Copyright © 1985, American Society forMicrobiology
Polyhedrin Gene of Bombyx mori Nuclear Polyhedrosis Virus
KOSTAS IATROU,* KENICHI ITO,AND HALINAWITKIEWICZ
Department ofMedical Biochemistry, Faculty of Medicine, The University of Calgary, Calgary, Alberta T2N4NJ, Canada
Received 18October 1984/Accepted 14January 1985
Aportion of thegenomeof thenuclearpolyhedrosisvirusofBombyx morihas beencloned.Thispartof the viralgenomecontainsthegeneencodingthe viral occlusionbody protein, polyhedrin. The polyhedringenehas beensequencedinitsentirety togetherwithsomeof its 5' and3'flankingsequences.Theprimarystructureof polyhedrin predicted from the nucleotidesequenceof thegene wasfoundtobe somewhatdifferent fromtheone reported previously for the authentic protein (E. A. Kozlov, T. L. Levitina, N. M. Gusak, and S. B. Serebryani, Bioorg. Khim., 7:1008-1015, 1981; S. B. Serebryani, T. L. Levitina, M. L. Kautsman, Y. L. Radavski, N. M. Gusak, M. N. Ovander, N. V. Sucharenko, and E. A. Kozlov, J. Invertebr. Pathol., 30:442-443, 1977). Comparisonof theprimarystructuresofthepolyhedrinof the nuclearpolyhedrosisvirus of B.moriwith that ofAutographacalifornicasuggeststhatconsiderable selectivepressurehas been exercised attheproteinlevelduring evolution. Nucleotidesequencecomparisonsof the twostructuralgenesrevealthat the coding sequences have diverged significantly through the accumulation of silent and replacement substitutions. In contrast, a remarkable degree ofsequence conservationwas foundto existinthe domains corresponding tothe 5' and3' noncoding regionsof thepolyhedrin mRNAs.
Nuclear polyhedrosis viruses(NPVs) representagroupof insect viruses (family Baculoviridae, genus Baculovirus) whosevirions areembeddedinto polyhedron-shaped
occlu-sion bodies or polyhedra in the nuclei of host cells (for extensive reviews see references4, 13,and 19). Thegenetic information of baculoviruses is stored in the form of a relatively large molecule of covalently closed double-stranded DNA ranging in size from 50 x
106
to 100 x 106daltons.
NPVshave been recognized formany years as the
infec-tious agents that cause acute diseases in a wide range of
insects, including the silkmoth, Bombyx mori (4), and the first tissue culture propagation of the NPV of B. mori (BmNPV) was reported as early as 1935 (47). Occlusion bodieswereshowntobemade almostexclusively ofasingle protein, polyhedrin, which can be solubilized by treatment
ofthepolyhedra withweakalkalinesolutions(3). In B. mori,
about350nucleocapsids enveloped singly orinmultiplesof two tofiveare occluded in each occlusionbody.
AlthoughNPVs mayinfectinsectsother than their normal
hosts, their host
range
is usually restricted to differentspecies or (at most) families within an order. Only very
rarelycanthehost range extend to differentinsectordersor to different arthropod classes (9). There hasonly been one case in which infection of a vertebrate cell culture was caused byan NPV (21), but thatcase has not been studied
anyfurther. In contrast, repeated attemptsbyother
investi-gators to propagate NPVs in a wide variety of vertebrate cells were unsuccessful (18, 25, 46). Because of their re-stricted host range and their effectiveness ininfecting their hosts, baculoviruses have beenconsidered as potential
bio-logical pest control agents(33, 45). This has in turn stimu-lated the molecular characterization ofsome NPVs, partic-ularly theNPV ofAutographa californica (AcNPV) (13).
Anumber of mutations have beenfound in AcNPV which result in the formation of abnormal, few, or no occlusion
bodies in the nuclei of infected cells (7, 12, 27, 53). In
addition, it has been established that extensive serial
pas-*Corresponding author.
sageof NPVs in cell cultures results inadramatic decrease in virus occlusion (14, 22, 31). In suchcases the production of nonoccluded virususually remains unaffected, and non-occluded virus becomes the predominant viral form in the infected cells.Onthebasis of theseobservations it has been concluded that occlusion body formation and, presumably,
polyhedrin synthesisarenotrequired forviralpropagationin
laboratorytissue cultures.
