0022-538X/80/09-0812/12$02.00/0
Restriction
Endonuclease Mapping
of
Unintegrated
Viral
DNA of B- and N-Tropic BALB/c
Murine
Leukemia
Virus
ERIC RASSART"2AND PAULJOLICOEUR' ;
Institut deRecherchesCliniques de Montreal, Montreal H2W1R71;Departement deMedecine2 and Departement deMicrobiologie et d'Immunologie,3 Universite de Montreal, Montreal, Quebec, Canada
Unintegrated linear and closed circular DNAs of B- andN-tropic endogenous
BALB/c murine leukemia virus (MuLV) were extracted from newly infected
mouse cells and cleaved with EcoRI, XhoI, PvuI, HindIII,
SalI,
XbaI, KpnI,SmaI, and PstIrestriction endonucleases. The DNAfragments wereseparated by electrophoresis and
analyzed
by the Southern blothybridization
procedure. EcoRI didnotcleavethe two genomes. Aphysicalmap of15cleavage
sitesonB-andN-tropicgenomes wasconstructed with the other restrictionendonucleases.
Identicalcleavage sites ofB-andN-tropic MuLV DNAswerefound with all these
enzymes. However,the N-tropiclinear genome was foundtolack about 75 base
pairsateach end of the molecule.PstI,KpnI,andSmaIrecognizeacleavagesite atboth ends ofthe linear molecules. Andsequencesderived from the 5' end of
the RNAgenome werefound in the third left end of the linearDNAandatits
extremeright-endterminus, suggesting thepresence of redundantsequences. Two
speciesofclosed circular viral DNA were observed. The largerspecies has the
samesize as the linearmolecule and appearsto bea circularized form of linear
DNA. Thesmallerspecies containssequences commontoboththelinear andthe
larger circular viral DNA but seems to be deleted from sequences present at
eitherone orboth endsofthelinear DNA.
Therefore,
thegeneral
structureof thelinear and circularDNAspecies of these B- and
N-tropic endogenous BALB/c
MuLVappearsanalogoustothestructurefound with otherretroviruses.
Naturally occurring
ecotropic
murine leuke-mia viruses(MuLV)canbeclassifiedasN-tropic
orB-tropic withrespecttotheir hostrange(13). N-tropic viruses infectNIH cells
(N-type)
moreefficiently
thanBALB/c
cells(B-type),
whereas B-tropic virusesgrow moreefficiently
onBALB/ccells thanonNIHcells. Thissusceptibility of
murine cells to the
naturally occurring
MuLVhas been shown to be determined
by
asingle
allelicgenetic locus,
the Fv-1gene(28,
for reviewsee 20, 27). The restriction of N- or
B-tropic
MuLV inFv-1-resistantcellsoccurs
early
inthevirus cycle, after
synthesis
of viral DNA butbefore itsintegration into the cell genome (23,
37). Our recent results indicate that accumula-tionof circular viralDNAispreventedin
Fv-1-resistant cells (24). The viraltropism
determi-nant which is required for this restriction has
been shown to be aprotein because ofitstransfer
by phenotypic mixingtoother viralgenomes(3,
19, 25, 29). Evidence obtained with recombinants
of N- andB-tropicviruses(16, 32)andwith
NB-tropicviruses derived from
B-tropic
viruses(17)indicated thatp30virionproteinmightcarry the
tropismdeterminant.
N-andB-tropicendogenousviruses obtained from BALB/c mice have been studied
exten-sively.Theirgenomesare
highly
homologous
asshownbymolecular hybridization(5,22, 30) and
by Ti oligonucleotide fingerprintanalysis (7, 8).
N-tropic specific oligonucleotidesofN-tropic
re-combinantvirusesand NB-tropic specific
oligo-nucleotides ofB-- NB-tropic viruses were
iden-tified, and bothwereshown to be located at the
5'endof the genome (7, 8). Sequencingof these
specific oligonucleotides revealed only few
dif-ferences, asexpected for allelicsequences (31).
To determine the exact location of the gene
coding for theviraltropism determinantand to
study the integrationof B- and N-tropicMuLV,
wehaveconstructed a physical map of the
cleav-age sites oftheirDNAwith several restriction
endonucleases. We have purified unintegrated
linearand closed circular viral DNAs of B- and
N-tropic MuLV from acutely infected mouse
cells. Duringtheseexperiments, we found a
de-leted form ofclosed circular viral DNA. Since
then, reports have been published on the
ex-istence of asmallercircular form ofviral DNA
duringreplication of other retroviruses (11, 18, 33, 34, 43).
MATERIALS AND METHODS
Cellsand viruses. TheoriginsofJLS-V9,Sim-R,
and NIH/3T3 cells have been described (21). Cells weremaintained inDulbecco-modified Eaglemedium 812
on November 10, 2019 by guest
http://jvi.asm.org/
supplemented with 10% calfserum (GIBCO Labora-tories,Grand Island, N.Y.). The cloned N-tropic (N-C1-35) and B-tropic (B-Cl-11) endogenous BALB/c viruseshave been described (19). Both viruses were grownonFv-I-permissiveNIH/3T3(N-CI-35)or JLS-V9andSim-R (B-Cl-11) cells (21, 23).
PreparationofviralDNA.N-Cl-35 andB-Cl-11
unintegrated viral DNAs were obtained by infecting NIH/3T3 orSim-R and JLS-V9 cells, respectively, at
amultiplicity of infection ofapproximately2,inroller bottles in the presence of 8 ,ug ofPolybreneperml. At 16 to 24hafter infection, a Hirt extraction (15) was performed, and the Hirt supernatant DNA was sub-jected to propidium iodide-cesium chloride centrifu-gation as described (24). DNA from the lower band containing theclosed circular molecules (FormI)and DNAfrom the upper band containing linearmolecules
(Form III)wereextracted withNaCl-saturated
isopro-panoltoremovethepropidium iodide and precipitated with 2volumes of ethanol at-20°C. Form I DNA was suspended in water and used as the substrate for restrictionendonucleases. DNA from the upper band wassuspended in0.01M Tris(pH 7.8)-0.02 M EDTA
andlayeredon a 15 to30%(wt/vol) sucrosegradient
in0.01 M Tris-hydrochloride (pH 7.5), 0.1 M NaCl,
and5mMEDTA.Centrifugationwasperformed in an
SW41 Spinco rotor at 23,000 rpm for 16 h at 40C.
Fractions of0.4mlwerecollected from the bottom of the tube (9,40). A sample of each fractionwasscreened by agarose gelelectrophoresis, and DNAwas
trans-ferredto nitrocellulose. Viral DNA wasdetected by hybridization to 32P-labeled complementary DNA (cDNA). Fractions containing DNA sedimenting at about 20S werepooled and precipitated with2
vol-umes of ethanol at -20°C and kept as full-length
double-strandedlinearviral DNA.
Synthesis and purificationof viral
[32P]cDNA.
Representative3P-labeledB-Cl-il DNAwas
synthe-sizedbyincubating bandedB-Cl-li virions with calf
thymus oligonucleotide primers, as described
previ-ously(24). Itsspecificactivityvariedbetween2 x 108
and4x
10W
cpm/ug.[32P]cDNA5'wassynthesized under the same con-ditions except that calf thymus oligonucleotide primerswereomitted (14). The reactionwasstopped after1hbyphenolextraction.[32P]cDNAwaspurified
on aSephadexG-50 column (1.5 x 35cm) and
incu-bated at 370C for 18 h in 0.3 N NaOH (24). After neutralization and ethanol precipitation,
['P]cDNA
chainswereseparatedbyelectrophoresison10% acryl-amidegelasdescribed (14).The strong stop cDNA 5' ofaround140baseslongwasdetectedby autoradiog-raphy. It constituted the major short cDNA species made. Itwaseluted from thegelasdescribed (14) and
usedas[32P]cDNA5'.
