JOURNAL OF VIROLOGY, Dec. 1980,p.639-651
0022-538X/80/12-0639/13$02.00/0 Vol.36, No. 3
Mapping of Adenovirus 12 mRNA's Transcribed from the
Transforming Region
YUKIHARU SAWADA AND KEI FUJINAGA*
Department ofMolecular Biology, Cancer Research Institute, Sapporo Medical College, Sapporo060,
Japan
Adenovirus12mRNA's transcribedfromthetransformingregion were analyzed and mapped on viral DNA by the nuclease
Si
gel and diazobenzyloxymethyl paperblot techniques in cells transformedby the EcoRI-C (leftmost 16.5%) andHindmI-G(leftmost 6.8%) fragments ofadenovirus 12 DNA and in cells early and late after lytic infection with adenovirus 12. Two initiation sites of mRNA transcription andtwokinds of splicingwerefound in each of early regions1Aand 1B. For early region 1A mRNA's, four species were found in lytically infected cells.Three of themwerecommonly foundincellstransformed by either HindIII-G or EcoRI-C. Cells transformed by HindllI-G contained two additional 1A transcripts, which could be the 3' portions ofchimericmRNA's ofcellular-viralor viral-viralsequences. Transcriptionin the 1Bregiondiverged among the above cell lines. Early afterlytic infection, noappreciable amount of 1B mRNA was detected, whereas two species of mRNA's, one corresponding to protein IX mRNA and another having splicing, were found at the late stage. In cells transformed by EcoRI-C, adistinct mRNA species with splicing was observed. Cellstransforned by HindIII-Gcontainedatranscript from the leftmostpartof the 1B sequence at the 5' portion of chimeric mRNA species, suggesting the presenceof tandemintegration of viral DNA in thecells. OthermRNA species in the 1A and 1B regions were also detected in both transformed cell lines. The results are discussed in relation to the nucleotide sequence of the HindIII-G
fragment of adenovirus12DNA.
Thetransforming ability of human adenovirus has beenlocalized in the leftmost7 to8% of the viralgenomeby cell transformation studies with restriction DNAfragmentsof human adenovirus 2 (Ad2), Ad5, Ad7, and Adl2 (23, 25, 29). Cell transformationexperiments with smaller DNA
fragmentshave revealedaminimumsizefor the
transformingfragments;the leftmostfragments AccI-H (BpaI-H; 4.7%, Adl2) (26), BglII-H
(4.5%, Ad7) (8), andHpaI-E (4.3%, Ad5) (A.J. Van der Eb, H. VanOrmondt, P. I. Schrier, J. H.Lupker,H.Jochemson, P. J. Van den Elsen, R. J. Deleys, J. Maat, C. P. Van Beveren, R. Dikjema, andA.DeWaard,ColdSpringHarbor
Symp. Quant.Biol.,inpress)stillcantransform
cells,althoughthe resultant cell lines show
in-complete transformation phenotypes. In the earlystage oflytic infection, before viral DNA replication, at leastfour regions ofviral DNA
are transcribed (18, 24). The leftmost region (11.5% of the viral DNA), which contains the entiretransforming ability, is dividedinto two
regions: 1A (0 toabout 4.5 map units) and 1B (4.5to 11.5mapunits),withspecificpromoters (31).Thus, region1A contains theinformation for the basic conversion of cells into a
trans-formed cellline,butregion1B codes forproteins
which alsoplay important roles in cell transfor-mation (26; Van der Ebetal.,in press).On the other hand, characterization ofthe host range deletion mutantsofAd5has offered inferences into functions encoded in the transforming re-gion; region 1Aproduct(s)regulates expression
ofotherearlyviralgenes(2,13).
AnalysisofearlymRNA'sfromearly region 1 of Ad2 andAd5bythenuclease S1gel technique
(2-4) showed that at least two mRNA's were
coded inregion 1A and thatone was coded in region 1B, allwithspliced structures. Electron
microscopicstudies identifiedadditional mRNA
specieswithdifferentsplicings (6).Adl2(17, 27)
hasessentiallythesametopography (9) asAd2 (7, 18,24), but no detailedmapping of mRNA
species has been available. In this paper, we
describethemappingofregion1mRNA's
tran-scribed in Adl2-infected cells and in cell lines transfornedbytheAdl2EcoRI-C(theleftmost 16.5%)andHindIII-G (theleftmost6.8%ofviral DNA)fragments.Anotable differencefrom Ad2 (4), Ad5 (2), and Ad7 (33) wasobserved upon expression of 1B mRNA's in cells early after lytic infection.Intransformed celllines,several unique mRNA's which were not detected in productively infected cells were found. In the 639
on November 10, 2019 by guest
http://jvi.asm.org/
640 SAWADA AND FUJINAGA
nucleotide sequence of theHindIII-G fragment ofAdl2 (K.Fujinaga,Y.Sawada,Y.Uemizu,T. Yamashita,H.Shimojo,K.Shiroki,H.Sugisaki, K. Sugimoto, and M. Takanami, Cold Spring HarborSymp.Quant.Biol.,inpress;H.Sugisaki, K.Suginoto,M.Takanami,K.Shiroki,I. Saito, H. Shimnojo, Y.Sawada, Y. Uemizu,S. Uesugi, and K. Fujinaga, Cell, in press), specific se-quences were found at the sites of initiation, termination, and splicing of RNAs mapped in thisstudy.
MATERIALS AND METHODS
Cells and virus.TransformedcelllinesCY1-1and GY1-1 used in this study were established from cell line 3Y1(establishedfrom a Fischer rat embryo) trans-fected with the EcoRI-C andHindIII-Gfragmentsof Adl2 DNA,respectively, as described previously (25, 32).Cells were grown in monolayers in Eagle minimum essential medium containing 0.1 mM Ca2" supple-mentedwith 10% fetal bovine serum (GIBCO Labo-ratories). Adl2(Huie)waspropagated in KBcells,and viral DNA was isolated from purified virions as de-scribed (11, 12).
Enzymes.HindIIIwaspurifiedin ourlaboratory.
AccI and HaeIII weregifts from MituruTakanami,
KyotoUniversity, Kyoto, Japan. EcoRI,Escherichia coli DNA polymerase I, and nuclease Si were pur-chased from Boehringer Mannheim Corp. Pancreatic DNase Iwas aproduct ofWorthington Biochemicals
Corp.
Preparation of 32P-labeled restriction frag-mentsofAdl2.32P-labeledvirus was prepared from
infected cellslabeled with 100 ,Ci of
'Pi
per ml inphosphate-free minimum essential medium supple-mented with 2%dialyzed calfserumfor 48 to 72 h at
37°C.Viral DNA hadaspecificactivityof1 x 106to 2 X 106 cpm/,ug and was digested with restriction
endonuclease.DNAfragmentswereseparated by 1.4% agarosegel electrophoresisandpurified as described
previously(23).Highlyradioactive DNA fragments (5 x 107to2x108cpm/4ug) wereprepared with
[a-32P]_
dATP and[a-32P]dCTPby the nick translation pro-cedure aspreviously described (21).
