minuteto
3H
countsper minute intrypticdigestimmediatelybeforeseparation ofpeptides.cNumber ofpeptides for which the IS/3H ratio
wasdetermined.
dPeptides for which the 35S/3H ratiowas greater than twice the initial ratio.
tonsfound in infectedcellswaschangedto
NVW1Y
from V2 because this polypeptide migrates
slightly ahead ofV2 fromvirions when subjected
toelectrophoresisinsodiumdodecyl sulfate-con-taining polyacrylamide gels (13, 15, 16a). The maps ofFig. 3 also imply that V2 is a slightly largerproteinthanNV1'/2 and showclearlythat V2contains the aminoacidsequence ofNV1Y2.
A comparisonof Fig. 3A andC enables vali-dation of the ratio methodusedfor
discriminat-ing between host-coded and virus-coded
pep-tidesinmapsofpolypeptidesfrominfectedcells.
Inthiscase,onlytwoofthelikelygenuine viral
peptides in
NVW1X
areexcluded(oneimmediately above thetop markerand one inthetopright-hand group ofthree), possibly because of the
presence of overlapping 3H-labeled host
pep-tides. This confirmsthe
findings
with V3:thatthe exclusion technique based on the
35S/3H
ratio is conservative and does not mistakenly
include hostpeptidesin the netviralmap (17).
Comparisons with V3. Inspection of the
peptidemapsofV3(Fig.2F and3B) and
NV2Y,
(Fig. 2A and B) suggests that the peptides of
NV2½2 could be common with those of V3. Therefore, a mixture ofdigests of V3 from
in-fected cells and NV2/2was analyzed (Fig. 4E),
and,
although
the seven NV21/2 peptides weretentatively identified (Fig. 4F), they were not
completely resolved from those of V3. The
ex-periment wasrepeatedseveraltimes, but inno case was resolution improved. However, it is
clear by analysis ofthe mixtures that the
pep-tides of NV2 (four offour; Fig. 4B) andNV11/2
(eightofeleven;Fig. 5A) are distinct from those ofV3(Fig. 2Fand3B).
The two major peptides ofV1 (Fig. 2E) and
somepeptidesof V3 (Fig. 2Fand3B) migrated
to similar positions in the maps. The pair of
peptidesin the topright of the NV1 map (Fig.
3F) is in a position correspondingto a heavily
labeled area of the V3 map (Fig.3B). The
pep-tide at the extreme left ofFig. 3Fcorresponds
to one found in V3 of infected cells (Fig. 3B).
The positions of the three otherNV1peptides
werecompared withpeptidesin similarpositions
in the V3 map (Fig.3B) byinspectingpatterns
of mixed NV4 and V3 from infected cells (17)
and mixed NV4 and NV1 (see Fig. 5F), thus
using NV4peptidesasmarkers. These
compar-isons demonstratedslightdifferences in
migra-tion between the three NV1peptidesand
possi-blecorrespondingV3peptides.Becauseatleast
threepeptides (ofthe six identified in Fig. 3F)
ofNV1 were resolved from those ofV3,we can
tentativelyconclude that NV1 isnotderivedby cleavage of V3. However, since
cleavage
of aprecursorproteinmay generateatleast two new
peptides in the product, and changes in the
glycosylationlevel(V3is aglycoprotein)of pep-tides may occur, altering their mobility during separation, ourresults havenotcompletely
ex-cludedaprecursor-product
relationship
betweenV3of infectedcells and NV1.
Thus, NV2 and NV1Y4 are not derived by cleavage ofV3; NV1 andpossibly
NV2Y;4
seemunlikely to be derived from V3, but the two
majorpeptides of V1 could be derived by
cleav-age of V3. This
possibility
is discussed morefully below.
Comparison
with NV4. The peptide mapsofan NV4 tryptic digest (Fig. 5B) mixed with digests of
NV2Y;,
NV2, NV1Y2, and NV1 aredisplayed in Fig. 5C through
F,
respectively. Peptidesthat werefound inthe netviralmapsof the smallest proteins and were resolved in
the mixtures are indicated by arrows. No
evi-dence was obtained that theseproteinsare
de-rived by cleavage of NV4.