In vitro synthesis of double-stranded viral
DNA.Single-strandedviral DNAwasfirst synthesized
in vitroessentiallyasdescribed (42).Banded
B-Cl-il
virions wereincubated for 14 to 16hat
370C
in 50 mMTris-hydrochloride (pH 8.5); 2mMeachdATP,dCTP, dGTP, dTTP; 50,uCi of [3H]dTTP (53 Ci;
mmol);6mMmagnesiumacetate;2mMdithiothreitol;
0.01%NonidetP-40;40
Mg
ofactinomycinDper ml inafinalvolume of1ml. The mixturewas extractedwith phenol, and theaqueousphasewasloadedona
Seph-adexG-50 column, as above. Fractionscontaining viral DNA werepooled, adjustedto0.3NNaOH,incubated at370C for 12 h, neutralized and ethanol precipitated. The cDNA recovered by centrifugation waslayered on a 15to30%alkalinesucrosegradient (0.9 MNaCl, 0.3 N NaOH, 5 mM EDTA) and centrifuged in an SW41 rotorfor 16h at 35,000 rpm. Fractions of the gradient corresponding tothe largest cDNA species werepooled and ethanol precipitated in the presence of100Mg of calf thymus DNA primers (39).
This (-) strand cDNAwasmadedoublestranded (42) by incubation at 170Cfor 6 h in 20 mM Tris-hydrochloride (pH7.5); 4mM dithiothreitol; 10mM magnesium acetate;60mMNaCl; 1mMeachdATP,
dCTP, dGTP,anddTTP;50,uCiof[3H]dTTP; 200Mug
of calf thymus DNA primers (39); and Escherichia coli DNApolymerase Iinafinalvolume of0.05ml. Viral DNA was phenol extracted, treated with Si nuclease (24), layered on a 15 to30%neutralsucrose
gradient, and centrifuged as described above. Frac-tionsof the gradientwerecollected,andasampleof each fractionwasscreened for the presence of viral DNA by agarose gel-DNA transfer procedure and hybridizationto[32P]cDNAasdescribed below. Frac-tionscontainingviral DNAmigratingasless than full-length molecules (between1x106 and3x106daltons)
werepooledand ethanolprecipitated.
Restriction endonuclease cleavages.
Restric-tion endonucleases SmaI, XhoI, BgII, BgIII, PvuI, XbaI, andKpnI werepurchased from NewEngland Biolabs (Beverly, Mass.); HpaI and Sail were pur-chased from Miles Laboratories (Elkhart, Ind.). EcoRI,HindIll, BamHI,andPstIwerefrom Boehrin-gerMannheim Corp.,Montreal, Canada. Restriction endonucleasedigestion conditionswerethose
recom-mendedbythesupplier.Completionof each reaction
was monitored by including 0.5Mug of lambda DNA
(Boehringer) in the reactionmixtures.
Agarose slab electrophoresis and transfer.
Samplesof25
p1
(2to 5Mgof cellDNA)werelayeredinto separatechannels ofaverticalslab gel (20 by20
by0.3 cm) castwith 1.4% agarose (SigmaChemical
Co.,St.Louis, Mo.).DNAwasseparatedby
electro-phoresis in 40 mM Tris (pH 8.3), 50 mM sodium acetate, and 1 mM EDTAfor about 18h at 35mA/ gel. Afterelectrophoresis, thegelswerestainedwith
0.5,ugof ethidium bromide per ml andphotographed with Polaroid filmby UV illumination. The DNAwas
then denatured, neutralized in situ, and transferred
onto a nitrocellulose membrane (0.45Mum; Millipore
Corp.,Bedford, Mass.)asdescribedbySouthern (36).
Hybridization procedure. After transfer of the
DNAtothenitrocellulose,the membraneswerebaked
at800Cfor2hand then soaked for10 to 12hatroom
temperaturein 25to 100mlof3x SSC solution
con-taining 50%formamide, 100Mugof calfthymus DNA,
and supplementedwith 0.02% each ofFicoll,
polyvi-nylpyrrolidone, and bovine serum albumin, as
de-scribed elsewhere(33). Thewetmembraneswerethen incubatedat390Cfor 30 to 48 h inplastic bagswith 0.5 to 1ml of thesamesolution containing3x 10W to
6 x 106 cpm of
[32P]cDNA.
After annealing,mem-braneswerewashedin 6xSSC-0.5% sodium dodecyl sulfate solution for12hat670Cand thenin the same
solutionat370Cfor6 to 8h(33). Membraneswereair
on November 10, 2019 by guest
http://jvi.asm.org/
driedand exposed at -20°C to Kodak-RP-Royal X
Omat filmwithaCronexLightningPlusintensifying
screen(DuPont Co., Wilmington, Del.) (33).
RESULTS
Mapping of in vitro B- and N-tropic
MuLV DNAs with EcoRI, HindIII, XhoI, Sall, PvuI,and XbaI. Toanalyzethe size and the structure of B- and N-tropic DNAs, we
partially purified linear and supercoiled viral
DNAmoleculesbycentrifugationonpropidium
iodide-cesium chloride gradients and further
purified theupperbandon sucrosedensity
gra-dients. The selected 20S linear viral DNA
mi-gratedas adiscrete band of 5.7x 106daltonson
agarose gel, as expected for a genome-length
molecule of double-stranded structure. DNA
from the lower band of the propidium
iodide-cesium chloridegradient contains closed circular
viral DNA which could be resolved into two
faster-migrating species byagarose gel
electro-phoresis,asreported for other retroviruses.
The linear and circular forms of B- and
N-tropic MuLV DNAweredigested byavarietyof
site-specific endonucleases.The DNAfragments
generatedbyenzymedigestionwereresolvedby
agarose gelelectrophoresisand detectedbythe
Southern procedure with a [32P]cDNA probe.
Molecularweightsweredeterminedby
compar-ison with Eco RI- or HindIII-digested lambda
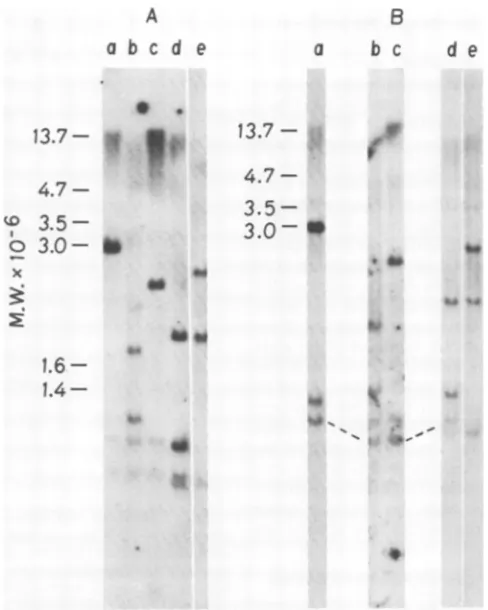
DNArunonparallel lanes. Eco RI didnotcleave
the linear nor the circular forms of B- and
N-tropicMuLV DNA(Fig. lg, Fig.
