0022-538X/80/08-0306/08$02.00/0
Multiple
mRNA
Species
for Adenovirus
Type
2
Polypeptides
III
and
pVII
CHARLES B. LAWRENCE
Department ofCellBiology, Baylor College of Medicine, Texas MedicalCenter, Houston,Texas77030,* and
The SalkInstitute, Tumor
Virology Laboratory,
SanDiego,California
92138Fractionationofmessengeractivities isolated from thecytoplasmofHeLacells
late in infectionwith adenovirus type 2 reveals that viral polypeptides III and
pVIIareeach synthesizedfrom two different-sized mRNA's. Themajormessenger
activity for each protein has the same sedimentation rate as that previously reported byAnderson et al. (Proc. Natl. Acad. Sci. U.S.A. 71:2756-2760, 1974). The minormessengeractivities for III andpVII sediment morerapidlyandare
notaggregatesof themajormRNA's for theseproteins.Thetwo minormessenger
activities cosediment with two polyadenylated RNA species which are labeled
late in infection with 32Pand whosemolecularweights areestimatedtobe 2.9 x
106 and 2.4 x 106. Both of these species hybridize to adenovirus type 2 DNA
specific for the mRNA family that is 3' coterminal at adenovirus type 2 map
position49.5 and the mRNAfamilythat is 3' coterminalat 62.0. This isconsistent
with the possibilitythat these RNAs have 5'-terminalsequencesidentical tothose
of the normal mRNA's for III andpVIIbutare3' coterminalatmapposition 62,
the normal 3' terminus of the mRNA's forpolypeptidesIIandpVI.Thesespecies
arenotfound inpolyadenylatedRNA isolated from the nucleus,suggestingthat
the minor mRNAspeciesarecytoplasmicRNAs.
Thelate mRNA'sof adenovirus type2 (Ad2)
coded tothe right of Ad2 map position 16 are
arranged in five families whose members have
overlapping 3'-terminal sequencesanddifferent
lengthsof sequencesin the5'regions (6, 16, 19,
24). These mRNA's are synthesized by
differ-ential processing of a single unique transcript
which is initiatedatposition 16and transcribed
rightward to theendof the genome (7, 8, 23, 25).
Cleavage and polyadenylation of the nascent
transcript occur at one of five possible sites which correspond to the 3' termini of the five mRNA families (20). The partially processed
polyadenylated transcript is then further
proc-essedby thesplicingof sequencesat16, 19,and
27 toform a 200-base 5' leader sequence (3, 5,
10) which is subsequently spliced to one of a limited number of acceptor sites coded within
the family that lies to the 5' side of the poly-adenylation site. The slicing of the leader to the mainbody of the mature mRNA may occur by way of a series of intermediates in which the leader is transferred to various acceptor sites along the transcript between the leader and the mainbody of the mRNA (18, 20).
In the course of fractionating cytoplasmic RNAs isolated from HeLa cellsduring the late phase of Ad2 infection, we observed two size classes of messenger activities for each of the viralproteinsIIIandpVII. The major messenger
activities corresponded in size to those
previ-ously reported for these proteins. The minor messenger activities for these proteins were,
however, much larger and may correspond to
molecules which have a 3' terminusatthe site
normallypresentinthemRNAfamily
immedi-ately downstreamfrom thefamily that encodes
IIIand pVII.
MATERIALS AND METHODS
Cells. HeLa S3 cellswere grownin suspension in
Joklik-modified Eagleminimalessentialmedium
sup-plemented
with 5%calfserum.Virus. Stocks of Ad2were obtained from Gernot
Walter. Cells were infected at a multiplicity of 20
PFU/cell. Virus was adsorbed for 30 min at room
temperatureandataconcentrationof4x10'cellsper
nml. Cellswerethendilutedto aconcentration of4 x
I105/mlwithfresh medium, andinfection was allowed
tocontinuefor24h.
Labelingof cells. Cellswerelabeledat24 hafter
infectionwith32p; (ICNPharmaceuticals, carrier free). Infectedcells (100 ml) were collected by centrifugation at 1,000 rpm and washed once with Eagle minimal essential medium without phosphate. Cellswere
re-suspended in 50 ml of this phosphate-free medium
supplementedwith2%dialyzedcalf serum;25mCi of
32p;wasadded,andthe cells were incubated for 3 h at
370C.