Comparison
of themap of V1 (Fig. 2E) with that of NV4(Fig. 5B)
does not exclude the
possibility
that V1 isde-rivedfromNV4.
Comparison
with NV5. Ananalysis
of amixture (Fig. 6B) ofNV5 (same preparationas
in Fig. 6A) and
NV2Y/,
(Fig. 2A andB) revealsno
relationship
between thesepolypeptides.
Thesamplesof NV2(Fig.6C)andof NV1 andNVW1Y (Fig. 6D) used forcomparisons with NV5 were
prepared in 13% discontinuous gels and
con-tained apartlyalteredbackgroundofhost
poly-peptides compared with preparations from 8%
continuous gels. NV1 and NV1 migrate as a
single band in the discontinuous system (17).
Viral peptidesasidentified inFig. 2D, 3D, and
3F forNV2,
NVJA4,
andNV1, respectively, aremarked in Fig. 6C and D. In mixed analyses
with NV5, the peptides of NV2 were resolved
from those of NV5 (Fig. 6E) as were those of
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
-*0
ak
6
io
i_tr
_0
0 E V3+NV2I
9.
CF-1 a
.Ol
Is
Sb
r0
.0
0
00c
e
f0
.2
7
0
o
0
0 O
C
FIG. 4. Tryptic peptidemapsofrS~methionine-labeledproteins. (A) Mixture ofNV2Y, (20,000 cpm)and
NV2(20,000cpm). Viralpeptides ofNV2(four of four, from Fig. 2D) identifiedin the mixturearemarked withanarrow. (B)MixtureofV3(20,000 cpm) from infectedcells andNV2(11,000 cpm). Viral peptides of NV2(four of four)aremarked. (C)MixtureofNV2Y4 (5,600 cpm) and NV1V,(3,200 cpm). (D) Viral peptides of
NVIP4 (5 of 11, from Fig. 3D) identifiedin themixturein (C). (E) Mixture ofV3(26,000 cpm) from infected
cells(samepreparation asinFig. 3B) andNV2-" (24,000 cpm). (F) Viralpeptides ofNVZ4 (seven ofseven,
from Fig. 2B) identifiedin themixture in(E).
667
B V3+NV2
S
A NV2+NV2!
2
C
NV212+NV1I
0
D
0
F
e6
a
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.500.65.459.59.601.2]AS
V3
1~
BV3tNV12 NV4
.~
~
~~0
_~~ a
* ~ t 0
C
D~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~D
NVN 24 NV4+NV2
._
E ~ F
NY4+NYI; NV4 +NYI
2~~~~ ~ ~ ~ ~
a
~1. 9
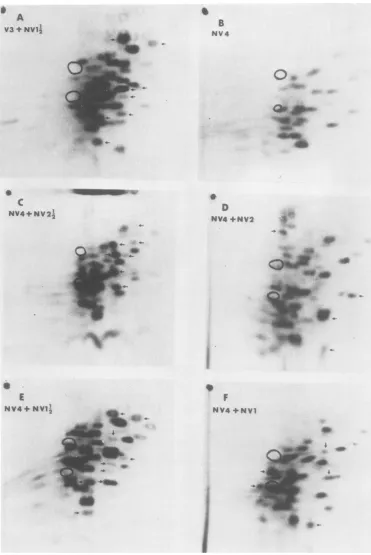
FIG. 5. Trypticpeptide maps of r3S~methionine-labeled proteins. (A) Mixture of V3 (20,000 cpm) from
infectedcells and NVZ½ (9/00) cpm). Viralpeptides of NV1½ (8 of 11,
fr-om
Fig. 3D) are identified witharrows.(B)NV4fr-ominfectedcells. Fordesignation ofhost and viralpeptides,seereference17. (C)Mixture
ofNV4(8,300cpm)and NV2½(5,600cpm). Viralpeptides NV2½2 (seven ofseven,fromFig. 2B) aremarked.