Isolation ofcytoplasmicRNA. KB cells in
mono-layerculture were infected withAdl2at amultiplicity
ofapproximately20PFU per cell.Cells were harvested at8h(withoutcytosinearabinoside)or12h(with 20
,tg
ofcytosine arabinoside perml)afterinfection for preparation ofAdl2 early RNA, and at 23 h after infection for preparation ofAdl2delayed early RNA. The cytoplasmic fractions ofGYl-1, CY1-1,and in-fected KB cells were prepared by hypotonic Nonidet P-40lysisinthe presence ofN-ethylmaleimide, sperm-ine, anddiethylpyrocarbonatebythemethod of Zieve and Penman(34). Thecytoplasmic fraction was made 0.5% in sodiumdodecyl sulfate, 10 mM in EDTA, and 0.1 M in NaCl and extracted by phenol andchloro-form. RNA in the aqueous phase wasprecipitated by adding 2 volumes of ethanol and rinsing with 70% ethanol. Polyadenylic acid-containing RNA was se-lected byoligodeoxythymidylicacid-cellulose
(Collab-orativeResearch, Inc.)column chromatography. NucleaseSIgelanalysis.Hybridizationsfor
nu-J. VIROL. clease Slgel analysiswerecarried out inhybridization
buffercontaining80%formamide,0.4 MNaCl, 0.04 M PIPES [piperazine-N,N'-bis(2-ethanesulfonic acid)] (pH 6.4),and0.001 M EDTA (5), essentially bythe method ofBerk andSharp(3)aspreviouslydescribed (33).Themeltingtemperature(Tm)irreversible in this solventwas46to47°Casdeterminedbysusceptibility
ofthe Adl2 EcoRI-C fragment to nuclease S1 (Y.
Sawada, unpublished data). Hybridizations were
therefore carried outat53°C,6 to7°Chigherthan the
Tm ofAdl2 EcoRI-C. RNA-DNA hybrids were
di-gestedbynucleaseS1 (30)asdescribedpreviously (3, 33)withslightmodifications.
Alkalinegel electrophoresis (15)wascarried out in 1.4% agarose slabgels(20cminlength)at40 V for 14 h.Gelswereneutralizedby soakingin0.05 M Tris(pH 7.2)for 30min, dried,andautoradiographedat-70°C with Kodak XR-1orXRP-1 film and Du Pont
tung-state intensifying screens. Lengths innucleotides of
cotranscriptswereobtained in alkalinegels.In neutral
gels, slightly largervalueswereobserved forrespective cotranscriptsthanwereobserved in alkaline gels(4).
Two-dimensionalelectrophoresis(8a)wascarried out
as follows. After afirst electrophoresis ofa neutral agarose slab gel, the geltrack containing respective
RNA-DNAhybridswascut outandequilibratedfor alkaline electrophoresisbuffer. Thegel stripwasset
ontopofasecondalkaline agarose slabgel and
elec-trophoresed.
Preparation ofdiazobenzyloxymethyl paper. 1-[(m-Nitrobenzyloxy)methyl]pyridinium chloride
wassynthesized essentially by the procedure previ-ouslydescribed (1). (m-Nitrobenzyloxy)methyl chlo-ride(yellow liquidwithaboilingpoint of139 to1440C at2mmofHg)wasmadefromparaformaldehyde,
m-nitrobenzyl alcohol,anddryHCI inbenzene andwas added slowly to ice-cold pyridine by stirring. The
crystalline pyridiniumsaltwascollectedon asintered
glassfilter,washed withpyridine,washed with petro-leumether,anddried under reducedpressure. The 1-[(m-nitrobenzyloxy)methyl]pyridinium chloride was stored at -20°C. Theaminobenzyloxymethyl paper derived fromasheet ofToyo514paper(14 by 20cm) wasconvertedtodiazobenzyloxymethyl(DBM) paper
bytreatmentwithafreshlyprepared solution
contain-ing40 mlofwater, 80 ml of 1.8 MHCI,and 3.2ml of
NaNO2(10 mg/ml)for 30minat0to4°C. The DBM paperwaswashed five times for 5 min each time with coldwater and thentwice for 10 min with ice-cold sodiumphosphatebuffer(25mM, pH 6.8); it was used within15minafter the finalwash.
DBM paper blotanalysis.Polyadenylic
acid-con-tainingRNAsweredenatured with1Mglyoxal in 50%
dimethylsulfate and6.7mMsodium phosphate (pH 6.8) for 60 minat500C aspreviously described (16). RestrictionfragmentsofAdl2 DNA for size markers werealsoglyoxalated. After addition of tracking dye
containing bromophenolblue andglycerol, denatured
sampleswereelectrophoresedona1.5% agarose slab
gelin10mMsodiumphosphate buffer (pH 6.8). After electrophoresis, the gelwasimmersed in 50 mM NaOH containing ethidium bromide (0.5 ,ug/ml) for 40 minto 1hto dissociate the RNA-glyoxal compound andto staintheRNA bands. The gel was thenneutralized by
washingtwice in 100mMphosphate buffer (pH 6.8) for 20 min, followed bytwowashes of25mM
on November 10, 2019 by guest
http://jvi.asm.org/
Adl2 mRNA's FROM THE TRANSFORMING REGION 641 phate buffer for 5 min each. The RNA was transferred
by blotting tothe DBM paper. The RNA blot was treated overnight at420C with hybridization buffer (50% formamide; 0.75 M sodium chloride; 75 mM sodium citrate; 0.02% [wt/vol] each of bovine serum
albumin,Ficoll, and polyvinylpyrrolidone;and 1.5 mg of sonicated denatured calf thymus DNA per ml) containing 1% (wt/vol) glycine (1). Hybridizations were carried out for 20 to 40 h at420Cinplastic bags containing hybridization buffer (50 to 100
Ail/cm2
of paper surface area) and the single-stranded 3P-labeled probe (3x 10Wto 5 x 104cpm/cm2of papersurface area). The paper was washed once in a plastic bag at420C and then again on a suction funnel at room temperature with asolution containing 50% formam-ide, 0.75 Msodiumchloride, and 75 mMsodiunrcitrate
and finallywith asolution containing 0.3 M sodium chloride and 30 mM sodium citrate. The paper strips wereblotted to remove excess solution and air dried, andthe RNAs were made visible by autoradiography asdescribed above.
RESULTS
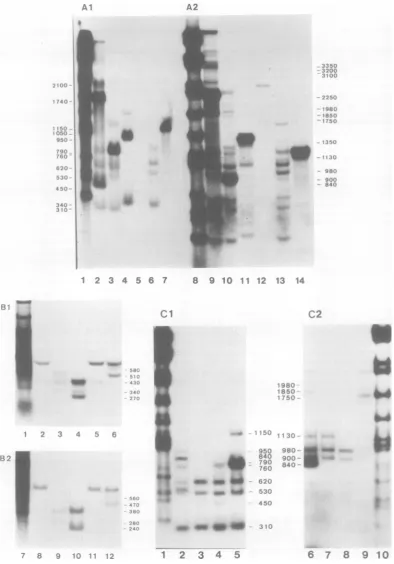
MappingofmRNA's in the CYl-1cell line
bythe nucleaseSi gel technique. The early region 1 sequences (0.9 to 11.2 map units [see below]) of Adl2 DNAare fully included in the EcoRI-Cfragment (0to 16.5mapunits). CYl-1 cells contain the EcoRI-C fragment of Adl2 DNAat 5to 6copiesperhaploidcell (21). Four
majorand three minorvirus-specificmRNA spe-ciesweredetected inCYl-1 cytoplasm, and all sevenmRNA's hadsplicedstructures (Fig. 1E). Averydense bandatabout2,250nucleotidesin aneutral gelwas observed after hybridization with the EcoRI-C fragment (Fig. 1A, lane 9). This apparently dissociated into two bands at 450and1,740nucleotides inanalkaline gel (Fig. 1A, lane 2). Portions of this long mRNAwere detected as bands by using the HindIII-G,
HindIII-I, AccI-J, and AccI-F fragments as probes (Fig. 1A). These bandswere assigned in
region 1B as shown inFig. 1D. Thus, the only 1B mRNAin CYl-1cells has its3'endat posi-tion 3,820 and 5' endatposition 1,530,
approxi-mately.It hasthesplicedstructure,having
splic-ing pointsapproximatelyatpositions 3,270 and 3,370.