Preparationof RNA. At24hafter infection with
Ad2,4 x 10' HeLa cellswerecollectedby centrifuga-tionat 1,000 rpm, washedoncewith cold saline,and
306
on November 10, 2019 by guest
http://jvi.asm.org/
Ad2 mRNA 307
suspended in 1.7ml of cold isotonic lysis buffer (0.15
MNH4CI, 10 mM Tris[pH6.8],2mMMgCl2, 50
Ag
ofdextransulfate per ml). Then 0.2 ml of 10% Nonidet
P-40(Shell Chemical Co.) was added, followed by brief
blending in a Vortex mixer;0.1 ml of 10% deoxycholate
was added, and the suspension was blended for 30 s.
Nuclei werepelleted by centrifugation at 2,000 rpm for
2min, and then 90
AI
of 0.5 M.EDTA (pH 7.0), 80ydof 4 MNaCl, 0.2 ml of 10% sodium dodecyl sulfate,
and 20[L of,B-mercaptoethanol were added to the
supernatant. To makecytoplasmic RNA, the
super-natant waswarmed to30°C and extracted with2 ml
of phenol. Chloroform (2 ml) was added, and the
mixture was reextracted. Phases were separated by
centrifugationat 10,000 rpm for 20 min at25°C. The
aqueous phase wasreextracted with 2 mlof
chloro-form, and the phases wereseparated as above. The
aqueousphasewasthencollected, and RNAwas
pre-cipitated by the addition of 2 volumes of ethanol.
Precipitated RNA was stored in aqueous ethanol at
-20°C. Polyadenylic acid [poly(A)]-containing
cyto-plasmic RNA was obtained by chromatography on
oligodeoxythymidylic acid-cellulose (T-3;
Collabora-tiveResearch, Inc.).
To make nuclear RNA, nuclearpellets were
sus-pended in1.0mlof0.5MNaCl-50 mMMgCl2-0.01M
Tris (pH 7.5), 60
A1
of0.5 M EDTA, 50t1I
of20%sodiumdodecyl sulfate, and 100 1Iof proteinase K (5
mg/ml)wereadded, and the suspensionwasincubated
at roomtemperature for 15min.Water (1.0 ml) and
BO-mercaptoethanol
(20tlI)
were added, and thesus-pensionwasagitated briefly. The suspensionwas
ex-tractedwithphenol and chloroformasdescribed above
forcytoplasmicRNA.
Poly(A)-containing nuclear RNA was selectedby
mixing the aqueous phase from theextracted nuclei
with anequal volume of0.5M LiCl, 1 mM EDTA,
and10mMTris(pH 7.5).Oligodeoxythymidylic
acid-cellulose (0.2 g)wasadded andkept in suspensionby
intermittentagitationat4°Cfor30min, thencollected
bylow-speedcentrifugation and washed three times
with 5 ml ofice-cold0.5MLiCl-1 mM EDTA-10 mM
Tris(pH 7.5). Bound RNAwaselutedbywashingthe
oligodeoxythymidylicacid-cellulosetwotimeswith1.0
ml of sterile water at roomtemperature. The eluted
RNA was adjustedto 0.2 Msodium acetate(pH5.2)
andprecipitated with2volumes ofethanol.
Sucrosegradientcentrifugation.Nuclearor
cy-toplasmic RNAs in a total volume of0.5 ml were
layeredon10.6-mlgradients of10 to30%sucrose(wt/
wt) in 10 mMsodiumacetate(pH5.2)-imM
EDTA-0.1%sodiumdodecylsulfate. Centrifugationwas
per-formed at-20°C for4.5 hat 40,000 rpm for nuclear
RNAs and for 15 h at 30,000 rpm for cytoplasmic
RNAs in aBeckmanSW41rotor.
Agarosegelelectrophoresis. [32P]RNA was
re-acted with glyoxal according to the procedure of McMaster and Carmichael (17). After reaction with
glyoxal,thevarioussamplesweremixed with anequal
volumeof 1 mM EDTA(pH7.0)-50%glycerol-0.001%
bromophenol blue and applied directly to a
4-mm-thick 1%agarosegel(Sigma Type V)containing Leon-ingbuffer Eplus0.1% sodiumdodecyl sulfate.
Electro-phoresiswas for 18 h at 75 V. After electrophoresis,
the gels were dried onto Whatman 3 MM paper.
Radioactive RNAspecies werevisualized by
autora-diography enhanced with Du Pont Cronex
Lightning-Plusintensifying screens.
Cell-free translation. RNAs were translated in
the messenger-dependent rabbit reticulocyte lysate
systemdescribedby Pelham and Jackson (21) in the
presenceof[35S]methionine (Amersham Corp., 1,000
Ci/mmol).
Polyacrylamide gel electrophoresis. Proteins synthesized in vitro were resolved by diluting reaction
mixtures with5volumes of sample buffer and applying
5
p1
to eachlane ofa 1-mm-thick,9.5-cm-longpoly-acrylamideslab gel according to the system described
by Laemmli and Maizel (12, 15). Electrophoresis was for4.5hat a constantcurrentof 15 mA.