2f).
XhoI,PvuI, HindIII,SalI,and XbaI all cleaved both linearviral DNAsonce,thusgeneratingtwofragments,
the molecularweights of whichaddeduptothat
offull-length linear DNA. XhoI generatestwo
fragments of 2.9x 106 and 2.8x106 daltons(Fig.
la); HindIII generates two fragments of 3.8 x
106 and1.9x106 daltons(Fig. lc);SalIgenerates
two fragments of 3.05 x 106 and 2.65 x 10"
daltons(Fig. le). PvuIgeneratesonedetectable
fragment of 5.3 x 106 daltons (Fig. lb). The
other fragment of 0.4 x 106 daltons was not
detected in this experiment, but since the
en-zyme cleaves the circular DNAonlyonce (Fig.
2b), its existence is deduced. XbaIgenerates two
fragments of4.95 x 106and 0.75 x 106 daltons
(Fig.
lf),
but thesmallerfragment whichappearsas averyfaint bandherewasstudied further.
Amore precise comparison of the molecular
weights of the shorter fragments derived from
B- and N-tropic viral DNAs revealed that the
smaller PvuI fragment (0.4 x 10' daltons) and
thesmallerXbaIfragment (0.75x 106daltons)
generated from theN-tropic MuLV DNAwere,
in fact, both smaller in size than their
corre-sponding fragments derived from B-tropic
MuLV DNA(Fig. 3a-d).This size differencewas
estimated to be 0.05 x
10'
daltons (approxi-mately 75 base pairs) in each case. This small difference couldnotbe observed in theprevious experiment (Fig.1)
because B- and N-tropic DNAfragments were not run on thesame gel. Since Pvu Iand Xba Icleave, respectively,atthe left andattheright ofthe linear viral DNA(Fig.4),such adifferencecould either result froman
additional cleavage site in each of these regions for both restriction endonucleases or from the absence ofa75-basepairsequence atboth ends of the linear molecule (seebelow).
As shown in Fig. 1, the presence of other
discrete bandswasalsoobservedafter digestion with various restrictionendonucleases, andmore
especially with XbaI
(Fig.
1B, f). These addi-tional bands were not detected with all DNA preparations (Fig. 4c), and their presence wasmarkedly reduced indigested linear DNA
prep-arationsthat had been selected onsucrose
gra-dients. The sum of the molecular weights of these additional XbaI fragments does not add
up tofull-length linear DNA. It isunlikelythat
they
are the products of partial digestion, ascomplete digestion of lambda DNA was
ob-served for each reaction. Theymostlikelyarose
eitherfrom the cleavage of contaminating cel-lular DNA, subgenomic linear viral DNA of
discrete size, or from partially single-stranded viral DNA.
The evidence that these enzymes cleave
B-and N-tropic MuLV DNAsat onlyone site was
confirmed by digesting the circular forms of
these DNAs. As shown in Fig. 2, all these
en-zymes transform the two circular species into
twolinear forms. Theupperband ofthe doublet
migratedas agenomic linearDNAmolecule of
5.7 x 106daltons.Thelower band has a
molec-ular weight of5.35x
106,
indicatingthat the twocircular species really differ in size and not in
theamountofsuperhelical twist.
Mapping
of in vivo B- and N-tropic MuLV DNAswith
KpnI,SmaI,
andPstI.
SmaI endonucleasecleavage
oflinear B-tropic MuLV DNA yields three detectable fragments of 2.95X
106,
1.25 x106,
and 1.15 x 106 daltons (Fig.5B,b). Cleavageoflinear N-tropic MuLV DNA
with the same enzyme alsogenerates three
de-tectablefragmentsof 2.95x
106,
1.20x106,
and1.15 x 106 daltons (Fig. 5B, d). Two of these
fragments (2.95 x
10'
and 1.15 x10'
daltons)from B- and N-tropic MuLV DNA comigrate.
The otherfragments(1.20 x
10;
and 1.25 x.106
daltons)did notcomigrate as verified by a
diges-tion of a mixture of B- and N-tropic MuLV
linear DNAs. SmaI endonuclease cleavage of
circular B- andN-tropicMuLV DNA generates
the same fragments as those obtained after
cleavageoflinear DNA (Fig. 5B,aand c),
on November 10, 2019 by guest
http://jvi.asm.org/
A
a b c
d
e
13.7-
II
i4.4
of4.7
-3.5
-3.0
-a
S
,,
law
01*
f
g h
a b c d e
Ii
5.3-3
98-.3ji. i*
11:
;:
I
0.75- 1.6-1.4
-0e
4
FIG. 1. Digestion of B- and N-tropic linear viral DNAs with XhoI, PvuI, HindIII, Sall, XbaI, and EcoRI.
In vivo linearunintegratedviral DNAsofB-andN-tropicMuLVwerepurifiedonapropidium iodide-cesium
chloridegradientanddigested with different restriction endonucleases tocompletion. After digestion, the
samples were run on 1.4% agarosegels, and the DNA fragments were transferred onto a nitrocellulose
membrane and annealed to[32P]cDNA. Fragment size was estimated by running EcoRI- and
HindIII-digestedlambda DNAsasmarkers. (A) B-tropicDNA. (B) N-tropicDNA.DNA samplesweredigested uwith
XhoI (a), PvuI (b), HindIlI (c), HindIII and SalI (d), Sall (e),XbaI (f), EcoRI (g), and nondigested (h). Nondigested B-tropicDNAwasfromadifferent preparation than theB-tropic viral DNAused for digestion.
cating that these fragmentsareinternal and that
theenzymealso cleaves closetooneendorboth
ends of thelinearmolecule. Evidence thatSmaaI
cleaves B- and N-tropic linear MuLV DNAs
close to the left end was provided by double
digestion of linear viral DNA with
SmaI-HindIII and SmaI-Sall endonucleases. The
smaller HindIII fragment (1.9 x 10" daltons)
wascleaved by Sma Itogenerateafragment of
1.65 x
10'
daltons (Fig. 6b; Table 1). Similarly,SmaI cleaved the smallerSailI fragment (2.65
x
10'
daltons) to generate a shorter fragment(2.4x 10" daltons) (Fig.6c; Table 1). Evidence that SmaI cleaves B- and N-tropic linear MuLV
DNAsclosetothe right end of the moleculewas
provided by double digestion with XbaI-SmaI
(Table 1). ThesmallerXbaI fragment(0.75x 10"
daltons)wascleaved bySma Itogeneratea0.65
x
10'-dalton
fragment, indicatingthatSmaIrec-ognizes acleavage site close to this end of the
viral DNA.
Cleavageof linearB-tropic MuLVDNA with
KpnI yields three detectablefragments by
hy-bridization, which have molecular weights of
2.45x
106,
1.8 x 106,and 1.1x 10" (Fig. 5,panelA(b)). Cleavageof linear N-tropic MuLVDNA
with thesameenzyme alsogeneratesthree
de-tectablefragmentsof 2.45 x
10W,
1.8 x106,
and1.05 x
10'
daltons (Fig. 5A, f).The twolargestfragmentscomigratedwith those of theB-tropic
MuLV DNA, but the small one (1.05 x 10"
daltons)wasslightlysmaller than that of the
B-tropic (Fig. 5A, candd). However, these three
fragmentsdonotaccountcompletelyfor thesize
of the linear DNA and add up to 5.35 x 10"
daltons. Digestion of the circular DNA yields
the same three corresponding fragments (Fig.