In region 1A, three mRNA's were found as major species (Fig. 1E). Theseweredetectedas bands in common with thefragmentsEcoRI-C, HindIII-G (0 to6.8map units), and AccI-H (0
to4.7mapunits) (Fig. 1A). Each of them had a
splicedstructure(760plus310, 620plus310, and 530plus310nucleotides inlength)asidentified bytwo-dimensionalanalysis (seeFig. 4A).These 1A mRNA's were mapped in detail
by
using
small DNA fragments produced byrecleavage
of AccI-H (positions 0 to 1,596) withHphI:
HphI-A (positions0 to692),-BD
(positions
692to 1,214), and a mixture of -B (positions 692 to 1,105) plus -EC (positions 1,214 to 1,596) (Fig. 1D).
A band appearing at about 500 nucleotides with the B fragment in a neutral gel almost disappeared inanalkaline gel (Fig. 1B, lanes 3 and9),whereas other bands didnotalter. This indicates that allsplicing of the three mRNA's in the region of0 to 4.7mapunitsoccursaround position 1,000. As two bands at 380 and 280 nucleotideswereobserved with theBDfragment aswell aswith theprobecontaining the Bplus EC fragments in alkaline gel (Fig. 1B, lanes 9 and 10), there should be two donor sites for splicing approximately at positions 970 and 1,070. One band at380nucleotides is observed with theAfragment, indicating thatatleastone mRNA starts approximately at position 310. These assignments areschematized inFig. 1D.
Hybridization with the AccI-H/HaeIII-A fragment (positions442 to 1,596)producedonly
twobandsatabout620and530nucleotides(Fig.
1C, lane 3). Thisclearly showed that thereare twopromotersites inregion 1Aof Adl2DNA. Theforegoingoneshouldstarttranscription ap-proximately atposition 310 as mapped above. The relativedensity of the bandat620 nucleo-tides as comparedwith thatat530 nucleotides inlane3 (with theAccI-H/HaeIII-A fragment)
ishigher than in lane4 (withthe AccI-H
frag-ment). Thus, the 760-nucleotide-long cotran-scriptgivesabandatabout620nucleotides after hybridization with the Afragment, resujtingin adoublet band with another
620-nucleotide-long
cotranscript. As the donorsites forsplicingare mapped approximately at positions 970 and 1,070 asabove,the530-and620-nucleotide-long
cotranscripts start approximately at position
450, very near the HaeIII site on the AccI-H fragment(Fig. 1D).
Three minorspecieswerealso found and iden-tifiedaslongmRNA's with thesame5'endsas
1AmRNA's and thesame3' endas1B mRNA's (Fig. 1E).Theycontained the 2,130-nucleotide-longcotranscripts,whose leftportionswere de-tectedasbandsat450nucleotides with the AccI-Hfragment (Fig. lAl and 1C, faintbands) and at 1,150 nucleotides with the HindIII-G
frag-ment (Fig. lAl) in alkaline gels. Two-dimen-sional analysis using the AccI-H fragment
re-vealed,afterextensiveexposure, minor dots
cor-responding to 670-, 310-, 760-, 620-, 530-, and three450-nucleotide-longcotranscripts.Eachof the 760-, 620-, and 530-nucleotide-long
cotran-scripts was spliced togetherwith a 450-nucleo-tide-longcotranscript,consistentwith the above assignmentof minor bands. ThesplicedmRNA
of 670- and
310-nucleotide-long
cotranscripts
wasmappedatits 5' and 3' endsatthecommon VOL. 36, 1980
on November 10, 2019 by guest
http://jvi.asm.org/
4.
xF-r'f.1
.0 .<.
d
0
0
W.K...
S8
__
U
U
,.
l-&
U
..
.-
*--er
340
..
1,
bel"_
iK- _
2343
FIG. 1. Autoradiograms ofnuclease SI-digested RNA-DNA hybrids of CY1-1 cytoplasmic RNA and diagramsofthe deduced RNAstructures.Alkaline(Al)and neutral(A2)agarosegelsofnucleaseSi-digested CY1-1 RNArestrictionfragment hybrids.RestrictionfragmentsusedareEcoRI-C(lanes2and9),HindIII-G (lanes3and10),HindIII-I(lanes4and11),HindIII-F(lanes5and12),AccI-H(lanes6and13),and AccI-J (lanes 7and14). The reannealedfragmentcanbeseenineach lane. Lanes1and8aretheHindIIImarker digestsofAdl2DNA. Thelengths(in kilobases) oftheAd12HindIIIrestrictionfragmentsare:A,5.40; B, 4.75;C, 3.70; D,3.66(CandDcomigrate);E, 3.30;F,2.51; G,2.32;H,1.80;I, 1.39;J,1.10;K,1.03;L,0.98;M,
0.68;N, 0.54;0,0.48;andP,0.36.SinceexcessreactionofnucleaseSiin theseexperimentscutthrough RNA-DNAhybridsatsplicing points, severalbandscorrespondingtoeachcotranscript of splicedRNAsarealso seenintheautoradiogramofthe neutralgel (for example,the bandsatabout1,200 nucleotidesin lane 10 and atabout1,000nucleotides in lane 11).Neutral (B1)and alkaline(B2) agarosegels ofnucleaseS1-digested
642
40'I"
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.496.65.460.29.594.2]D
1 A 450 980 1140 1450
0///7/ 280 10 2
310
310~~~
V////,///:l 380 2A402
380 1 380
A BD EC AccI-H/hphI
0 A1596
EIB ]1214
692 1105
530 10706 10
310 620 10
V//A 620 3
A AccI-H/HaeIII
442 1596
1 B
1530 3270 3370 3820
L 790|S- o"S}yv l ,ln5Zu
S G I I _ FV,
A A
2320 3710
HindIII
E
530 " O
620
~310_
760 1740 450
620 2130 450
760 A 2130 450
0 2
i3
4Kilo
bases0 4 6 10
Map
unitsA
IDE
CB I A
H
Accl-H/Hphl Accl-H/HaelIl
J I I, F Acci
G I ,F
Hindill
C EcoRI
hybrids of HphIrestrictionfragmentsandcytoplasmicRNAsfrom CY1-1 (lanes2,3, and 4), GY1-1(lane 6) andinfected earlycells(lane 5).Restrictionfragments usedareAccI-H/HphI-A (lanes 2, 5, 6,8, 11, and12), AccI-H/HphI-BD(lanes3and9)andAccI-H/HphI-Bplus EC (lanes4and10) (see Fig. ID and IE for the
physical map).Lanes 1and7 aretheHindIII markerdigestsofAdl2 DNA. Alkaline(Cl) and neutral (C2) agarosegels ofnuclease Si-digested hybrids ofrestriction fragments andpolyadenylic acid-containing
cytoplasmicRNAsfromCY1-1 and GY1-1 cells. RestrictionfragmentsusedareHindIII-G(lanes2, 5,6, and 9),AccI-H(lanes4and7),andAccI-H/HaeIII-A (lanes3and8) (seeFig. IDforthephysical map). Lanes1 and10 arethe HindIII markerdigestsofAdl2 DNA. (D) Assignment of main bands detected in CYI-1 cells
bythe nucleaseSlgel procedure.Detectedportions ofeachcotranscriptarerepresented by open boxes with numbers in nucleotidesof correspondingbandlengths.Hatched boxes representportionsnotidentified by the relevant restrictionfragments.Caretsymbols represent splicing, and the 3' endofanRNA is indicatedby an arrowhead. Numbers above verticalarrowsrepresent theassigned positionsof RNA initiation, termination andsplicingsites.Restrictionfragmentsrelevanttotheassignmentsarerepresentedbelow the RNAstructure. (E) Diagrams ofthestructureofAdl2-specificRNAsinCY1-1cytoplasmsdeducedfrom the data in (A), (B), and(C). Viral sequences present inanRNAarerepresented byaline above thecorresponding region of the
Adl2genomemap. Thegenome map is markedoffin kilobases and map unitsfromtheleft end. Caret symbols representsplicingofcolineartranscripts intooneRNAmolecule,and the 3' endofanRNA is indicatedbyan arrowhead. The more abundant cytoplasmic RNAs are represented by heavy lines, and less abundant
cytoplasmicRNAsarerepresented bythin lines. Numbers above the lines represent thecotranscript lengths (in nucleotides). Restriction maps relevanttotheanalysisarerepresentedbelow thegenome map.