DNA-paper andhybridization protocols.
Puri-fied plasmid DNA was coupled to
diazobenzyloxy-methyl-paper by the method of Stark and Williams
(22). Each coupling reaction contained 100
,Ag
ofsheared, denatured plasmid DNA and a
1.0-cm-diam-eterpapercircle.
Hybridizationswereperformed in 50%
formamide-0.4 M NaCl-20 mM PIPES
[piperazine-N,N'-bis(2-ethanesulfonicacid)] (pH 6.4)-5 mM EDTA-0.2%
so-diumdodecyl sulfate at37°C for 5h. Each reaction
contained one DNA-paper circle, 200 ,ug of yeast
tRNA, and 100,000 to 500,000 dpm of 32P-labeled
poly(A)-containing RNA in afinal volume of140 Il.
The DNA-paperwaswashed three times (15 min each
time) in50%formamide-0.1 MNaCl-10 mM PIPES
(pH 6.4)-5 mM EDTA-0.2% sodium dodecyl sulfate at
32°C, andtwotimes (5mineach time) in 0.2 M
NaCl-20mM PIPES (pH 6.4)-5 mM EDTA-0.1% sodium
dodecyl sulfateat55°C. Hybridized RNA was eluted
by washing the DNA-papertwotimes with 100ulof
99%formamide-1 mM EDTA at55°C and one time
with 200
pl
of 1.0mM EDTA at55°C. Eluted RNAwasprecipitatedfrom the pooled washes by the
addi-tion of20 ug of yeast tRNA, 40
pl
of2 M sodiumacetate(pH 5.2) and1ml of ethanolat-20°C.
RESULTS
Two messenger activities for m and
pVH.
Totalcytoplasmic
RNAwasisolated fromAd2-infected HeLa cellsat 24hafter infection
and fractionated on a sucrosegradient.Aportion
of each fractionwastranslated in the
messenger-dependent rabbitreticulocyte
lysate
in thepres-enceof
[35S]methionine,
andtheproductswereanalyzed by
polyacrylamide
gel
electrophoresis.
Afluorogram ofthisgel isshown inFig. 1.The
major activitiesfor III andpVII sedimentedat
25S and21S,respectively,aspreviously reported
by Andersonetal. (1). Minormessenger
activi-ties for III and
pVII
werealsofound,
sediment-ing at 32S and30S,respectively (Fig.1, fractions 5, 6, and7).To examine the possibility that these
repre-sent aggregated forms of the major messenger
activities,
32P-labeled, poly(A)-containing
RNAVOL. 35,1980
on November 10, 2019 by guest
http://jvi.asm.org/
308 LAWRENCE
w 4 O N
15 ;
.iw.*...
...A,.XX
-_
..._____i
_______..v.::^:
__w.=E
_LZ===
_ __ _ _ ___Fr-111|
9-. X -_ Y ..
k . y*d
SLt-AR&.;&4s..5..ss#
us =33; ze...aw,."-4w.
FIG. 1. Translationof size-fractionated Ad2 mRNA's. A100-,.gsampleofpoly(A)-containingRNA isolated
from the cytoplasmofHeLa cells infected for24hwith Ad2 was fractionated on sucrose gradients as described
in the text. Thegradient was dividedinto 0.45-mlfractions, and RNA in each fraction was collected by
ethanolprecipitation and then dissolved in 50ytlof water; Itldof eachwastranslated in a 10-,Il reaction
mixtureasdescribed in thetext.One-tenthof each reaction mixture was analyzed on a 12.5% polyacrylamide
gel, and35S-labeledproteinswerevisualizedby fluorography.
wasisolatedfrom HeLa cellsat24hafter
infec-tion with Ad2. The RNA was sedimented
througha sucrosegradient, andaportion of each
fractionwas translated and analyzed as shown
inFig. 1.Thefaster-sedimentingmessenger
ac-tivitiesfor III andpVII werepooled, aswellas
themajor peak ofactivityfor III.ThetwoRNA
poolswereprecipitated, dissolvedinbuffer, and
heatdenatured; each was sedimented through a second gradient. The peak fractions of
radioac-tivity from each gradient were again collected
andsedimentedthrough a third gradient as
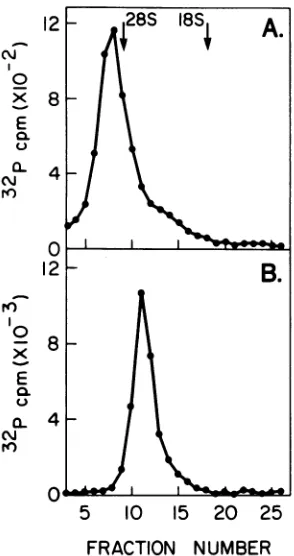
de-scribed above. The radioactivity profiles from the thirdgardientareshown inFig.2.[32P]RNA
associatedwith thefast-sedimenting messenger activities for III and pVII (Fig. 2A) still sedi-mented faster than did the[32P]RNAassociated
withthemajorIII messengeractivity (Fig. 2B).