(D) MixtureofNV4(12,000 cpm) andNV2 (11/KY)cpm). Viralpeptides ofNV2(four of four, from Fig. 2D) aremarked. (E)Mixture ofNV4(8,300cpm) andNV1½%(3,200 cpm). Viralpeptides ofNVJ½1 (9of11,
fr-om
Fig. 3D)aremarked. (F)MixtureofNV4(12,000cpm)# and NV1 (12,000cpm). Viralpeptides ofNV1 (fiveof
six,from Fig. 3F)aremarked.
668
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.500.71.442.42.597.2]V,, .#
i.I.; (
i
9'.
.-D
co
I
D
NVI+NV11
Oe
0...
a9
w-aI
L
I
.-0do
l -P.o
w* 040
0
F
NYS+NV1
+NVIj
40
0-.-t_.
p4-
0-I
4-0
[image:8.500.70.429.29.627.2]v
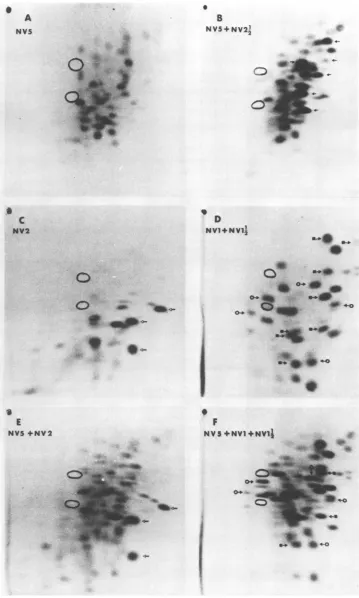
FIG. 6. Trypticpeptidemapsof[fS~methionine-labeledproteins. (A)NV5from infected cells.For
desig-nationofhost andviralpeptides, seereference17. (B)Mixture of NV5 (17,000cpm)and NV2½ (15,000cpm).
Viralpeptides of NV2% (sixofseven,from Fig. 2B) are indicated. (C) NV2from infected cells (different preparation fromthat inFig.2C);A, viral peptides.(D) Mixture ofNVJ andNVIJ½;0,peptidesof NVJ; E.
peptides ofNV1½h. (E)MixtureofNV5(17,000cpm) and NV2(14,000 cpm);samepreparationasin (C); NV2
peptidesaremarked. (F)MixtureofNV5(17,000 cpm) and NVJ plusNV½1h (14,000cpm);samepreparation asin(D);peptidesmarkedasin(D).
669
S
A
NV5
B NV5+NV222
.
c
NV2
-4-g
S
E NYV5 NV2
C
*ra
d
V
010
on November 10, 2019 by guest
http://jvi.asm.org/
670 WRIGHT AND WESTAWAY
NV1 (Fig. 6F). Because at least threepeptides
of NV1W? were identified in a mixed run with
NV5 (Fig. 6F), we tentatively conclude that
there isnorelationshipbetween these two
pro-teins. The mapsofV1 (Fig. 2E) and NV5 (Fig.
6A) have peptides in similar positions at the
right of the top marker. Thus, NV5 isapossible
precursorof V1.
DISCUSSION
ThepeptidemapsofNV2i4, NV2,NV1W,,NV1,
and V1 demonstrate thatnoneofthese
polypep-tides was derived from another by proteolytic
cleavage. However,NV1 and NV2weredelayed
in appearance during pulse-chase experiments
(16a), and evidence has been obtained that
NV1i2 and NV2/2 are products of
post-transla-tional modification of newly synthesized
pro-teins in infectedcells synchronized by removal
ofahypertonicblock(14a). V1is found invirions
and SHA butnot in infected cells(7, 13).
There-fore,toobtain more informationonthe
forma-tion ofNV2Y2, NV2, NV1i/2, NV1, and V1, the
peptidemaps of these proteinswere compared
with thoseofthelarger virus-specified proteins,
NV5, NV4, and V3, eachofwhich hasaunique
peptidemapandsoisindependently coded (17).
The results of the mapping experiments
pre-sented in thispaper aresummarized inTable2.