With the demonstration that polyhedrin is a protein en-codedbyaviral gene (50) and with the advent of recombi-nant DNA technology, it became apparent that the poly-hedrin gene might represent a site of the viral genome amenable togenetic manipulation, suchasthe introduction offoreigngenes intothe DNA of the virus. Experiments in which the structure of the polyhedrin gene from spontane-ous, chemically induced, or in vitro-constructed mutants wasexaminedshowedmutational changes inthe gene orits
immediatevicinity(8,11, 40),thussupportingthehypothesis that the polyhedringene representsa convenient target for
geneticengineering.Finally, polyhedrinissynthesizedin the host cells late in infectionin large amountsandthis in turn
reflects acorresponding abundance in the amount of accu-mulated polyhedrin mRNA (1, 42), probably achieved
throughhigh ratesofmRNA synthesis. This suggestedthat
heterologous genes might be appropriately introduced into the genomes of baculoviruses and expressed effectively
under the control ofthe polyhedrin promoter. The use of
AcNPVas avectorfor theexpressionof
foreign
genesunderpolyhedrin promoter control was
successfully
attempted recently and resulted in the production of human ,B-interferon and Escherichia coli 3-galactosidase in largeamounts in insect tissue culture cells(34, 41).
Unfortunately, themolecular characterization of BmNPV has lagged far behind that of AcNPV.
Although
somecontroversialreportsonthenatureof the viral genome have
appeared in the literature (reviewed in reference 19), no accurategeneticmapor measurementsofthesize of BmNPV DNA have beenreported duetothelack of
published
work basedon currentcloning methodology. Weareinterested in the mechanisms regulating chorion gene expression in the436
on November 10, 2019 by guest
http://jvi.asm.org/
BmNPV POLYHEDRIN GENE 437
silkmoth B. mori(forarecent review see reference 16) and
feel that BmNPV may be used effectively as a vehicle for introducing authentic or mutagenized silkmoth genes into
thetissuesinwhich they arenormally expressed inorder to
study theirexpression invivo. To testmodels of differential
regulation
of choriongeneexpression (24),wewould liketo recombine well-characterizedchoriongenesintothe genome ofBmNPV, using the vicinity of the polyhedrin gene as a target site, and to infect follicular cells in vivo with therecombinant virus. As afirststepin this directionwereport here the localization, cloning, and complete nucleotide se-quence ofthe
polyhedrin
gene ofBmNPV and its vicinity.The primary structure of polyhedrin predicted from the
nucleotide sequence of the gene is not identical to that
derivedfrom the protein itself (26, 39). Comparison of the polyhedrin genes of BmNPV and AcNPV indicates that
although a large degree of point mutations were fixed in someparts of the genesduring evolution, other domains of
the genes and theproteins themselves have been subjected
toconsiderable selective pressure.
MATERIALS ANDMETHODS
Cells and virus. B. moritissue culturecells(Bm-5 [17])and
BmNPV passedonce in Bm-5 cellswerekindly supplied by
J. L.
Vaughn,
InsectPathology Laboratory,
U.S.Depart-mentofAgriculture, Beltsville,Md.Cellsweremaintainedat
28°C
incomplete
IPL-41 growth medium (52) supplementedwith 0.24,uM
ZnSO4
and 16 ,uMAIK(SO4)2 in the absenceof antibiotics. The cells were subcultured weeklyat aseedingdensity
of0.2 x 106 cells per ml (usuallya 1:5 dilution ofa 1-week-oldculture). Infection ofthecells with BmNPVwascarriedout
by
removing
themediumfrom2 x107
cells and replacing it with 5 ml ofa nonoccluded virus inoculum of tissue cultureorigin containing
1 x107
to 5 x107
PFUper ml. After 1 h the viralinoculum wasremoved and replaced with25mlof fresh mediumcontaining
gentamycin (50,ug/ml).
Cellswerecollectedby low-speed centrifugation
3to4days
later whenmore than98% ofthemexhibiteda
large
number ofviralocclusion bodiesintheir nuclei. Cellularpellets
wereused as thesource of occluded
virus,
whereasthe superna-tants were used as the source of nonoccluded virus and asinoculafor further infections.
A. californica polyhedrin gene probe. A cloned HindIII
fragment
ofthe polyhedrin gene ofAcNPV (HindIII-V) inpBR322 (clone pM5 HindV) waskindly supplied by Eric B.
Carstens, Department
ofMicrobiology
andImmunology,
Queen's
University, Kingston, Ontario,
Canada.Thisclonedfragment
is 937 base pairs long and contains the codingregion
ofthe gene downstream from amino acid number 83(nucleotide
251ofthepolyhedrin
gene sequencepublishedinreference23)and almosttheentire 3' noncodingpartofthe gene
(E.
B.Carstens, unpublished data).