5A,aande), indicatingthat thesefragmentsare
internal and that,in addition tocleaving twice
inside thelinearmolecule, KpnI alsocutsclose
B
f
q
h
f
'- _- _
-
13.7
-
4.7
-
3.5
3.0
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.510.73.433.66.380.2]A
a b c d e f
g
B a b c d e f
g
5.735-a
5.35/-FIG. 2. Digestion ofB- andN-tropic circular viral DNAs with XhoI,PvuI, HindIII, SalI, XbaI, andEcoRI.
Circular viral DNAsofB- andN-tropicMuL Vwereincubated withdifferentrestrictionendonucleases.After
digestion, the samples were run on a 1.4%agarose gel, and the DNA fragments weretransferred onto a
nitrocellulose membrane and annealed with [32P]cDNA. Closed circular viral DNA (Form I) wasnicked
duringstorage,and therelaxedform (FormII) migratedslower than the linear viral DNA.(A)B-tropic DNA. (B) N-tropicDNA. DNA digested with XhoI (a), PvuI (b), HindIII (c), XbaI (d), SalI (e), EcoRI (f), and nondigested (g).
a b c d
0.75-35 0.4
-FIG. 3. Detectionof thesn
atedby cleavageoflinearanc tropicMuLVDNAswithPvu]
Circularandlinear viralDP
MuLVwerepreparedfrom
in)
inthe text.After digestion u
cleases, the samples were ru
DNAfragmentsuweretransfe)
membrane and annealedwith tropic (a)andN-tropic (b) DI LinearB-tropic (c) and N-tr with XbaI. Circular B-tropii DNAdigestedwithSmiaI.Ci
N-tropic (h)DNAdigestedwi
e f g h to oneendorboth ends of the linearmolecule.
Evidence thatKpnIcutsclosetothe left endof
linear DNA was provided by double digestion
with HindIII-KpnI (Table 1). The smaller
HindIII fragment (1.9x
10'
daltons)wascleavedby KpnIto generate a new fragment of 1.65 x
106daltons. ThisKpnIcleavagesite must be at
0.35- A Z the left end of theHindIII fragment. A similar
conclusionwasreachedbydoubledigestionwith
SmaI-KpnI, which leaves the 1.8 x 106-dalton
KpnI fragment intact(Fig. 6d; Table 1). If this
nallerfragmentsgener- fragment were terminal, it should have been
d supercoiledB- and N- cleavedbySmaI, which cleaves close to theleft
I,XbaI,SmaI,andPstI. end of the molecule. Therefore,thisKpnI
frag-VAsof B- and N-tropic ment is internal, and a KpnI cleavage site is
fectedcellsasdescribed present atthe left end ofthe molecule. Evidence 'ith restriction endonu- thataKpnIcleavagesiteispresentattheright
in on agarosegels, the end of linear viral DNA was also provided by
rredontonitrocellulose double digestion withKpnI-SmaI (Fig. 6d; Ta-i
12P
PJcDNA.LinearB- ble1).Thisdoubledigestionleaves the 1.1 x106*opic (d) DNA digested dalton KpnI fragment intact. If this fragment
c (e) and N-tropic (f) were terminal, it should have been cleavedby
ircular B-tropic(g)and SmaI (which recognizes a cleavage site at the
thPstI. right endoflinear DNA) to give rise toa
frag-(0
0
5.7,_'
5.35""
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.510.146.382.76.357.2] [image:5.510.66.255.439.543.2]a b c
de f
4.95-
1.1
-
0.75-FIG. 4. Orientationof B-tropicMuL V DNA frag-mentsproducedby differentrestriction enzymes with respect tothe3' and 5' endsof the RNA molecule. Double-stranded viral DNAwassynthesizedin vitro
asdescribed in thetext. Less than full-length viral DNA wasselectedon sucrosedensitygradientand
usedfordigestion withdifferentrestriction
endonu-cleases.After digestion, sampleswere run on a1.4% agarosegel,and the DNAfragmentsweretransferred
onto nitrocellulose membrane and annealed with
[32P]cDNA.
In vitro-synthesized viral DNAfrag-mentsnondigested (aand
/)
anddigestedwith XbaI(b)orKpnI (e).Asacontrol,invivo-extracted linear
B-tropic viral DNA was cleaved with XbaI (c) or KpnI(d).
ment shorter
by
0.1 x 106 daltons as shown abovewith XbaI andSmaI.
Therefore,
this 1.1 x 106-dalton KpnI fragment isinternal,
and aKpnI
cleavage
site ispresentattheright
end of themolecule.PstI restriction endonuclease
produces
only
one detectable
fragment
of 5.35 x 106 daltonsafterdigestionof linear B- and
N-tropic
MuLVDNA (Fig. 5C) and thus seems to cleave
only
once. However, cleavage of the two circular
DNAspeciesgenerates
only
onedetectablefrag-mentof5.35x
10W
daltons instead ofproducing
theexpecteddoublet of5.7 x
106
and5.35x10W
daltons (which is whatoneshouldexpect from
circular DNA withan enzymethat cleaves once).
Thisfragment comigrates withthesmaller frag-mentobtained after digestionof the two circular
species with HindIII (Fig. 5C,a),indicating that
PstI cleaves the smaller circular species once
and the larger circular viralDNA species at least
twice. Evidence that PstI cuts the linear viral DNA at both ends was provided by double
diges-tion with HindIII-PstI.Asshown(Fig. 7; Table 1), PstI cleaves the smaller HindIII fragment
(1.9 x
10'
daltons)to generate a fragment of 1.85x
10'
daltons and the largerHindIIIfragment(3.8x
106
daltons) togive riseto afragment of3.5x
10'
daltons. Thesmallfragments generatedby digestion of linear DNA with PstI are too
smalltobedetected in this experiment.
A more detailed analysiswas alsoperformed
todetect thesmaller DNA fragments generated fromsupercoiled viralDNAby SmaI,Kpn I, and PstI. As shown, these three restriction
endonu-cleases cleave linear viralDNA atbothends and
generateidentical largeinternalfragments from
linear and circular viral DNA. Therefore, the
smaller DNA fragment generated by these
en-zymes in circular viral DNA should represent
the fused ends of the linear molecule. After
digestion ofB-andN-tropic circular viralDNAs
with SmaI (Fig. 3e and f) and PstI (Fig. 3g and h), we could observe a fragment of0.35 x
106
daltons undetected in previous experiments. We found that thisfragment derived from N-tropic viral DNA was approximately 75 base pairssmaller than the B-tropic fragment. A similar resultwasobtained with Kpn I (data not shown).
Digestion of in vivo B- and N-tropic
MuLV DNA with otherrestriction endonu-cleases. We have useda number of other
re-striction endonucleases, including BglI, BglII,
HindII,
HpaI, and HpaIItocleave B- and N-tropic MuLV DNA. All these enzymes yield multiple fragments which have not beenor-dered. Cleavage of linear MuLV DNA with BamHIgives three detectable fragments of2.0
x
106,
1.25 x106,
and 1.20 x106
daltons. Thesmallestfragment (1.20 x 106 daltons) appears
in someexperiments as adoublet and, in fact,
represents two fragments of almost identical
size.Usingdigestion of circular MuLV DNA and doubledigestion with
HindIll,
these fragments have been ordered. This gives the orientationfor the
BamHI
DNA fragments of5'-1.2-1.25-2.041.2-3'.