643
on November 10, 2019 by guest
http://jvi.asm.org/
644 SAWADA AND FUJINAGA
siteswiththesplicedoneof 760plus310 nucleo-tidesinlength (see Fig. 3D).
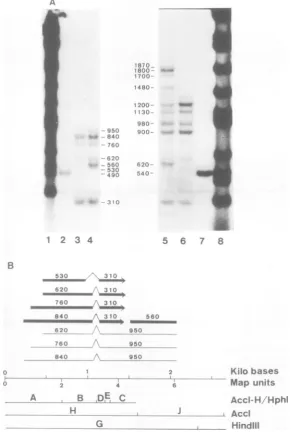
Mappingof mRNA's inGY1-1celllineby
nuclease Si gel technique. Three
major
1A mRNA's detectedinCY1-1cellswerealsotran-scribed in GY1-1cells. Anadditional 1Aspecies
wasfoundas abandat840nucleotides with the AccI-H and HindIII-Gfragmentsinalkalinegels
I
80C
950
840
6?C'
* 560
530
490C
200
SC0
98?
[image:6.496.107.400.169.602.2]C48 b.,2Cl
...'... .
.. 4
(Fig.2Aand 1C) anda bandat470nucleotides withtheAccI-H/HphI-A
fragment
(Fig. 1B,
lane 12). This longer cotranscript was alsospliced
together with the310-nucleotide-long
cotran-script, as wasresolved
by
two-dimensional,
nu-clease
Si
gel analysis (see Fig. 40). In the 1Bregion, onecotranscript
specific
toGY1-1 cells was mapped. After hybridization with the5 6
3I.;JLi 3 1O X... V 3 1°
____ __ 31
'60 3 10
840 310 560
,-: 9C55
.. I 950
4 6
E
D.E
CH
G
Kilo bases
Map units AccIt-H Hphl
..J Acc
HindllT
FIG. 2. Autoradiograms ofnuclease Sl-digested RNA-DNA hybrids of GY1-1 cytoplasmic RNA and
diagrams ofthe deduced RNAstructures.(A) Alkaline (left) andneutral (right)agarosegelsof nucleaseSl
-digestedGYI-IRNArestrictionfragment hybrids. RestrictionfragmentsusedareHindIII-G(lanes4and 5),
AccI-H(lanes3and6),andAccI-J (lanes2and 7). Lanes1 and 8aretheHindIIImarkerdigests of Adl2
DNA.(B)Diagrams ofthestructureofAdl2-specificRNAs inGYl-1cytoplasms deduced fromthe data in (A)
andFig.I(BandC). RepresentationisasinFig. 1.
Ai
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
Adl2 mRNA's FROM THE TRANSFORMING REGION 645 HindIII-G fragment, aband at 560 nucleotides
wasfoundin an alkaline gel (Fig. 2A, lane 4). Its right portionwasdetected with theAccI-J frag-ment as a band at 490 nucleotides. This 560-nucleotide-long cotranscript was not found in either CYl-1 or KB cells after infection with Adl2,and isconsideredaGY1-1-specific cotran-script.
Threeminor mRNA specieswere detected in GY1-1 cytoplasmic RNA after hybridization with theHindIII-G fragment. Thebandat950 nucleotidesinalkalinegels (Fig. 2A,lane 4, and Fig. 1C, lane2) isattributedtothe 3' portion of these minor species. These were mapped through the 1A and 1B regions (positions230 to 2,090, 310 to 2,090and450 to2,090)andhad the samesplicedstructure (positions1,070 to 1,140) as was determined for the major 1A mRNA's (Fig. 2B). There was another minor species whose 5'portionwasdetectedas abandatabout 560 nucleotides with the AccI-H/HphI-A frag-mentin an alkalinegel (Fig. 1B, lane 12). It is also considered to bea GY1-1-specific species, startingapproximatelyatposition120with the same spliced structure as other minor species (notshownin Fig. 2B).
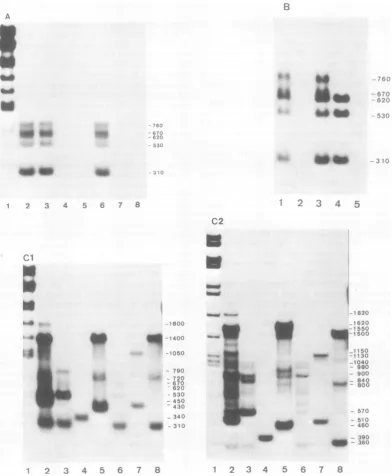
Mapping ofmRNA's inlytically infected
cells bynuclease Si gel technique. Incells
earlyafterlyticinfection withAdl2, four major mRNA species were found for the 1A region
whether
l-p-D-arabinofuranosylcytosine
was present or not (Fig. 3A and B). These were mapped in detail with the AccI-H/HphI frag-ments (showninpartinFig. 1B) and theAccI-H/HaeII
fragment (Fig.3B). Three of the fourmRNA'sof infectedearlycellscorrespondedto
the three 1AmRNA's transcribed inCY1-1 and GYl-l cells. The fourth startedatthesamesite asthelongestoneof the threeandsplicedasthe shortest (Fig. 3D). Their spliced structure was resolved bytwo-dimensional analysis (Fig.4B). IncontrastwithCYl-lorGYl-lcells,no appre-ciable bandwasobservedonautoradiogramsof
nuclease Si gel analysisfor 1B mRNA's from
infectedearlycells.Longspecies throughthe1A and1Bregionswere notfound,either.