Preparationof occlusionbodies.Atthe endoftheinfection
period
cellswerepelleted by
centrifugation
andwashed oncewith phosphate-buffered saline (10). Cellular pellets were
solubilized with 0.4% sodium
dodecyl
sulfate-10 mM Tris-HCI(pH 7.8) by gentle rocking for2 h(approximately5mlofsolution per4 x 107 cells),and the cellularsuspension was
layeredona30-ml cushion of
65%
(wt/vol)sucrosein 10mM Tris (pH7.8)-10
mM EDTA. Aftercentrifugationat110,000x g (24,000 rpm) in the SW27 rotor of a Beckman
ultracen-trifuge
for4h at 15°C, the supernatant was removedpartlyby aspiration
and finally by decanting, and the pelleted occlusion bodies were suspended in a small volume ofdistilledwaterwith theaidofaPasteurpipette. Finally,the occlusion
body suspension
wasmade 0.25MwithrespecttoNaCl andcentrifugedat 17,000 x g (12,000 rpm)in the SS-34 rotorofaSorvall centrifuge. Thepelleted occlusion bodies were washed twice with 0.25 M NaCl as described above, and the finalpellets were stored at -20 to -70°C until used for the extraction of viral DNA.
Viral DNAisolation.Occlusion bodies were solubilizedin 0.1 MNa2CO3-10 mMEDTA-0.1M NaCl (pH 10.8)
(occlu-sion bodies from2.5 x
107
cells per ml ofbuffer)withgentleswirlingatroomtemperature overaperiodof 2 h. At the end of theprocess the volume of the solution was increased by 50%by the addition of distilledwater, and the solution was
finally made 1% with respect to sodium dodecyl sulfate. A small amountof insoluble matrix was removed by
centrifu-gation,and the supernatant was extractedexhaustivelywith
phenolandchloroform-isoamylalcohol(24:1, vol/vol). After
the extractions, the aqueous phase was concentrated to a volume equal to that ofthe Na2CO3 solution used at the
beginning of the process, made 0.25 M with respect to
ammoniumacetate, and spun at30,000x g(17,000 rpm)in a Sorvall centrifuge. The supernatant was precipitated with 2.5 volumes of ethanol. After being washed with 70% ethanol, the DNApelletsweredissolved in 5 mM Tris-HCl
(pH 7.8)-0.1 mM EDTA and stored at4 or
-40°C.
The Na2CO3-insoluble matrix was also partially
solubil-ized inamixture ofequal volumes of0.1 M
Na2CO3-10
mM EDTA-0.1 M NaCl (pH 10.8) and 7 M urea-0.135 MNaCl-10 mMTris-(pH
7.5)-i
mM EDTA-2% sodiumdod-ecyl sulfate (occlusion bodies from 5 x 107 cells per ml of
buffer) for3 to4h atroomtemperature with
swirling.
Afterextractions andprecipitationsasdescribedabove, the DNA was pelleted bycentrifugation and storedin thecold.
The distributions of DNA in the fraction solubilized in
carbonatesolutionalone and in the
partially
insoluble matrix were 66% and33%, respectively. The yield ofDNA was in the rangeof1.2 ,ug per106cells.Othermethods. Restriction enzyme
digestions
wereper-formed with the buffers recommended
by
thesuppliers.
Afterelectrophoresis
on agarosegels,
the resolvedfrag-ments were immobilized on a membrane support
(Gene-Screen;
NewEngland
NuclearCorp., Boston, Mass.)
aspreviously described (44)except thattwo15-mintreatments
of the
gels
with 0.25 N HCl were included before alkalinedenaturation to
depurinate
the DNA into sizes ofapproxi-mately
1 to 2 kilobases(kb) (51).
Nick translations andSouthern
hybridizations
were as describedpreviously
(24). Probeswerenicktranslatedtospecific
activities of2 x107
to 4 x107
cpm Cerenkov per pLg. Hybridizations wereper-formedat
70°C
for 14to16hwith400,000cpmCerenkov ofeach
probe
per mlofhybridization
solution.DNAfragments
were isolated from gel slices by electroelution (20), but no
bovine serumalbuminwas used.
Cloning
of various restric-tion fragments ofthe viral genome was done as described previously (24) byusing equimolar
mixtures oflinearized
and dephosphorylated
plasmid
pUC9 (48) andpurified
re-striction
fragments
isolatedby
electroelution. After thetransfection of competent Escherichia coli cells (a strain
derivative ofE. coli K-12 RRI
having
the genotype leupro thistrA hsdr- m- lacZ AM15 F'lacIQ
pror [38]), transfor-mants were selected on ampicillin-containing plates.Ampi-cillin-resistant colonies were further plated on plates
con-taining
5-bromo-4-chloro-3-indolyl-,-D-galactoside
andisopropyl-p-D-thiogalactoside
(30), and colorless colonieswerepickedfrom thoseplates forfurther biochemical
anal-ysis.