Orientation of B- and N-tropic linear
MuLVDNAs. We haveperformeddouble
diges-tions on both B- and N-tropic viral DNAs to
orient thecleavagesites of the restriction
endo-nucleases with respect to each other. Double
digestionswere performed on B- and N-tropic
linearviral DNAs with HindIIIand each of the
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.510.85.226.72.369.2]A
a b c d e f
B
a b c d
C
a b c d e f
a
IN as
5.357-2.95- t-_
1.25
1.15- -1.20
1.1- -,
FIG. 5. Digestion ofcircular andlinearforms ofB- andN-tropic MuLVDNAs with Kpn I,SmaI,andPst I.
Circular and linear viral DNAsofB-andN-tropicMuL Vwerepreparedfrom infectedcellsasdescribed in
the text. After digestion with restriction endonucleases, the samples were run on agarosegels; the DNA
fragmentsweretransferredontonitrocellulosemembraneand annealedwith[32PJcDNA. (A)KpnI digestion ofB-tropiccircular DNA(a)andlinear DNA(b, c);andofN-tropiccircular DNA (e)andlinear DNA (d, f). (B)SmaIdigestion of B-tropiccircularDNA(a)andlinear DNA (b);andofN-tropiccircular DNA(c) and linear DNA (d). (C) PstIdigestion. B-tropic circular DNA digestedwithHindIlI (a) and PstI(b), B-tropic
linear DNAnondigested (c),anddigestedwith PstI(d).N-tropiccircular DNAdigestedwithHindIII (e)and PstI(f).
other restriction enzymes. Double digestions
with SmaI-KpnI,SmaI-SalI, SmaI-PvuI, and
SmaI-XbaI were also performed. The results
are summarized in Table 1. Double digestions
with SmaI-HindIII, SmaI-SalI, and
SmaI-KpnIareillustrated inFig. 6, and with Hin
dIII-SailIinFig. ld. For example, the double
diges-tionswithHindIII-SailI endonucleasesresulted
inanaltered mobility of the smaller SailI
frag-ment (Fig. ld). A new fragment of 0.75 x 106
daltons was generated which must originate
fromthecentralpartof thegenome. Thus, the
smallerHindIllfragment (1.9 x 106 daltons) is
located at the same end as the smaller SailI
fragment (2.65 x 106daltons). (See mapin Fig.
8.)
Orientation ofthe DNAfragments produced
by different restriction endonucleases with
re-specttothe 5'and 3'endsof the RNA molecule
was accomplished by restriction endonuclease
digestion of less thanfull-length in
vitro-synthe-sized double-strandedDNAasdescribed (42).It
has been shown that in vitro (-) strand DNA
synthesis initiates at the 5' end of the RNA
molecule and extends toits 3' end (for review,
see2). In themurinesystem,less thanfull-length
in vitro DNA represents the first 135 to 140
bases derivedfrom the 5' end of the RNA
mol-ecule, followed bysequencesrepresentingthe3'
end of the RNA molecule (2, 14). We used this
ordered synthesis to map the DNA fragments
relativetothe3' end of the RNA. Less than
full-length cDNAwasmade double stranded in vitro
with E. coli DNA polymerase I and used as
substrate forrestriction endonucleases.
Restric-tion endonuclease cleavage of less than
full-length, double-stranded in vitro-synthesized
DNA ofheterogeneous size should generate a
discretefragmentderivedfrom the 3' end ofthe
RNA, plus fragmentsofheterogeneoussize.
As showninFig. 4,cleavageof less than
full-lengthinvitro-synthesizedDNAwithXba I
pro-(x
x
2.45-
-1.8- N
J. VIROL.
I
I I I
I I i
ll
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.510.86.446.70.353.2]A
a
b c d e
S.
1
3.7
-:...iq
4.7
3.5-3.0-
S
B
a
b c
d
e
13.7
...I
4.7
-3.5
-3.0
-e A
1.6-1.4
-.ff!!.,
> .4'
FIG. 6. Double digestion oflinearB- andN-tropicMuLV DNAs with restriction endonucleases. Double digestion oflinear B- andN-tropicMuLV DNAs with various restriction endonucleases was carriedout sequentially byaddingthe secondenzymewith its recommendedbufferto the reactionmix.After digestion, sampleswere run on a1.4%agarosegel;the DNAfragmentsweretransferredonto nitrocellulosemembrane and annealed with[32P]cDNA. (A) B-tropicDNA. (B) N-tropicDNA. (a)SmaI, (b)SmaI and HindIII (c)
SmaI andSall, (d)SmaI andKpnI, and (e) KpnI. The twoHindIII fragments (3.8 x 10 and 1.9 x 10'
daltons)and thetwoSalIfragments (3.05x106and 2.65x10 daltons)arenotshown,buttheywerealsorun
onthesamegeltolocalizeappropriatefragments.
duced thesmallerfragment (0.75x
10'
daltons)whichcomigrated with the fragment generated
by thesameenzymefrom invivo-extracted viral
DNA.KpnIcleavage of this in vitro-synthesized
DNA generatedonly the smallerfragment (1.1
x
10'
daltons)whichagain comigratedwith thefragment derived from in vivo-extracted viral
DNA.Similar digestion with SmaI only
gener-atedthetwosmallerfragments (1.15 x
106
and1.25 x
106
daltons) (data not shown). Theseresults indicated that the smaller XbaI and
KpnI fragments and thetwosmallerSmaI
frag-mentsofB-tropic MuLVrepresentthe 3' end of
MuLV genomic RNA (Fig. 8). The ordering of
thefragmentsontheN-tropic MuLV DNA
rel-ativetothe 5'and 3'end of the N-tropic MuLV
wasnotdone but is presumedtobethesame as
the B-tropic MuLV because of their high
ho-mology.
Studies ofterminal sequences in B- and
N-tropic MuLV DNA. Unintegrated linear
DNA of aviansarcomaviruseshas been shown
to possess a direct terminal sequence
redun-dancy of about 300 nucleotides (34, 38) derived
from both the 3' and 5' endsofviral RNA (34).
The largerclosed circular avian sarcoma virus
DNAwasshowntocontaintwocopiesintandem
of this repeat sequence whereas the smaller
closed circular form contains only one copy of
thissequence (18, 34). Linear (4, 10-12, 18, 26,
33, 35, 43) and closed circular (12, 33, 43) viral
DNAs withasimilarrepeatstructurehave also
beenobserved in cells infected with other
retro-viruses.