The overall conclusion is that the amino acid
sequencesof theKunjin virus-specified proteins,
NV5, NV4, V3, NV2, and NV1i2, are unique.
These proteins, with a total molecularweight
of250,000 (13), account for 60% ofthe coding
potential of the Kunjin virus genome of 4.2 X
106 daltons (1). The peptides of NV21/2, NV1,
andV1, possibly representing afurther 10% of
the total coding capacity,were not adequately
resolvedfromthoseof V3 (for NV2Y2, NV1, and
V1) or NV5 and NV4 (for V1) to rigorously
excludeNV2/2, NV1, and V1asproducts of
post-translationalcleavage.
Since thepeptides of NV2i4, NV1, and V1 do
not overlap and arenot resolved from those of
V3, itmaybe thatNV2YI, NV1, and V1 are all
derived by cleavage of V3. However,in infected
cells treated with proteolysis inhibitors,the
for-mation of NV2½2 andNV1isnotinhibited(16a).
During a 1- or 2-min pulse of [35S]methionine
with cells reinitiatedintranslationafter reversal
of a hypertonic block, label was incorporated
intoV3, NV2½2, andNV1,withsimilaramounts
of label being incorporated into NV2'/2 and NV1
(14a). If NV2Y2 and NV1 are derived from one precursor,thepolypeptide arisingnearerthe
N-terminal end of thatprecursor should be more
heavily labeled. Further evidence on the
sug-gested cleavages from V3 comes from
experi-ments with puromycin. The presence of
puro-mycin (160
tg/ml)
inhibits the production ofNV212, NV1, and infectious virus (V1 is found onlyin virionsorSHA) despite ongoing
synthe-sis of V3 (10, 16a). After removal of the
puro-mycin, infectious virus is again produced but
does not incorporate V3 labeled with amino
acids during the inhibition period (16a). These
findingsmaybeinterpreted inatleasttwoways.
They are compatible with the suggestion that,
undernormalconditions,someV3moleculesare
cleaved to NV2Y2, NV1, and V1, but that the
cleavage is inhibited in the presence of
puro-mycin, perhaps becausearequired unstable
pro-tein is not made. Because of the pulse-label
incorporationinreinitiationexperiments and the
lack of effect ofproteolysis inhibitors,wefavor
the notion that NV2Y2 and NV1 synthesis is
unrelated to that of V3 and that puromycin
affects V3 incorporation into virions in some
othermanner.Toconvincingly demonstrate the
presenceorabsence ofanyrelationship between
V3 andNV21/2, NV1, orV1, it will benecessary
tofurtheranalyze the tryptic peptides of these
proteins. Peptides must be labeled with amino
acids other than methionine and separated by
differenttechniques.
Shapiroetal. (8) proposedthat NV2 is cleaved
to produce V1, based on the presence of NV2
in particles (T forms) released from LLC-MK2
cellsmaintained inmedium containing Tris and
infected withdengue-2orJapanese encephalitis
TABLE2. Uniqueness of Kunjinvirus-specifiedproteins smaller than V3
Possible Excludedasproducts
precursor NV2½/2 NV2 NV1½/2(V2) NV1 V1
NV5a Yes,6Bb Yes,6E Yes, 6F Yes, 6F Uncertain, 6A, 2E
NV4 Yes,5C Yes,5D Yes,5E Yes, 5F Uncertain, 5B, 2E
V3 Uncertain,'4E Yes,4B Yes, 5A Uncertain,d3B, 3E, 3F Uncertain, 3B, 2E NV5,NV4,and V3 are unrelated to each other (17).
Source(figure)ofdata.
Peptide mappingwasinconclusive.However,puromycin (160 ,ug/ml) inhibits the synthesis of NV5butnot that ofNV1 /2 (10, 16a).
dPeptide mappingwasinconclusive.Puromycin (160 ,ug/ml) inhibits the synthesis ofNV2'/2andNV1 but not that of V3
(10,16a).