The RNAs in the peak fractions from each
gradient were precipitated, and a sample of
eachwastranslated invitro in the presence of
[35S]methionine and analyzed on a
polyacryl-amidegel. The translation productsareshown inFig. 3.The peaks of minor messenger activi-ties for III and pVII (Fig. 3A) and the major
peak of III mRNA activity (Fig. 3B) cosedi-mented with their respective peaks of
radioac-tivity (Fig. 2A and B). The minor messenger
activity for III sedimented slightly faster than
did the minor messenger activity for pVII, sug-gesting the presence of two distinct RNA spe-cies. It was apparent that the minor messenger activities for III and pVIIstillsedimented more
rapidlythan did the major activity for III, even
after3cyclesofheat denaturation and
sedimen-tation.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.504.68.446.69.404.2]Ad2 mRNA 309
C\j
0
x E
a.
CM N
0 x E a.
C-NtL
5
10
15
20
25
molecular-weight speciesais theminormRNA
for
III and thatspecies b is the minor mRNAforpVII. The peak fraction from Fig. 2B (the
major mRNA for III) contains three labeled
bands(species c,d, ande), whichmigrated more
rapidly than did species a and b (Fig. 4, lanes 2
and5). Since the peak fraction from Fig. 2B is
associated withmessengeractivitiesfor the III,
lOOK, and pVI proteins (Fig. 3B) specified by
4 5 6 7 8 9 O 11 12 13 14 15
A
-III
-PVII
FRACTION NUMBER
FIG. 2. Sedimentation of32P-labeledAd2 mRNA's
afterthree cycles ofsedimentation and heat
denatur-ation.32P-labeled,polyadenylatedRNA was isolated
asdescribed in the text. Minor mRNA's(30S and 32S)
forpolypeptides III and pVII (A) and the major
mRNAforIII(B) were isolatedfrom a sucrose
gra-dient andsubjected to two subsequent cycles of heat
denaturation and sedimentation. The figure shows
radioactivity present in each fraction of the third
sucrosegradient.
4 5 6 7 8 9 10 11 12 13 14 15
-B
m
-100K
- III
Toconfirm that therapidly sedimenting
activ-ities for III and pVII were not aggregates of
smaller RNAs, the
peak
fraction ofradioactivityfromeachgradientwastreated withglyoxal (17)
andanalyzedon anagarosegel. The peak
frac-tion of radioactivity from Fig. 2A (which has
messenger activity for both III and pVII)
con-tainedtwo
species
of RNA(Fig.
4,lanes1and4)designated a and b. Ithas recently been
dem-onstrated that the only polyadenylated RNAs
transported
to the cytoplasm late in infectionare adenovirus specific (2). It is demonstrated
below that the 32P-labeled species a and b are
cytoplasmic and contain sequences from the
mRNA familylocated at map positions 39.0 to
49.5, strongly suggesting that they do in fact
correspond tothe minormRNA's for
polypep-tides III andpVII. Since the minor mRNA for
IIIsedimented
slightly
faster than did the minormRNA for
pVII,
it is likely that the [image:4.504.80.227.66.344.2]higher-_ _-iUbam -pVI
FIG. 3. TranslationofRNAsisolatedasdescribed in thelegendtoFig.2.RNA ineachfractionofthe
gradientsinFig.2wascollectedbyethanol
precipi-tation and dissolvedin 10,ilofwater,1,ld fromeach
fraction was translatedinvitro, and 35S-labeled
pro-teinswereanalyzedon a10%polyacrylamidegeland
visualizedbyfluorography, asdescribed inthetext.
Fractions in(A) and(B) arethesameasthosefrom
Fig.2(A)and(B),respectively.
VOL. 35,1980
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.504.262.457.139.567.2]50.1)
restrictionfragment
immobilized ondia-zobenzyloxymethyl-paper.
Hybridized
RNAwaseluted,
treated withglyoxal,
andanalyzed
on anagarose
gel.
Anautoradiograph
of thisgel
is shown inFig.
5. Twomajor
32P-labeledspecies
which
migrated
faster than did 28S RNAhy-bridizedtothisDNA
(Fig.