The restriction analysis presented above
showsthat linear viral DNAs of B- andN-tropic
MuLV containonePstI,KpnI,andSmaIsiteat
bothends of the molecule. Inaneffortto
estab-lish whetherthegeneralstructure of B- and
N-tropic BALB/c MuLV might be similar tothe
(0 0
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.510.130.374.76.381.2]structure of other retroviruses, we used strong stop [32P]cDNA 5' as a probe to determine whether 5'-derived sequences were present at
both ends of the linear molecule. As shown in
Fig.7B, a,thetwoHin dIII fragments of B-tropic
MuLV linear DNA hybridized tocDNA 5', in-dicating that this sequence is present on both halves of the DNA molecule. Thesame conclu-sion could be reached after annealing ofboth SaII fragmentswithcDNA 5'(datanotshown). However, when double digestionwith HindIII-PstI (whichcuts atbothends of the linear DNA)
wasperformed, only thesmallerfragment (1.85
x106daltons) couldstillbedetected with cDNA
5' (Fig. 7B, b). This indicates that the larger HindIII fragment, which represents the 3'
se-quence of genomic RNA, contains at its right
terminus asequence homologoustothe 5' end of the RNAgenomeand which iscleaved off by PstI. Since the smaller HindIII fragment can
still be detected with cDNA 5' after cleavage with PstI, it appears that the 5'-derived
se-quence present on thisfragment isnotlocated
at theextremeterminus. Thesameexperiment
has also been done with the N-tropic linear MuLV DNA, and the results (not shown) are
TABLE 1. Sizeof fragments fromlinearB-tropic DNA ab
Restrictionendonu- Size
(X1Ob
daltons) clease5'-a
3'end of RNAHindlIl 1.9 3.8
SmaI 2.95 1.15 1.25
HindIII+SmaI 1.65 1.3 1.15 1.25
SaiI 2.65 3.05
HindIII+Sai 1.9 0.75 3.05
SmaI+
Sail
2.4 0.65 1.15 1.25KpnI 1.8 2.45 1.1
HindIII+KpnI 1.65 2.45 1.1
SmaI+KpnI 1.8 1.23 1.15 1.1
XbaI 4.95 0.75
HindIII+XbaI 1.9 3.05 0.75
SmaI+XbaI 2.95 1.15 0.5 0.65
XhoI 2.9 2.8
HindIII+XhoI 1.9 1.0 2.8
PvuI 0.4 5.3
HindIII+PvuI 0.4 1.5 3.8
SmaI+PvuI 2.8 1.15 1.25
PstI 5.35
HindIII+PstI 1.85 3.5
BamHI 1.2 1.25 2.0 1.2
aThe size of some fragments is given with two decimals.Although the second decimalisnotaccurate,
ithas been included to distinguish some fragments fromoneanother.
bThe underlinedfragmentsare shorterby0.05 x 106 daltons for theN-tropic linearDNA.The size of all other fragments of B- and N-tropic linear DNA wasidentical.
A
B
a
b
a
b3.8
-x
1.9-FIG. 7. Analysis of 5' derived sequences. B-tropic linear viral DNAwascleaved with HindIII (a) and
withHindIII-PstI (b).After digestion, samples were
run on a1.4% agarose gel; the DNA fragments were transferred onto nitrocellulose membrane and annealed with representative [32P]cDNA (A) or
[32P]cDNA
5'(B) prepared as described in the text.5' END OF RNA 3' END OF RNA
B-TROPIC DNA
Hindm Sal I XhoI XbaI PvuI PstI KpnI SmaI
P.1 1 .
.1 I I L
I I I
N-TROPIC DNA
o-,
_-FIG. 8. Restrictionendonuclease mapsoflinear
B-andN-tropic MuLV DNAs.Cleavagesitesonlinear B- andN-tropic DNAsproduced byHindIII, SalI,
XhoI, XbaI, PvuI, PstI, KpnI, and SmaI. Linear DNA isoriented with respecttothe5'and3'endsof
the RNA. Molecular weights ofthe fragments are giveninTable1.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.510.299.443.85.326.2] [image:9.510.70.260.380.601.2] [image:9.510.272.466.411.670.2]identicaltothoseobtainedwithB-tropic MuLV
DNA. Inaddition,ourstudyofB- and N-tropic
MuLV DNA showed that the N-tropic linear
molecule,ascompared to the B-tropic molecule,
is shorter at both ends byapproximately75 base
pairs.Taken together, these results suggest the presence of terminal repeated sequences on lin-ear B- and N-tropic MuLV DNAs as found in
otherretrovirus DNA.
DISCUSSION
In this
study,
we havemapped
thecleavage
sites ofeight
restriction endonucleases onBALB/c
endogenous
B- andN-tropic
MuLV DNAs, using the Southern agarosegel-DNA
transfer
procedure.
We have concentrated onrestriction
endonucleases thatrecognizeahex-anucleotide
sequenceand henceareexpected
tocleaveaDNA
molecule
of about6x 106daltons onlyafew times. The DNAfragments
have been ordered relativetothe 5' and 3' ends of the viral RNA molecule. Thegeneral
structure of both linear andcircular B- andN-tropic
MuLV DNA has been studied with sucharestriction endo-nucleaseanalysis.
Wehave found three enzymes
(PstI,
KpnI,
andSmaI) abletorecognize
acleavage
siteatboth
ends
of the linear viral DNA within afragment of0.35 x
10'
daltons(approximately
550base
pairs).
We havealso shown that the 5'-derivedsequences presentattheright
terminus oflinear DNA arerepeated
somewhere within the left third end of the linear DNA. The pres-ence of a5'-derived
sequence at the extremeright
end of linear viral DNA has been foundonother
retroviral
DNA(11,
34,35).
Theprecise
organization
of the distal ends of B- andN-tropic
MuLV DNAs has not beenfully
elucidated. However, our restriction enzymeanalysis
and cDNA 5'annealing
dataareconsistentwith thepresence of terminal redundant sequences on
the linear viral DNA of B- and
N-tropic
MuLVas
found
with other retrovirus DNA(4, 10-12,
18, 26, 33-35, 38, 43). Definitive evidence willrequire
sequencing
data.We havealsodetectedtwocircular DNA
spe-cies in B- and
N-tropic
MuLV-infected cells. Aftercleavage
of these circular DNAs withre-strictionenzymeswhichcleave thegenome
only
once
(Sall,
HindIII, XbaI,
XhoI, PvuI),
oneofthe linearmoleculesgeneratedhadthesamesize
(5.7 x
10W
daltons) as the linear moleculeex-tracted fromcells(Fig. 2).Thissuggests that the
largerof the twocircularDNAspeciesof B-and
N-tropic MuLV is a circularized form of the
linear viral DNA. The smaller circularspecies
appearsto berelated tothe
larger
species.
In-deed, cleavage of thetwocircular species with
enzymes which recognize more than one site
(SmaI,
KpnI, and PstI) generates fragments identicaltothose obtainedafter digestion of thelinear molecule (Fig. 5). This result indicates
that thesefragments areinternalandthat they
arecommon to both circular species as
well
asto the linear form. Therefore, if the
smaller
circular speciesis derived from the linear viralDNAorfrom the larger circular species, it must
be deleted from sequences outside the
internal
SmaI, KpnI, and PstI fragments.PstI
restric-tionendonuclease analysissuggests, in fact, thatthe smaller circular species is deleted from
se-quences present at one or bothends of the
linear
viral DNA because PstI cleaves linear viral
DNAclosetoboth ends of themolecule. From
thetwocircular DNA species, PstIgenerates a
single 5.35 x
106-dalton
fragment. Most likely,PstI
cleaves thelarger
circular DNA species twice in regions correspondingto both ends oflinear DNA and the
smaller
circularDNAspe-cies once to generate a 5.35 x
106-dalton
frag-ment.Therefore, the
smaller
circular DNAspe-ciesseems tobedeleted fromasequence present
at one (orboth) end(s) of the
linear
viralDNA.Thesedata
confirm
and extend previousresultsonthestructureofcircular DNA ofretroviruses
(12,18, 33, 34, 43).In eachcase,the
smaller
viralDNA species appears to lack one copy of the redundantsequence.