Incells late afterlyticinfection withAdl2,all
of the four 1A mRNA's found inthe
early
stage were also detected (Fig. 30). In addition, twomRNA'swerefoundin the1Bregionatthe late stage. One was detected at 1,150 nucleotides withEcoRI-Cinaneutralgel (Fig. 3C2,lane2)
andat530nucleotideswithHindIII-Gand340
nucleotides with HindIII-I in an alkaline gel
(Fig. 3C1,lanes3and4).Thismappedat
posi-tions1,530to3,820andsplicedat
positions
2,060and3,370.Asecondwasdetectedat450 nucleo-tideswithEcoRI-C andat340nucleotideswith
HindIII-I in an alkaline
gel (Fig.
3C1, lanes 2and 4) and mappedatpositions 3,370 to 3,820. Data presentedin Fig. 3C1 andC2alsoshow at least four mRNA species comprising four cotranscripts of 1,400, 720, 150, and 80 nucleo-tides withtwodifferent splicings. These can be mapped in theregion which is apparently out-side early region1and correspondsto theregion onthe Istrandassigned for protein IVa2 in the case of Ad2 (14). A prominent band at 1,400 nucleotides was commonly found with the EcoRI-C, HindIII-F, and AccI-F fragments in alkaline gels (Fig.
301).
In neutral gels, two bands at 1,620 and 1,550 nucleotides were de-tected with the HindIII-F fragment, one was detectedat1,550nucleotideswith theEcoRI-C fragment,and one was detected at 1,500 nucleo-tides with the AccI-F fragment(Fig.3C2).Thus, there are twomajorspecies having the 5' ends atpositions approximately5,600 and 5,670, near the AccI (position 5,530) and theEcoRI (posi-tion 5,600) sites. They presumably havea com-mon 3' endnearposition 3,850 anda splice at positions 5,250 and 5,520. Two minor species were composed of the 720-nucleotide-long co-transcripts (presumably havingthe common 3' end as the 1,400-nucleotide-long cotranscripts) (data not shown). A band at 430 nucleotides found with the HindIII-Ffragment inan alka-linegel (Fig.3C, lane 5,correspondingto aband at 460 nucleotides in a neutral gel) can bemappedatthe
right-hand
end of thefragment.
It could be the 3' portion of mRNAspecies
corresponding to the early region 5 species of Ad5, where the ts36genehas beenmapped (10). Analysis of mRNA's byDBMpaper blot
hybridization. Theprocedure of Alwineetal. (1)wasappliedtocharacterize mRNA's. DBM papers carrying polyadenylic acid-containing
RNAs from CYl-1, GYl-l, and infected
early
cells were hybridized with 32P-labeled AccI-H and AccI-Jfragments
specific
tothe 1A and 1B regions, respectively (Fig. 5). In CYl-1 cells,mRNAspeciesof about3,200 and1,000to1,200 nucleotides inlength were found in region 1A. For the 1B region, mRNA's with lengths of about 3,200 and2,300 nucleotidesweredetected. Longspeciesof about3,200nucleotides inlength apparentlyseem tobetranscribed from the 1A
and 1B regions
contiguously.
Thiscorresponds
well with the three minorspecies detected by
nuclease
Si
gelanalysis. lA-specific species of 1,000to1,200 nucleotidesandlB-specificspeciesof2,300nucleotidesinlengthcomparewellwith thoseidentifiedbynucleaseSLgel
analysis.
InGYl-1 cells,however,mRNA specieswere
foundwith sizesunexpectedfrom the resultsof nuclease S1 gel analysis. Species with sizes of about3,600,2,500,1,900,1,700,and1,000to1,200
nucleotides were detected with the
32P-labeled
VOL. 36, 1980
on November 10, 2019 by guest
http://jvi.asm.org/
I
_
_0
sue.0
_n._
..4 '3
--l
-0
w
N
!=..40
.4
...Wt
4
...*
_p
C2
_N.w -# >.
1800.
--1400 -- _
'§0-1050 9 C.
670 _
620
530
450
430 _ _
:340
I
.. BC),. 62"C
-15D l.
I - 15r;ki
l., -t
_N 1i. 3 c
.104C
98C;
90C
84.-;,):f'
40 .3
aFf
[image:8.496.60.453.56.533.2]4 -6 7 8 12 34 5 -D 8
FIG. 3. Autoradiograms ofnucleaseSl-digestedRNA-DNAhybrids of early region1cytoplasmicRNA in
cellslytically infectedwithAdi2,anddiagrams ofthe deduced RNAstructures.(A)Alkalineagarosegel of
nucleaseSI-digested earlyRNA restrictionfragment hybrids.Adi2earlyRNAwasprepared from lytically
infectedKB cells incubatedinthepresenceof20pgofI-,8-D-arabinofuranosylcytosinepermlofculturefluid. RestrictionfragmentsusedareEcoRI-C(lane2),HindIII-G(lane 3),HindIII-I(lane4),HindIII-F(lane5),
AccI-H(lane 6),AccI-J(lane7),and AccI-F(lane 8).Lane1is theHindIIImarkerdigest ofAdi2 DNA.(B)
Alkalineagarosegel ofnucleaseSl-digested earlyRNA restrictionfragment hybrids. i-f3-D-Arabinofurano-sylcytosinewasnotpresentin thecourseofAdi2infection.RestrictionfragmentsusedareHindIII-G(lane 1),
HindIII-I(lane2),AccI-H(lane3),AccI-H/HaeIII-A(lane4),and AccI-J(lane 5). (ClandC2) Alkalineand neutralagarosegels, respectively, ofnucleaseSl-digested delayed earlyRNArestrictionfragment hybrids. CytoplasmicRNAwasprepared fromcells 23 hafter lytic infection.RestrictionfragmentsusedareEcoRI-C (lane 2),HindIII-G(lane 3),HindIII-I(lane 4),HindIII-F(lane 5),AccI-H(lane 6),AccI-J(lane7),and
AccI-F(lane 8) forbothCl and C2. Lane1inCl and C2 is theHindIII markerdigest ofAdl2 DNA. (D) Diagrams
ofthe deducedstructuresof early region1mRNA's in cellslytically infectedwith Adi2. The mRNA's detected
attheearlystagearerepresented byboldlines, andthose detectedonlyatthelatestagearerepresented by openboxes; representationis otherwise thesame asinFig.1.