5,
lane3).They
mostlikely
correspond
to themajor
mRNA's for III andpVII.
The minorspecies
wehavedesignated
aand b also annealed
specifically
toHindIIIDDNA, demonstrating
the presence ofsequencesfrom map
positions
41.0to50.1 in thesespecies.
Because theradiolabeled
species
a and bco-sedimented with the minor messenger activities for III and
pVII
and contained sequencesfrom theregion
oftheAd2 genomeknowntocodefor theseproteins,
we conclude that thesespecies
are in fact the minor mRNA's for these two
proteins.
Sinceonly
5'-terminal genesonpoten-tially
polycistronic eucaryotic
mRNA's aretranslated
(11),
it follows that thecoding
se-quences for III and
pVII
arelikely
tobe onthe 5'terminus ofspecies
aand b and thatadditional sequencesnotnormally
presentin III andpVII
mRNA's are present on their 3' termini. One
possibility
isthat theseadditional sequencesarecoded
by
aregion
of the Ad2 genomeimmedi-ately
downstream from the III andpVIImRNAfamily, namely,
theregion
whichnormally spec-ifies II andpVI
mRNA's. Totestthishypothesis,
32P-labeled,
cytoplasmic,
poly(A)-containing
RNAisolated fromAd2-infectedHeLa cellswas
fft..
FIG. 4. Agarose gel electrophoresis of
[32P]-mRNA's.32P-labeled RNAsweretreatedwithglyoxal
andanalyzedon a1.0%/agarosegel asdescribedin the text. Lane 1, Fraction 8 from Fig. 2A; lane 2,
fraction 11 from Fig. 2B; lane 3, total 32P-labeled,
poly(A)-containing RNA from HeLa cells infected
withAd2for24h;lanes 4to6, same asI to3except
exposed longer; lane 7, ['4C]rRNA's; Lane 8, same aslanes 3 and 6.
adenovirus,
it was notsurprising
to find three32p-labeled
RNAspecies
in this fraction. Total32p-labeled, poly(A)-containing,
late Ad2 RNAwas also run in a
parallel
lane(Fig.
4, lanes 3 and 6).Species
a, b, c, and dareevident inthis RNA.Species f
and gareidentified belowasthe mRNA forhexon and the 215 mRNAforpVll,
respectively.
32S and 30S
32P-labeled species
havese-quences from Ad2 map
positions
41.0 to50.1 and 56.5 to
63.7.
Toprovethat thehigh-molecular-weight
32p-labeled
RNAspecies
con-tained sequences from the
region
of the Ad2genome
coding
for III andpVIL, 32p-labeled,
cytoplasmic,
poly(A)-containing
RNA from Ad2-infected HeLa cells washybridized
to acloned Ad2 HindIII D
(map position
41.0 toa.
-:*
I.
FIG. 5. Selectionof 32P-labeledcytoplasmic RNAs
on restriction fragments ofAd2 DNA. 32P-labeled
cytoplasmic,poly(A)-containingRNA washybridized
to cloned Ad2restriction fragments asdescribedin
thetext.SelectedRNAwastreatedwith glyoxal and
analyzedon a1.2%oagarosegel.Lane 1,
["4C]rRNA;
lane2,total32P-labeled,
cytoplasmicpoly(A)-contain-ingRNA isolatedfromHeLa cells infected with Ad2
for 24 h; lane 3, 32P-labeled RNA hybridizing to
cloned Ad2HindIIIDDNA;lane 4,32P-labeled RNA
hybridizing tocloned Ad2 PstE DNA; lane 5, same
aslane4exceptlongerexposureofautoradiograph.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.504.59.250.69.387.2] [image:5.504.282.426.401.543.2]Ad2 mRNA 311
hybridized to aclonedAd2 PstE (map position
56.5 to63.7)restrictionfragment immobilizedon
diazobenzyloxymethyl-paper.
Two major spe-cies of RNA hybridized to this DNA fragment (Fig. 5, lanes 4 and 5), corresponding to the mRNA's for pVI and II. Both species a and balso hybridizedto this DNA,demonstratingthe
presence of sequences coded inthe 56.5 to 63.7 region in these RNAs. Species a and b do not
hybridizeto DNAspecific for fiber mRNA (data
notshown).