Out of14enzymestestedon B-and
N-tropic
MuLVDNAs,
only
one(Eco
RI)
failedtocleave thetwoMuLVDNAmolecules.Eight
restriction endonucleases(XhoI, PvuI, HindIII,
SalI,
XbaI,KpnI, SmaI,
andPstI) cleaveB- andN-tropic MuLV DNAsatidentical sites. This
sim-ilarityinthe restriction endonucleasepattern
of the two genomes wasexpected
since severalgroupshave
reported
thatBALB/c
B- andN-tropic MuLV genomes are
highly homologous
(5, 7,8, 22,30). However, ourdataprovide
evi-dence thatN-tropic viral DNA used inourstudy isnotidenticaltotheB-tropic
viral DNA. With four restriction endonucleases(PvuI, XbaI,
KpnI, andSmaI), wefound that theright
and leftterminal sequences ofN-tropic
linear viral DNA were shorter than theB-tropic
terminalsequences
(Fig.
3and5).
Wealso found that thesmaller fragment generated by digestion
ofN-tropic,
supercoiled
viral DNA withSmaI,
KpnI,andPstI wasshorter than thecorresponding
B-tropic fragment (Fig. 3). This
small
fragmentderived from
supercoiled
DNAislikely
torep-resent the fused fragments from both ends of
the linear molecule and, therefore, to contain
onecopy of the redundantsequence. This
differ-encebetween theterminalfragmentsof B- and
N-tropic viral DNA is unlikely to arise from
on November 10, 2019 by guest
http://jvi.asm.org/
additional cleavage sites, since itappears at both
ends of the molecule and is observed with five different restriction endonucleases.Mostlikely, this difference could be accounted forby dele-tion ofashortsegment (approximately 75base pairs) in the terminal redundantsequence. Ob-viously, suchasmall differenceinsizecouldnot
be detectedwith XhoI, HindIII, and SalI
be-causetheyproduceonly large fragments.
Physical mapping of theDNAof three
differ-entexogenousmurine retroviruses
(Mo-MuLV,
Mo-murinesarcoma virus, Ha-murine sarcoma
virus)has now beenreported (1, 4, 6, 10, 12,
41-43).Theavailability of thephysicalmapsof the
DNA of two MuLV endogenous to BALB/c
mouse should provide the basic information
needed tostudy the numerous endogenous
se-quences present in the mouse genome. Wehave
already found that restriction endonucleases able to cleave in the redundant sequences of BALB/c endogenous MuLV DNA give a very
simplified band pattern of virus-specific DNA aftercleavage ofmouseliver DNA from various strains (E.R. and P.J., unpublished data). This work should also allow comparisons of B- and N-tropic MuLV variants and give the informa-tion to map the tropism determinant onviral DNA. Thesize difference observed between the terminal sequences of N-Cl-35 and
B-Cl-il
MuLVDNAsisunlikelytoberelatedassuchto
thetropism determinant.Indeed, thispartof the
genome is notthoughttobecoding foraprotein,
and thetropism determinantseems tobea virus-coded protein (3, 17, 19, 25, 29). Moreover, an
endogenous B-tropicMuLV fromC57BL/6 mice hasrecently beenshown to havealso ashorter terminal sequence than
B-Cl-li
MuLV DNA(E.R. and P.J., unpublished data). Therefore,
thesequence
coding
for thetropism
determinanthasnotbeenidentified by thepresentrestriction analysis, since all the restriction sites studied
wereidenticalonB-andN-tropic viralDNA. A
finer
analysis with cloned DNAandconstruction ofspecific
recombinantswillbeneededtosolve thisproblem. Alongthis line and using the in-formation provided bythe present map, we have recently cloned both B- and N-tropic MuLV DNAmoleculesinCharon21A.Aftercompletion of this work, we learned that
aphysicalmap ofN-tropic MuLV DNA of AKR
mice had beenderived inWeinberg's laboratory
(R.A.Weinberg,personal communication).Five
restrictionendonucleasesusedin common
gen-erated identicalfragmentswith the same 3' to 5'
orientation, suggestingthat AKR N-tropic and
BALB/c B-tropic and N-tropic MuLV DNAs
share ahighly homologousstructure.Thisclose
homology of ecotropic MuLV from different strains is interesting in view of the fact that
AKR and BALB/c strains were derived from
differentancestors.
ACKNOWLEDGMENTS
Wethank Marcelle Bedard forpreparing this manuscript. This work was supported by grants from the Medical Research Council of Canada and the National Cancer Institute of Canada to P.J., and agrant from the Cancer Research Society Inc. to E.R.
LITERATURE CITED
1. Anderson, P., M. P.Goldfarb, and R. A.Weinberg. 1979.Characterization of in vitro synthesized Moloney sarcoma virus DNA: useof transfection in assignment of thetransforming functionto adefined subgenomic DNAfragment. Cell 16:63-75.
2. Baltimore, D., E.Gilboa, E. Rothenberg, and F. Yosh-imura. 1978. Production of a discrete infectious double-stranded DNAby reverse transcription in virions of Moloney murine leukemia virus. Cold Spring Harbor Symp. Quant. Biol. 43:869-874.
3. Bassin, R.H.,B.I.Gerwin, G.Duran-Troise, S. Gis-selbretch,and A. Rein. 1975.Murinesarcomavirus pseudotypesacquireadeterminantspecifying N- or B-tropism from leukaemia virus during rescue. Nature 256:223-225.
4. Benz, E.W., and D. Dina. 1979.Moloneymurine sar-coma virions synthesize full-length double-stranded DNA in vitro. Proc. Natl. Acad.Sci. U.S.A. 76:3294-3298.
5. Callahan, R.,R. E.Benveniste,M. M.Lieber, and G. J. Todaro. 1974. Nucleic acid homology of murine type-C viral genes. J. Virol. 14:1394-1403.
6. Canaani, E., K. C.Robbins,andS. A. Aaronson. 1979. The transforming gene of Moloney murine sarcoma virus. Nature282:378-383.
7. Faller,D.V.,and N.Hopkins.1978a.TIoligonucleotide mapsofN-, B-, and B - NB-tropic murine leukemia virusesderived from BALB/c. J. Virol. 26:143-152. 8. Faller, D. V., and N.Hopkins. 1978b.TI
oligonucleo-tides that segregate withtropism and with properties of gp70inrecombinants betweenN-andB-tropic murine leukemia viruses. J. Virol.26:153-158.
9. Gianni,A.M.,D.Smotkin,and R. A.Weinberg.1975. Murine leukemia virus: detection ofunintegrated dou-ble-stranded DNA forms of the provirus. Proc. Natl. Acad. Sci. U.S.A.72:447-451.
10. Gilboa,E., S.Goff,A.Shields,F.Yoshimura, S. Mi-tra,andD.Baltimore. 1979.In vitrosynthesisofa9 kbpterminallyredundant DNAcarryingtheinfectivity ofMoloneymurine leukemia virus. Cell 16:863-874. 11. Goldfarb,M.P.,and R. A.Weinberg. 1979.Physical
map ofbiologically active Harveysarcomavirus unin-tegratedlinear DNA. J.Virol. 32:30-39.
12. Hager, G.L.,E.H.Chang,H. W.Chan,C.F.Garon, M.A.Israel,M.A.Martin,E. M.Scolnick, andD. R.Lowy.1979. Molecularcloningof theHarvey sar-comavirus closed circularDNAintermediates:initial, structural, andbiologicalcharacterization.J.Virol.31: 795-809.