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
PEPTIDES OF KUNJIN PROTEINS. II. 671
virus,andonthepresence of NV2 in intracellular
virus (Iforms)obtained from chick cells infected
with Japaneseencephalitisvirus. In our
experi-ments, we compared the peptide maps of [3S]
methionine-labeled tryptic peptides of V1
de-rived fromKunjinvirusand fromSHA with the map of NV2 derived from infected cells. The two majorlabeled peptides of V1 are not
con-tained inNV2, indicating that V1 isnotderived from NV2 by proteolytic cleavage unless the
amino acidsequenceofV1 isnotNorC terminal in NV2 but is internal and includes only one
tryptic cleavage site. To rigorously exclude a
relationship between NV2 and V1, it may be
necessary to examine tryptic peptides of NV2
fromTforms,Iforms, and SHA becauseof the
considerable heterogeneity inmigration through
gels ofNV2 (2,9,16a). We have been unableto
obtain sufficient NV2 from SHA for peptide mapping, since the amount of this protein in
the
particle
issmallandvariable(7, 12, 14).In further search of a precursor of V1, we
compared the peptide mapsofNV5, NV4, and V3withthemapof V1.Quite obviously, V3 and V1 contain
peptides
migrating
to similarposi-tions. However,
defining
a precursor of V1 isnot easy because V1 has
only
twomajor [3S]methionine-containing peptides. The
larger
pro-teins, NV5, NV4, and V3, have
fairly complex
peptidemaps,increasing the
probability
of find-ingby chancespotsin thesameareaofthemapas one of the two V1
peptides,
but the spotsmay not
necessarily
representpeptides
of thesame structureas those ofV1. IfV1 is derived
from one of NV5, NV4, or V3
by proteolytic
cleavage, then the other
products
of thecleavage
havenotbeenidentifiedininfected cells; NV5, NV4, and V3 are stable in
pulse-chase
experi-ments
(16a).
IfV1 is cleaved fromV3, then V1isderived from V3 molecules other than those foundin
purified
virus, since thereis noloss of the "V1peptides"
from these molecules. If V1 is derived from NV5 or NV4, then the larger protein is almostcertainly
multifunctional. It has beensuggested
that NV5 and NV4 maybe polymeraseproteins
(14a). The existence of sucha
large
stableprotein
only
as aprecursor ofV1would beout of
keeping
with thegenetic econ-omy of the virus. In summary, although the tryptic peptide maps are consistent with a hy-pothesis that NV5 or NV4 or V3 could be aprecursor ofV1, no other supporting evidence
isavailable.
Clearly,
NV1W,
and V2 are relatedproteins,
the amino acid sequence of
NV1YW being
con-tainedinV2.Theextrapeptidein V2is
consist-entwith itslargersizebasedonits
migration
inacrylamide
gels
(15). Theonly
feasibleprecursorof
NV1Y, (and
V2) from themapping
experi-ments is NV5; however, because treatment of
infected cells with puromycin (160,g/ml)
in-hibits the synthesis ofNV5 but notthat of
NVW1Y,
(10,16a), we conclude that
NV1½
isnot derivedbycleavageof NV5.
Thus, theonlytwopost-translational cleavage
productssuggested by the tryptic peptide maps are
NV1W2
(from V2) and V1 (fromNV5, NV4, orV3).Thepossibility thatNV2Y;
andNV1arederived by cleavage of V3 also arises from the
peptide maps; however, this possibility arises
mainly
because oflack of peptide resolution inheavilylabeled areasof themaps, andthere is
considerable other evidence implyingthat such acleavage does not occur (see above). In
con-trast totheseexperiments with
flavivirus-speci-fiedproteins, trypticpeptide mapshave readily demonstrated precursor-product relationships between proteins specified by other positive-strand viruses, namely, the picornaviruses and alphaviruses. For example, post-translational cleavage of viral proteins has been confirmed
by
peptide mapping
forpoliovirus
(4), SemlikiForestvirus(11), and Sindbis virus
(6).