646
f,f
_^
S
be
'
40w_
_. .a
I f- tt 6 -.' 7 8
on November 10, 2019 by guest
http://jvi.asm.org/
Adl2 mRNA's FROM THE TRANSFORMING REGION 647 AccI-H fragment,andoneof about1,700
nucleo-tides was detected with the AccI-J fragment (Fig. 5). Very large species (more than 8,000 nucleotides) were also found with both probes. The 1,000- to 1,200-nucleotide-long species cor-respondtothe threemajor 1AmRNA'smapped inGYl-l cells by nucleaseSigel analysis.There arethreeothermRNA's(3,600,2,500,and1,900 nucleotides inlength) specificto the 1A region of the viralgenome,but these havelarger sizes than expected for any 1A mRNA species ana-lyzedby nucleaseSigel analysis, suggesting that theyarechizneric mRNA's of cellular and viral sequences.These should contain the GYl-l-spe-cificspecies (840plus310nucleotides inlength)
asviral components.The 1,700-nucleotide-long species detected with both probes could com-prise atleasttwo mRNA's. One could start at position 450 by one of the 1Apromoters, read
contiguouslythrough the 1B region, and termi-nate at acellularsignal,which is assumedtobe close after the viral-cellular DNA junction. A second could start atposition 1,530 by the 1B promoterand terminate atthe 1A termination signal, if the viral DNAfragmentsareintegrated
inatandemarray.Inthe nuclease Slgelanalysis
with theHindIII-G fragment (Fig. 2A, lane 5), a 1,480-nucleotide-long band was observed in theneutralgel,whichcannotbemappedonthe Adl2 DNA in theprevioussection. Thisspecies
could be explained by the second possibility
assumed above, comprising the 560- and
840-nucleotide-long cotranscripts. It could then splice with the310-nucleotide-long cotranscript
to be the 1,710-nucleotide-long mRNA. Such chimeric molecules of viralsequenceshave been identified in cells transformed byaDNA
frag-mentof Ad7(33).DISCUSSION
We have defined the structure of
Adl2-spe-cific mRNA's encoded in the transforming re-gion by the methods ofBerk and Sharp (3, 4) and Alwine et al. (1). The structures of these mRNA's as detected in transformed cell lines carrying only transforming DNA segments
(CYl-l and
GYL-L)
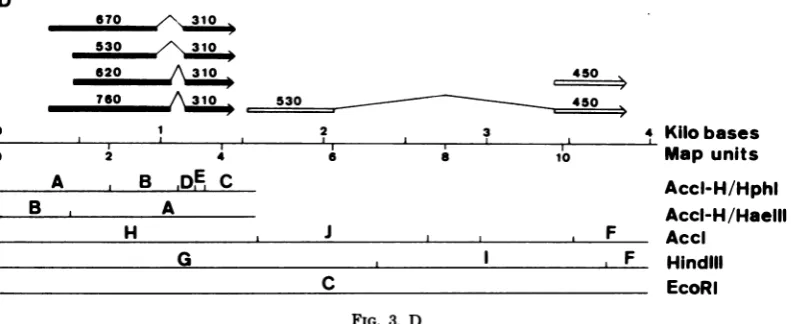
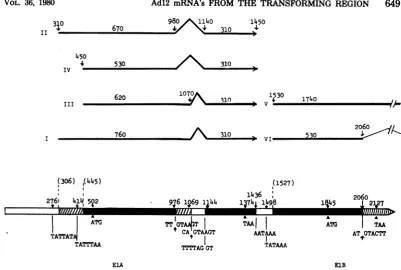
and in cells early and late after lytic infectionwithAdl2 aresummarized in Fig. 1E (CYl-l),2B (GYl-l), and 3D (early andlate).In thesefigures,the genomesequences represented inanmRNAaredepicted by a line above the genome map of Adl2. Intervening sequencesbetween continuouslytranscribed se-quences(cotranscripts),which arespliced out in thematuremRNA,areshownby a caretsymbol. Abundant mRNA's are represented by bold lines, and minorspeciespresent atlower concen-trationsare represented by thin lines. We can find specific sequences in the close vicinity of initiation,splicing, and termination sites of these mRNA species onthe nucleotide sequences of theHindIII-Gfragment (2,320-nucleotide-long)of Adl2 DNA(Fujinagaetal.,inpress;Sugisaki
etal.,inpress),assummarized in Fig.6. Incellslytically infected withAdl2, four 1A mRNA's(RNAsItoIV)werefoundtoasimilar extent.Thesearegenerated bycombinations of twoinitiation sites andtwosplicingsas summa-rizedinFig.3D. Thepresenceoftwopromoter sites was supported byour preliminary
experi-ments using exonuclease VII (kindly provided
by J. W.Chase,Albert EinsteinCollegeof Med-icine). InCYl-l andGYl-l cell lines, three of thefour 1A mRNA'swere found (RNAs I, III,
andIV). Thereare sequencescorresponding to
theHogness box (19) for thesemRNA's: TAT-TATAatposition 276andTATTTAA at
posi-tion414 (numbers in nucleotides from the left terminus of the viral genome). Assumed cap sitesarefoundatpositions 306and445, 30and 31 nucleotides downstream from the signal
se-quences, respectively (Fig. 6). The RNA
map-4
,620 450
760
-,
530-2
2
3
8 10
A B
DEI
CB , A
H J i F
G I I I F
C
FIG. 3. D
Kilobases Map units
Accl-H/Hphl Accl-H/HaelIl
.Acci . Hindlil . EcoRI
D
0
0
VOL. 36, 1980
II 4
1
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.496.52.448.506.668.2]648 SAWADA AND FUJINAGA
A
760 620 530
0 0
0
3100 00
B 760 670 620 530
310 0
aem
Incellslytically infected by Ad2 (4), Ad5 (2), and Ad7 (33), only two major species are de-tected for 1A mRNA's. The third 1Aspecies is detected inanappreciableamountin Ad2-trans-formedcells(J.Sambrook, R. Green, J. Stringer, T. Mitchison, S. L. Hu, and M. Botchan, Cold Spring Harbor Symp. Quant. Biol., in press).
Adl2seems tobe inasomewhat different situ-ation from the above three serotypes. We typi-cally detected four major 1A mRNA's, but in some cases twospecies initiated by the preceding Hogness box (RNAs I and II) decreased their relative abundance incytoplasmic RNA prepa-rations (Y.Sawada,unpublished data). Onlythe remainingtwospecies (RNAsIII andIV) were
detectedbySegawaet al. (K. Segawa,I. Saito,
K.Shiroki,and H.Shimojo, Virology,inpress).
Itispossible that thereareunknown controlson
thesynthesisorprocessingof theAdl2 1A
tran-scripts in lytically infectedcells. There should alsoexist different controlsontheexpressionof 1A mRNA'samonglytically infectedcells and transforned cell lines described in thisstudy.
Thereare distinct features tothe expression
of 1B mRNAamonglyticallyinfectedcellsand
transformed cell lines. InCY1-1cells, onelong
species of about 2,200 nucleotides with asmall
splice(RNAV)wasdetected inprominent abun-dance, and mapped approximately at position
1,530 to position 3,820. The relevant Hogness
box TATAAA isseenatposition 1,498and the
840 0
760-
-620 *
530
o
310 @00.
FIG. 4. Diagrams andautoradiograms of two-di-mensional analyses of nuclease Sl-digested RNA-DNAhybrids. The AccI-H fragment was hybridized withpolyadenylic acid-containing cytoplasmic RNAs from CYl-I cells (A), infected early cells (B), and
GYI-1 cells (C). Thefirst electrophoresis was from
lefttoright inaneutral gel, and the second wasfrom toptobottom in an alkalinegel. The numbers to the
left ofthe dots represent the lengths of corresponding cotranscripts in nucleotides. Cotranscripts derived from a given mRNA species fall on the respective vertical line.
ping dataontheinitiation sites described in this study are in good agreement with these two signals, whichareseparated by 138nucleotides. There is another Hogness box-like sequence TATTTA at position 292, but this is not as-sumedhere tobe the signal because it is only 122 nucleotides upstream from the expected Hognessboxat position 414. Thereare possible donor site sequences (22) TT/GTAAGT at po-sition 975and CA/GTAAGT atposition 1,068 andan acceptor site sequence (22) TTTTAG/ GT atposition 1,138. In the vicinity of an ex-pected termination site for 1A mRNA's, a se-quence AATAAA (20) is also presentat position 1,436(Fig. 6).