13. Hartley,J.W.,W. P.Rowe, and R.J.Huebner.1970. Host-range restrictions ofmurineleukemia virusesin mouseembryo cell cultures.J.Virol.5:221-225. 14. Haseltine, W.A.,D.G.Kleid,A.Panet,E.
Rothen-berg, andD.Baltimore.1976.Orderedtranscription of RNA tumorvirus genomes. J.Mol. Biol. 106:109-131.
15. Hirt,B. 1967.Selectiveextraction ofpolyomaDNA from infectedmousecell culture.J.Mol. Biol.26:365-369. 16. Hopkins,N.,J.Schindler, andP. D. Gottlieb. 1977.
Evidence forrecombinationbetweenN- andB-tropic murineleukemiaviruses.J.Virol.21:1074-1078.
on November 10, 2019 by guest
http://jvi.asm.org/
17. Hopkins, N., J. Schindler, and R. Hynes. 1977. Six NB-tropic murineleukemia virusesderived froma B-tropic virusof BALB/c have altered p30. J. Virol. 21: 309-318.
18. Hsu, T. W., J.L.Sabran,G. E.Mark,R. V.Guntaka, and J. M.Taylor.1978.Analysisofunintegratedavian RNA tumor virusdouble-stranded DNA intermediates. J. Virol. 28:810-818.
19. Ishimoto,A., J. W. Hartley, and W. P. Rowe. 1979. Fv-1 restriction ofxenotropicand amphotropic murine leukemia virus genomes phenotypically mixed with eco-tropic virus. Virology 93:215-225.
20. Jolicoeur, P. 1979. TheFv-l gene of the mouse and its control of murine leukemia virusreplication.Curr.Top. Microbiol.Immunol. 86:67-122.
21. Jolicoeur,P.,andD.Baltimore.1975.Effect of the Fv-1locus on the titration of murineleukemiaviruses. J. Virol. 16:1593-1598.
22. Jolicoeur,P.,and D. Baltimore. 1976a. Effect of Fv-1 gene product on synthesis ofN-tropic and B-tropic murineleukemiaviralRNA.Cell7:33-39.
23. Jolicoeur, P.,andD.Baltimore. 1976b. Effect of Fv-1 geneproductonproviralDNAformation and integra-tion incells infected with murine leukemia viruses. Proc.Natl. Acad. Sci. U.S.A. 73:2236-2240.
24. Jolicoeur, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA incelisinfected with murine leukemia virus. J. Virol.33:183-195.
25. Kashmiri,S. V. S., A.Rein,R.H.Bassin,B.L. Gerwin, and S. Gisselbretch.1977.Donation of N-orB-tropic phenotype to NB-tropic murine leukemia virus during mixed infections.J.Virol.22:626-633.
26. Keshet,E., J. J.O'Rear, and H.M.Temin.1979.DNA ofnoninfectious and infectiousintegrated spleen necro-sis virus(SNV)iscolinear with unintegrated SNV DNA andnotgrosslyabnormal.Cell16:51-61.
27. Lilly, F.,and T. Pincus.1973.Genetic control of murine viralleukemogenesis. Adv. Cancer Res. 17:231-277. 28.Pincus, T.,J. W.Hartley, and W. P. Rowe. 1971. A
major geneticlocua affectingresistance to infectionwith murineleukemia viruses. I. Tissue culture studiesof naturally occurring viruses. J. Exp. Med. 133:1219-1233.
29. Rein, A., S. V. S.Kashmiri,R.H.Bassin, B. I.Gerwin, and G. Duran-Troise. 1976. Phenotypicmixing be-tweenN- andB-tropicmurineleukemia viruses: infec-tiousparticleswith dualsensitivitytoFv-lrestriction. Cell 7:373-379.
30. Robbins,K.C., C.D.Cabradilla,J. R.Stephenson, and S. A. Aaronson. 1977. Segregation ofgenetic information for a B-tropic leukemia virus with the structurallocus for BALB: virus 1.Proc. Natl. Acad. Sci.U.S.A. 74:2953-2957.
31. Rommelaere, J.,H. Donis-Keller, and N. Hopkins. 1979.RNA sequencingprovidesevidence forallelismof
determinants of N-, B-, orNB-tropismof murine leu-kemia viruses.Cell16:43-50.
32. Schindler, J.,R.Hynes,and N.Hopkins. 1977. Evi-dence forrecombinationbetween N- andB-tropic mu-rineleukemia viruses: analysis of three virionproteins by sodium dodecyl sulfate-polyacrylamide gel electro-phoresis. J.Virol. 23:700-707.
33. Shank, P. R., J. C. Cohen, H. E. Varmus, K. R. Yamamoto, andG. M. Ringold. 1978. Mappingof linear and circular forms of mousemanmnary tumor virus DNAwith restriction endonucleases: evidence for alarge specific deletion occurringathigh frequency during circularization. Proc. Natl. Acad. Sci.U.S.A.75: 2112-2116.
34. Shank,P.R., S.H.Hughes,H. J.Kung,J. E.Majors, N. Quintrel, R. V.Guntaka, J. M. Bishop, and H. E.Varmus. 1978.Mapping unintegratedaviansarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in twospecies of circular DNA. Cell15:1383-1395.
35. Sherr, C. J., L.A.Fedele, L. Donner,andL.P.Turek. 1979.Restriction endonucleasemapping of unintegrated proviralDNA of Snyder-Theilen feline sarcoma virus: localizationof sarcoma-specific sequences. J. Virol. 32: 860-875.
36. Southern, E. M. 1975.Detection ofspecific sequences among DNAfragmentsseparatedbygel electrophore-sis.J. Mol. Biol. 98:503-517.
37. Sveda,M.M.,and R. Soeiro. 1976.Hostrestrictionof Friend leukemia virus: synthesis and integration of the provirus.Proc.Natl. Acad.Sci. U.S.A.73:2356-2360. 38. Taylor, J. M., T. W. Hsu, and M. M. C.Lai. 1978.
Restriction enzyme sites on the avian RNA tumorvirus genome.J. Virol. 26:479-484.
39. Taylor,J. M.,R. Illmensee, and J.Summers. 1976. Efficient transcription of RNA into DNA by avian sarcomaviruspolymerase. Biochim. Biophys. Acta 442: 324-330.
40. Varmus,H.E.,R. V.Guntaka, C.T.Deng,andJ.M. Bishop.1974.Synthesis,structureandfunction ofavian sarcomavirus-specificDNA inpermissiveand nonper-missive cells.ColdSpringHarborSymp. Quant.Biol. 39:987-996.
41. Verma,L.M.1979.Genome organizationofretroviruses. III.Restrictionendonucleasecleavagemapsofmouse sarcomavirusdouble-stranded DNAsynthesizedin vi-tro.Nucleic Acids Res. 6:1863-1867.
42.Verma, I. M., and M. A. McKennett. 1978. Genome organizationof RNA tumor viruses.II.Physicalmaps of invitro-synthesized Moloney murine leukemia virus double-strandedDNAbyrestrictionendonucleases.J. Virol.26:630-645.
43. Yoshimura,F.K., andR. A.Weinberg.1979. Restric-tionendonucleasecleavageoflinear and closedcircular murineleukemia viral DNAs: discovery of asmaller circularform.Cell 16:323-332.