The 32S and 30S mRNA's are not found
in the nucleus. The presence of
high-molecu-lar-weightmessenger activities for III and pVII
among cytoplasmic mRNA's suggested that
some
partially
processed nuclear RNAs mayhave been present in our cytoplasmic mRNA
preparations. To determine whether species a
and b werepresent in thenucleus,
32P-labeled,
nuclear, poly(A)-containing RNA was isolated
and sedimented through asucrose gradient. A
sample of each fraction in the 20S-to-45S size
range wastreated wtihglyoxaland analyzedon an agarose gel. Figure 6 shows an
autoradi-ograph of thisgel. A numberof prominent
la-beledbands canberesolved; however, none of
them comigrated with bandsaand b.One
prom-inent nuclear speciesmigratedveryclosetoband
a but was
consistently
found to migratejust
slower than a in a numberofdifferent
experi-ments.Nonuclear RNAspecies
comigrated
withband b.Thisexperiment demonstrates that the
presence ofspecies aand b in the
cytoplasmic
RNA is notdue to contamination ofour
cyto-plasmic preparations with nuclear RNA. The
possibilitythatsmall amountsof
species
aandbarepresent inthe
nucleus, however,
cannotbe excluded.DISCUSSION
The major messenger activities for III and
pVII havepreviously beenshowntosedimentat
approximately 25S and 21S,
respectively
(1).These mRNA's have been
mapped
onthe Ad2genome to the mRNA
family
which hasacom-mon3' terminusatabout map
position
49.5(4,
6, 13, 14). The 5'endsofthemain
body
of themRNA's for III andpVII are
probably
locatedat 38.8and 42.8,
respectively (4, 6).
These mapcoordinatesareconsistent with theobservedsize
of the mRNA's from these proteins as deter-mined bysedimentation
(Fig. 1)
andmigration
ongels (Fig. 4). Thus,thesetwomRNA's
overlap
in sequence, with III mRNA
having
additionalsequences presentonthe5'portionof the mol-ecule which arelikelytobe those that
actually
code for the IIIpolypeptide.The sequences that code forpVII are present
internally
in III mRNAbut arenot translated.
The present paper describes two minor
mRNA's codingfor III and pVII which are
con-siderably greaterinsizethanthe major mRNA
species for these proteins. Assuming that the
coding region for these proteins is at the 5'end
of these large molecules, then the additional
sequences mustbeatthe3'end of the molecule.
The large messenger activities cosedimented
with two 32P-labeled cytoplasmic
poly(A)-con-taining RNA species (designatedaand b) which
hybridizeto aregion of the Ad2 genome specific
for theIII andpVII mRNA familyand also toa
regionspecificfortheIIand pVI mRNA family.
The molecular weights of a and b have been
estimated by comparison of the migration of
glyoxal-treated species with the migration of
glyoxal-treated 18S and 28S rRNA's (Fig. 4,
lanes 7and9).Themolecularweights of a and
b were thus determined to be about 3.3 x
106
and 2.7 x 106, respectively. Using the same
standards, the molecular weight of hexon mRNA
(speciesf ) wasestimatedtobe about 1.5 x 106.
However, the heteroduplex analyses of Chowet
al. (4, 6) indicate that hexon mRNAspansabout
10.5 map units of the Ad2 genome (molecular
weight,23 x106)(9), whichwouldcorrespond to
amolecularweight of about1.3 x 106, including
leadersequence and poly(A). Thus,use of 18S
and28S rRNAasmolecularweight standards in
this gel system apparently results in a small
overestimation ofthemolecular weight of hexon
mRNA. Ifwe have overestimated thesize of a
and bbyasimilarproportion, their actual
mo-lecularweights would be2.9x 106 and2.4x 106
and would span 24 and 20 map units of the
genome, respectively. Therefore, these
mole-cules are each approximately 10.7 map units
longer than the major mRNA's for III and pVII
(1,4,6). This isquite close tothe
length
of themRNA family that lies
immediately
down-stream from the III and pVII
family.
Since aand b are composed in partofsequences from
this downstream region, it is
likely
that theseRNAspecieshave 5' sequences thatareidentical
to those of the normal mRNA's from III and
pVIIthatare3'coterminal with the mRNA's for
pVI and II
(hexon)
atmapposition
62instead ofthe normal 3' terminus for the III and
pVII
mRNA'sat49.5.
Howmightsuchmolecules arise? The
major-ityofmoleculeswith3'termini codedat62 are
destinedtobecomemRNA'sfor
pVI
orII(4, 6,
20).Thus,it is
likely
that thelarge
mRNA'sforIII and pVII arise
by
incomplete
or aberrantprocessingoftranscriptswhich
normally
wouldproducepVIorII mRNA's.