I
I.
-'' P-AfiC!0-r- x' *.--.
FIG. 5. DBM paper blot hybridization of mRNA's
from CYI-1,GYZ-I,and infected early cells. Polyad-enylic acid-containing cytoplasmic RNAs were puri-fied fromCYI-I, GYI-I,and KB cells early after lytic
infection withAdl2(in the presence of 20pgof
l-,B-D-arabinofuranosylcytosine per ml of culture me-dium). RNA blots prepared as described in the text werehybridized with32P-labeledDNAprobesspecific toearly regionsIA(AccI-H) andlB(AccI-J) ofAdl2 andautoradiographed.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:10.496.57.250.73.399.2] [image:10.496.264.450.400.569.2]Adl2 mRNA's FROM THE TRANSFORMING REGION 649 67o
530
620
98~~~~0
114 15V123~~0
1740 530(1527) 1436, * 9X6
1969
19;441;74_1*49,
206>
2060 1<,45 1
2V27
TTGT4T |CAGTAACT
+
TTTTAGGT
TAA
I
I
AATArA TATAAA
A A
ATG
ITAA
AT GTACTT
II
ElB
FIG. 6. Diagrams ofthetranscriptionmapand predicted signalsequencesin the HindIII-G fragment.All
ofthe main mRNA species, except chimeric ones, detected in CYI-1, GYI-I, and infected KB cells are
presented in theupperdiagram. Numbers above the linesrepresentRNA lengths in nucleotides. Caretsymbols representthesplicing ofprimarytranscripts.The3' endsofRNAsareindicated by arrowheads.In the lower
diagram, predicted regions composing mRNA'sareindicated by hatchedandfilledlines. Numbers above the
linesrepresentnucleotidepositions of assigned initiation and termination codons, splice sites, and signal sequencesfor transcriptionstartandstop. Theirsequences arealso shown belowthe lines. Numbers within
parenthesesshow the assumedcapsites.
presumed cap site is seen atposition 1,527 on
the nucleotidesequencesof theAdl2HindIII-G fragment (Fig. 6). This mRNA corresponds to
1B mRNA's detected incells lytically infected
with Ad2 (4), Ad5 (2), and Ad7 (33) and inacell
line transformed by Ad2 (Sambrook et al., in press). However, no appreciable amounts of
mRNAwasfound in cellsearly after lytic
infec-tionwithAdl2, byeither nuclease Si gel analysis
or DBM paper blot hybridization (this study; Segawaetal.,inpress).Thelong 1BmRNAwas
notclearly foundinthisstudy evenatthe late
stage,althoughitwasdetectedas aminor
spe-ciesinsomeexperiments (datanotshown).
In-stead,two 1B mRNAspecies,oneof about 980
nucleotidesinlength (530 plus 450)withsplicing (RNA VI)and another of about 450 nucleotides
in length, were found at the late stage,
corre-spondingtothe results ofpreviousworkonAd2
mRNA's(6, 7, 28, 31).Apresumeddonor site for
splicingof the former isAT/GTACTTat posi-tion2,059 (Fig. 6).The latterspecies corresponds
toproteinIX mRNA identified for Ad2 (7),and
couldhave itsownpromotersequences
preced-ing its5' end. Thesespecieswere notfound in
the CY1-1 cellline,asis thecasewith the
Ad2-transformed cellline(Sambrooketal., in press). Clearly, the expression of1B mRNAspecies of Adl2 is controlled by distinct mechanisms
be-tween lytically infected cells and transforned celllines. Inaddition, Adl2 is distinguished from
Ad2, Ad5,orAd7by the 1B mRNA expression
in lytically infected cells. The DNA sequences
of the1Bregion beyond the HindIII-G fragment of Adl2arenowunder currentinvestigation.
InGY1-1cells,onlya560-nucleotide-long
co-transcriptwasdetected in the 1Bregion,and it
mappedataboutposition 1,530toposition 2,090. This speciesis apparently initiated by the
pro-moter atposition 1,498, asisthecasein CY1-1
cells andinlyticallyinfectedcells.However,no
signal sequencesfor terminationorsplicingare
found around position 2,090. This cotranscript couldbe the 5'portionof chimeric RNAspecies containing viral-cellular and viral-viral
se-quencesassuggested by DBMpaperblot
anal-ysis. In the former case, mRNA transcription
startsby the 1Bpromoter, continuesbeyondthe
viral-cellular DNAjunction, and terminates at
anappropriatecellularsignal.In the lattercase, 310
450 IV
III
760 I
(306) ,(445) 276I 4*145+2I
ATG
TATTAT4
TATTTAA
ElA
I W&am SIMMUNT- M
VOL. 36, 1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:11.496.45.447.57.327.2]650 SAWADA AND FUJINAGA
mRNA
transcription
startsby
the 1Bpromoterand terminatesatthe 1A
signal
intheadjacent
viralDNA,which isintegrated
inatandemarray(33). The band at 1,480 nucleotides detected with theHindIII-G
fragment
in the neutralgel
by
the nuclease Sigel procedure
(Fig. 2A,
lane5) needsthepresence of tandem
integration
of viral DNA in GY1-1 cells as described above.The
840-nucleotide-long cotranscript specific
toGY1-1 cells should
comprise
the 3'portion
of the chimeric RNAspecies,
since nopromoter-likesequenceswerefound in the
vicinity
of its 5' end on the DNAsequence ofAdl2. The DBM paperblotanalysis
offered evidence forthepres-ence of at least three
species
of cellular-viral chimeric mRNA's. Suchspecies
were also de-tected in cells transformedby
Ad2(Sambrook
et
al.,
inpress).
InGYl-l
cells,
anotherchimericmRNA
species
containing
the840-and950-nu-cleotide-long
cotranscripts
issuggested
by
nu-clease
S1
gel mapping
(Fig. 2B).
These data reflect thecomplicated
integration
ofmultiple
copies
ofviral DNAinGYl-l
cells(21).
ACKNOWLEDGMENTS
We thank Mituru Takanami, Kyoto University, for the generous gifts ofAccIandHaeHl. We arealsogratefulto
TohruShibata,HokkaidoUniversity,for hishelpand advice in preparing 1-[(m-nitrobenzyloxy)methyl]pyridinium chlo-ride,andtoMichael R. Green for hisadviceonthenuclease Sigeland DBM paper blottechniques.
This workwassupportedinpartby grants-in-aidforcancer
research and for scientific research from the Ministryof Education, Science and Culture, Japan, anda grant-in-aid
from the Princess Takamatsu Fund for Cancer Research. LITERATURE CITED
1. Alwine, J. C., D. J. Kemp, and G. R. Stark. 1977. Methodfordetectionofspecific RNAs in agarose gels bytransfertodiazobenzyloxymethyl-paperand hybrid-izationwith DNAprobes.Proc. Natl. Acad. Sci.U.S.A. 74:5350-5354.