Peptide
mapsof
Kunjin
virus-specified proteins
demon-strate alack of
precursor-product relationships
among them
(with
thepossible exceptions
notedin Table 2); they are consistent with the
pro-posal,
based onexperiments
with inhibitors ofproteolytic
cleavage
and onexperiments
using pactamycinorhypertonic
saltto controltrans-lationevents, that
during
thereplication
of fla-viviruses thetranslation of themajority
of viral polypeptidesisinitiatedinternally
onviral RNAasthe
single
template (14a).ACKNOWLEDGMENT
This work wassupported by a grant from the National
Healthand Medical Research Council of Australia.
LITERATURE CITED
1. Boulton,R.W.,and E.G.Westaway.1972.
Compari-sonsoftogaviruses:Sindbis virus(group A)andKunjin
virus(groupB). Virology49:283-289.
2. Boulton,R.W.,and E.G.Westaway.1976.Replication
of the flavivirus Kunjin: proteins, glycoproteins and maturation associated with cell membranes.Virology
69:416-430.
3. Hershko,A.,and M.Fry.1975.Post-translational cleav-age ofpolypeptidechains:role inassembly.Annu.Rev. Biochem. 44:775-797.
4. Jacobson, M. F., J. Asso, and D. Baltimore. 1970. Furtherevidence on the formation ofpoliovirus pro-teins.J. Mol. Biol.49:657-669.
5. Laemmli, U. K. 1970. Cleavage of structural proteins duringtheassemblyof the head ofbacteriophageT4. Nature(London)227:680-685.
6. Schlesinger, M.J., and S. Schlesinger. 1973. Large-molecular-weightprecursors of Sindbis virusproteins.
J. Virol.11:1013-1016.
7. Shapiro, D.,W. E.Brandt, R. D.Cardiff,and P. K. Russell. 1971. TheproteinsofJapanese encephalitis
virus.Virology44:108-124.
8. Shapiro, D.,W. E.Brandt,and P. K. Russell. 1972. VOL. 24,1977
on November 10, 2019 by guest
http://jvi.asm.org/
672 WRIGHT AND WESTAWAY
Change involvingaviralmembraneglycoprotein during morphogenesis of group B arboviruses. Virology 50:906-911.
9. Shapiro, D.,K. A.Kos,and P. K.Russell.1973.
Japa-neseencephalitis virusglycoproteins. Virology56:88-94.
10. Shapiro, D.,K. A. Kos,andP.K.Russell. 1973. Protein synthesisinJapanese encephalitis virus-infected cells Virology 56:95-109.
11. Simons, K., S. Keranen, and L. Kaariainen. 1973. Identification ofa precursor for one of the Semliki forestvirus membraneproteins.FEBSLett.29:87-91. 12. Stollar,V. 1969. Studiesonthe nature ofdengue viruses. IV. The structural proteins oftype 2 dengue virus. Virology39:426-438.
13.Westaway, E. G. 1973. Proteins specified by group B
togavirusesinmammaliancellsduringproductive infec-tions. Virology51:454-465.
14. Westaway,E.G.1975.The proteins ofMurrayValley encephalitisvirus.J. Gen. Virol. 27:283-292.
14a.Westaway, E.G.1977.Strategyof the flavivirusgenome:
evidenceformultiple internal initiationoftranslation of proteins specified by Kunjin virus. Virology 80:320-335.
15. Westaway,E.G.,J.L.McKimm,and L.G. McLeod. 1977. Heterogeneity among flavivirus proteins
sepa-ratedinslabgels.Arch. Virol.53:305-312.
16.Westaway,E.G.,and B.M.Reedman.1969.Proteins of thegroupBarbovirusKunjin.J.Virol. 4:688-693. 16a.Westaway. E. G., and M.Shew. 1977. Proteins and
glycoproteins specified bytheflavivrusKunjin.
Virol-ogy80:309-319.
17. Wright,P. J.,D. S.Bowden, andE. G.Westaway. 1977.Uniquepeptidemapsof thethree largestproteins
specified bytheflavivirusKunjin.J.Virol. 24:651-661. 18. Wright, P.J.,andG. diMayorca. 1975.Virion
poly-peptide composition of the human papovavirus BK: comparisonwithsimianvirus40andpolyomavirus.J. Virol. 15:828-835.
J. VIROL.