Processing
of thesetranscriptswouldnormally involve the
complete
VOL. 35,1980
on November 10, 2019 by guest
http://jvi.asm.org/
312 LAWRENCE
FIG. 6. Analysis of20S to 45S
32p_labeled,
nuclear, poly(A)-containing RNAs on an agarosegel. 32P-labeled, nuclear,poly(A)-containingRNA wasfractionatedon asucrosegradient. RNA in the 20S to45Sregionofthegradient (fractions8to20asindicated) and totalpoly(A)-containing cytoplasmicRNA were
treatedwithglyoxalandanalyzedon a 1.0% agarosegelasdescribed inthetext.
removal of sequences betweenthe3' end of the leader sequence at map
position
27 and the 5' end of the body of pVI mRNA at 49.5. Theprocessing of the fiber mRNA precursor
appar-ently involves the removal ofRNA sequences
coded between the leader andmain
body
ofthe mRNA in a series of steps (18), which may involve a transfer of the 5' leader to variousacceptorsitesalongthelengthofthe precursor. If theprocessing of the mRNA precursor for II andpVImRNA's occurs in steps in which the 3' end of the leader is first spliced to 5'-mRNA sequences in the familywhich normally codes for III andpVII and then issubsequentlyspliced
to49.5 (pVI mRNA) or 51.2 (II mRNA), then
the large mRNA's for III and pVII might be
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.504.118.395.74.534.2]Ad2 mRNA 313
intermediates in this pathway which are
trans-ported to the cytoplasm before completion of
processing.
Alternatively, if removal ofsequencesbetween map positions 27 and 49.5 or 51.2 occurs
nor-mally in a single step, then the large mRNA's for III andpVII could arise byaprocessingstep
in which sequences at map position 27 are
splicedtosequencesat38.8 (III mRNA)or42.8
(pVIImRNA). This would beanabnormal splice
fora transcript witha3' terminusat62butan
entirely normal splice for molecules which have 3' termini at 49.5. In this event, the large mRNA's forIII and pVII would be formed from
anabnormalprocessing eventofa precursorto
pVIorIImRNA resulting inalarge mRNA for
IIIorpVII. Suchamoleculemaynotbe further processed becauseanappropriate nucleotide se-quence or RNA secondary structure which
would allow the leader to be spliced again to map position 49.5 or 51.2 may not be present oncetheleader has beensplicedto38.8or42.8.
Further analysis of the structure of interme-diates in the synthesis of pVI and II mRNAs should distinguish between these two possibili-ties. In theeventthatspeciesa and bproveto
be normal intermediates in thesynthesis of these mRNA's, they may be usefulas substrates for
studying the biochemistry of mRNAprocessing becausetheyarereadily isolated from
cytoplas-micextractsof infectedcells.
ACKNOWLEDGMENTS
I gratefully acknowledge thegenerous supportofTony Hunterand GemotWalter,in whoselaboratoriespartof this workwasperformed,and I thank SueBergetforproviding
cloned Ad2 DNA restrictionfragments.
Thisworkwassupportedin partbyPublic Health Service
grant CA 17096from theNational Institutes of Health to TonyHunter. The authorwassupported byPublic Health Service National Research Service award CA06037,a
fellow-shipfrom the LeukemiaSocietyofAmerica,and Public Health Service grantAI16484 from the National Institutes of Health.
LITERATURE CITED
1. Anderson,C.W.,J.B.Lewis,J. F.Atkins,and R. F.
Gesteland. 1974. Cell-freesynthesisofadenovirus 2-specific proteinsprogrammed byfractionatedmRNA:
acomparisonofpolypeptide productsize andmessenger RNAlengths. Proc.Natl. Acad. Sci. U.S.A.
71:2756-2760.
2. Beltz,G.A.,and S. J. Flint. 1979. Inhibitionofcellular
protein synthesis duringadenovirus infection. J. Mol. Biol. 131:353-373.
3. Berget,S.M.,C.Moore,andP. A.Sharp.1977.Spliced segmentsof the 5'-terminus of adenovirus 2 latemRNA.
Proc.Natl. Acad. Sci. U.S.A. 74:3171-3175.
4. Chow,L.T.,and T. R. Broker.1978. Thespliced struc-turesof adenovirus 2 fibermessageandtheotherlate
mRNAs. Cell 15:497-510.
5. Chow,L.T.,R. E.Gelinas,T. R. Broker,and R. J.
Roberts. 1977. Anamazingsequencearrangementat
the 5'-ends of adenovirus 2messengerRNA.Cell
12:1-8.
6. Chow,L.T.,J. M. Roberts,J. B.Lewis,and T. R.
Broker. 1977. AmapofcytoplasmicRNAtranscripts
fromlytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrids. Cell 11:819-836. 7.Evans, R. M., N. Fraser, E. Ziff. J. Weber, M. Wilson,
andJ. E. Darnell. 1977. The initiation sites for RNA transcription in Ad2 DNA.Cell 12:733-739.