2. Berk,A.J.,F.Lee,T.Harrison,J.Williams, and P. A.Sharp. 1979.Pre-earlyadenovirus5 gene product regulatessynthesisofearlyviral messenger RNAs. Cell 17:935-944.
3. Berk,A.J.,and P.A.Sharp.1977.Sizingandmapping ofearlyadenovirusmRNAsby gel electrophoresis ofSi endonuclease-digestedhybrids.Cell12:721-732.
4. Berk, A.J., andP. A.Sharp.1978. Structure of the adenovirus2earlymRNAs.Cell14:695-711.
5. Casey, J., and N.Davidson. 1977.Rates offormation and thermalstabilities of RNA:DNAduplexesathigh concentration of formamide. Nucleic Acids Res. 4:1539-1552.
6. Chow, L T., T.R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regionsofadenovirus-2. J.Mol. Biol. 134:265-303.
7. Chow, L. T.,J. M.Roberts,J. B.Lewis,and T. R. Broker.1977.Amap ofcytoplasmicRNAtranscripts fromlyticadenovirus type 2,determinedby electron microscopyofRNA:DNAhybrids. Cell11:819-836.
8. Dijkema,R.,B. M.M.Dekker,M.J.M. Van derFeltz, and A. J. Van der Eb.1979.Transformationof pri-mary ratkidney celisby DNAfragments of weakly oncogenicadenoviruses.J.Virol.32:943-950.
8a.Favaloro,J.,R.Treisman,andR. Kamen. 1980. Tran-scriptionmapsofpolyoma virus-specificRNA:analysis bytwo-dimensional nuclease Si-gel mapping. Methods Enzymol.65:718-749.
9. Flint, J. 1977. The topography and transcription of the adenovirus genome.Cell10:153-166.
10. Galos, R. S.,J. Williams,M. H. Binger,and S. J. Flint. 1979. Location ofadditionalearlygene sequences inthe adenoviral chromosome. Cell17:945-956. 11.Green,M., and M.Pina, 1963. Biochemical studieson
adenovirus multiplication. IV. Isolation, purification, andchemical analysis of adenovirus. Virology 20:199-207.
12. Green, M., and M. Pina. 1964. Biochemical studies on adenovirusmultiplication.VI.Propertiesofhighly pur-ified tumorigenic human adenoviruses and their DNAs. Proc. Natl. Acad.Sci. U.S.A. 51:1251-1259.
13. Jones,N., and T. Shenk. 1979. An adenovirus type 5 earlygene functionregulates expressionof otherearly viral genes.Proc.Natl. Acad. Sci. U.S.A. 76:3665-3669. 14. Lewis, J.B., C. W. Anderson, and J. F. Atkins. 1977. Furthermapping of late adenovirus genes by cell-free translation of RNA selected by hybridization to specific DNAfragments. Cell 12:37-44.
15.McDonnell, M.W., M. N. Simon, and F. W. Studier. 1977.Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis inneutral and alkalinegels. J. Mol. Biol. 110:119-146. 16. McMaster,G. K., and G. G. Carmichael. 1977.Analysis
of single- and double-stranded nucleic acids on poly-acrylamide and agarosegels by usingglyoxal and acri-dineorange. Proc. Natl.Acad. Sci. U.S.A. 74:4835-4838. 17. Ortin, J., K. H. Scheidtmann, R.Greenberg, M. West-phal, and W. Doerfier. 1976. Transcription of the genome of adenovirus type 12.HI. Maps of stable RNA fromproductively infected human cells and abortively infected and transformed hamster cells. J. Virol. 20: 355-372.
18. Pettersson, M., C.Tibbetts, and LPhilipson. 1976. Hybridization maps of early and late mRNA sequences on the Ad2genome. J. Mol. Biol. 101:479-501. 19. Proudfoot, N. J. 1979. Eukaryotic promoters. Nature
(London) 279:376.
20. Proudfoot, N. J., and G. G.Brownlee. 1976.3' Noncod-ing region sequences in eukaryotic messenger RNA. Nature (London) 263:211-214.
21. Sawada, Y., S.Ojima, H. Shimojo, K.Shiroki, and K. Fujinaga. 1979. Transforming DNA sequences in rat cells transformed by DNA fragments of highly onco-genichuman adenovirus type 12. J. Virol. 32:379-385. 22. Seif, I., G. Khoury, and R. Dhar. 1979. BKV splice
sequences based on analysis of preferred donor and acceptor sites. Nucleic Acids Res. 6:3387-3398. 23. Sekikawa, K., K.Shiroki, H. Shimojo, S.Ojima,and
K.Fujinaga. 1978. Transformation of rat cell line by anadenovirus 7 DNA fragment.Virology 88:1-7. 24. Sharp, P. A., P. H. Gallimore, and J. S.Flint. 1974.
Mapping of Ad2 RNA sequences in lytically infected cells and transformed cell lines. Cold Spring Harbor Symp. Quant.Biol.39:457-474.
25. Shiroki, K., H.Handa, H. Shimojo, S. Yano, S. Ojima, and K. Fujinaga 1977. Establishment and character-ization of rat cell linestransformed by restriction en-donuclease fragments of adenovirus 12 DNA. Virology 82:462-471.
26. Shiroki, K., H. Shimojo, Y. Sawada, Y.Uemizu,and K.Fujinaga. 1979. Incompletetransformation of rat cellsby a small fragment of adenovirus 12 DNA frag-ment.Virology 95:127-136.
27. Smiley, J.R., and S. Mak. 1978.Transcriptionmap for adenovirustype12 DNA. J.Virol. 28:227-239. 28. Spector, D. J., M.McGrogan, and H. J.Raskas.1978.
Regulation of the appearance of cytoplasmic RNAs J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
Adl2 mRNA's FROM THE TRANSFORMING REGION 651 fromregion1of the adenovirus 2genome.J. Mol. Biol.
126:395-414.
29. Vander Eb, A. J., C. Mulder, F. L Graham, and A. Houweling. 1977. Transformation with specific
frag-mentsof adenovirus DNAs. I. Isolation ofspecific
frag-mentswithtransforming activity of adenovirus2and5
DNA. Gene 2:115-132.
30. Vogt, V. M.1973.Purificationand furtherproperties of singlestrand-specificnucleasefromAspergillusoryzae.
Eur. J. Biochem. 33:192-200.
31. Wilson, M. C.,N. W.Fraser,and J. E.Darnell,Jr. 1979.Mapping of RNA initiation sites by high dosesof
uv irradiation: evidence for three independent
pro-moterswithinthe left 11% of the Ad2genome.Virology
94:175-184.
32. Yano,S., S.Ojima,K. Fujinaga, K.Shiroki,and H.
Shimojo. 1977. Transformation ofaratcellline byan
adenovirustype 12DNA fragment. Virology 82:214-220.
33. Yoshida, K., and K. Fujinaga 1980.Unique species of mRNA from adenovirustype 7early region1incells
transformed by adenovirustype 7DNAfragment. J. Virol. 36:337-352.
34. Zieve,G., and S. H. Penman. 1976. Small RNA species ofthe HeLa cell; metabolism and subcellular localiza-tion. Cell8:19-31.
VOL. 36, 1980
on November 10, 2019 by guest
http://jvi.asm.org/