8. Goldberg, S., J. Weber, and J. E. Darnell, Jr. 1977. The definition of a large viral transcription unit late in Ad2infection of the HeLa cells: mapping by effects of ultraviolet irradiation.Cell 10:617-621.
9. Green, M., M. Pina, R. C. Kimes, P. C. Wensink, L. A. McHattie, and C. A. Thomas, Jr. 1967. Adenovirus DNA: I. Molecular weight and conformation. Proc.
Natl. Acad.Sci. U.S.A. 57:1302-1309.
10. Klessig, D. F. 1977. Two adenovirus mRNAs have a
common5'-terminal leader sequenceencoded at least 10Kb upstream from their main coding regions.Cell
12:9-21.
11. Kozak, M. 1978. How do eukaryotic ribosomes select initiation regions in messenger RNA? Cell 15:1109-1123.
12. Laemmli, U. K. 1970. Cleavage of structural proteins
duringthe assembly of the head of bacteriophage T4. Nature (London)227:680-685.
13. Lewis,J.B., C.W.Anderson, and J. F. Atkins. 1977.
Furthermapping of late adenovirus genes by cell-free translationof RNA selected by hybridization of specific DNAfragments. Cell12:37-44.
14. Lewis, J. B., J. F. Atkins, C. W. Anderson, P. R. Baum, and R. F. Gesteland. 1975. Mapping of late adenovirus genes by cell-free translation of RNA se-lected by hybridization to specific DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 72:1344-1348.
15.Maizel, J. V., Jr. 1971. Polyacrylamide gel electropho-resis of viral proteins, p. 179-246. In K. Maramorosh and H.Koprowski (ed.), Methods in virology, vol. 5. AcademicPress,Inc., New York.
16.McGrogan,M., and H. J. Raskas. 1978. Two regions of theadenovirus2genomespecifyfamiliesoflate poly-somal RNAs containing common sequences. Proc. Natl. Acad. Sci. U.S.A. 75:625-629.
17.McMaster,G.K.,and G. G. Carmichael.1977.Analysis ofsingle- and double-stranded nucleic acids on
poly-acrylamideandagarosegels by usingglyoxal and acri-dine orange. Proc. Natl. Acad. Sci. U.S.A.74:4835-4838.
18. Nevins,J.R.1979.Processing of lateadenovirus nuclear RNAtomRNA. Kinetics of formation ofintermediates and demonstration thatalleventsarenuclear.J. Mol. Biol. 130:493-506.
19. Nevins,J.R., and J. E. Darnell.1978.Groupsof ade-novirus type2mRNA's derived fromalarge primary transcript: probable nuclearorigin and possible com-mon3'ends. J. Virol. 25:811-823.
20. Nevins,J.R.,andJ. E.Darnell,Jr.1978.Stepsinthe processing of Ad2 mRNA:poly(A)+nuclear sequences areconserved andpoly(A) additionprecedes splicing. Cell15:1477-1493.
21. Pelham,H. R.B.,and R. J. Jackson.1976. Anefficient
mRNA-dependenttranslation system fromreticulocyte lysates.Eur. J.Biochem. 67:247-256.
22. Stark, G.R., and J. G. Williams. 1979. Quantitative analysisofspecificlabeled RNAsusingDNAcovalently linkedtodiazobenzyloxymethyl-paper. Nuc. Acids Res.
6:195-203.
23. Weber, J., W. Jelinek,and J. E.Darnell,Jr. 1977. The
definition ofalargeviraltranscription unit lateinAd2 infection of HeLacells:mapping ofnascentRNA mol-ecules labeledinisolatednuclei.Cell10:661-616.
24. Ziff, E., and N.Fraser. 1978. Adenovirus type 2late
mRNA's:structural evidencefor3'-coterminalspecies.
J. Virol.25:897-906.
25. Ziff,E.B.,andR. M. Evans. 1978.Coincidenceof the
promoter and capped5'-terminus of RNA from the adenovirus 2 major late transcription unit. Cell 15: 1463-1475.
VOL. 35,1980

![FIG. 4.poly(A)-containingfractionexposedaswithmRNA's.andthe lanes Agarosegelelectrophoresisof[32P]- 32P-labeled RNAs were treated with glyoxal analyzed on a 1.0%/ agarose gel as described in text](https://thumb-us.123doks.com/thumbv2/123dok_us/1492952.101972/5.504.282.426.401.543/containingfractionexposedaswithmrna-agarosegelelectrophoresisof-labeled-treated-glyoxal-analyzed-agarose-described.webp)