The clones of the EcoRIfragment
of BmNPV DNA inserted inpUC9
in two orientations have beendesignated
Bmp/pR5
andBmp/pR8,
whereas thoseofthePstIfragment
VOL. 54, 1985
on November 10, 2019 by guest
http://jvi.asm.org/
438 IATROU ET AL. J.VIROL.
1 2 3 4 5 6 7 8 M 1 2 3 4 5 6 7 8 M 1 2
.485 *24.7S
j94 24
*742 * 6.56
.56e5 .458
.44
-372
.325
*285
.19
g
.08
.0 61 -053
1 2 3 4 5 6 1 2 3 4 5 6
--'
wr
soI
4oa_
[
-48 5
.2475
*21? *S42 *742
.6 56
. 565 .4 88 4 4 *372
.325 *285
.2 lb
-1.9
.1 I9
.708
*053
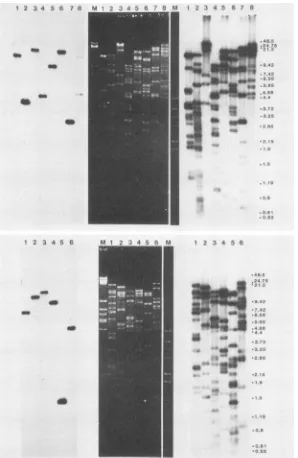
FIG. 1. Southern hybridizations ofBmNPVDNArestrictiondigests. BmNPV DNA (0.15 to 0.22 ,ug perdigest)wasdigested with avariety of restriction enzymes, and the digests were resolved on 0.7% agarose gels in the presence of ethidium bromide (0.5 ,ug/ml). After electrophoresis the gels were photographed (middle portion of each panel), and the digests were subsequently Southern transferred to membrane sheets, hybridizedtonick-translated clonepM5HindVcontainingpartofthe AcNPVpolyhedringene,andautoradiographed (left portion of each panel). After autoradiographyandwithoutanyprior melting of thehybridizedAcNPVpolyhedringene sequences,thesame filterswereprobed with total nick-translatedBmNPV DNA andautoradiographed (rightportion of each panel). The orderofthedigestswas
asfollows: (upperpanel)1, AvaI;2, BalI;3,BamHI;4,Clal;5,EcoRI;6,EcoRV; 7,HindlIl;8,NdeI;and(lowerpanel) 1, NruI;2, PstI; 3,PvuI;4,PvuII;5,SalI;6,SphI. For sizemarkers (M), a mixture ofHindlll-digestedandEcoRI-digestedA DNA wasusedoneach ethidium bromide-stained gel.Forsize markersonthe narrowstrips oftheethidium bromide-stainedgels,amixture ofHindIl-HindIll doubledigest of X-DNAandHaeIII-digestedpMB9 DNA wasused.The numbers at therightside of eachpanel indicate thelengthinkilobases ofsome
ofthesizemarkers described above. Asterisksonthe bandsoftheethidium bromide-stainedgels indicatethefragmentshybridizingtothe AcNPV polyhedringeneprobe.
have been designated Bmp/pP3 and Bmp/pP14. 32P end
labeling
ofplasmid DNA preparations with T4 polynucleot-ide kinase after linearization and dephosphorylation hasbeendescribed previously (24).Restriction enzymemapping wascarriedoutbythepartial digestionmethod with
single-end-labeledfragments of cloned viralDNA(43). Nucleotide sequencingwasdonebythechemical method of Maxam and
Gilbert (29), and chemical reactions were analyzed on
85-cm-long 6% polyacrylamide gels. 32p labeling at the 3' termini of restriction fragments was performed by repair
synthesis of3' recessed endsbyusingtheKlenow fragment
of E. coli DNA polymerase in a 20-,l reaction mixture
containing50 mMTris-HCI(pH 7.5), 5mM MgCl2, 10 mM
,B-mercaptoethanol, 70,uMeachofdATP, dGTP, andTTP,
gm am
40
de
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.163.461.73.531.2]BmNPV POLYHEDRIN GENE 439
1 kb
PstI R I Sal I Sal I Bam H I SalI
r
-Pst I R I
1 5'
S i
Sal I
I Coding
'11
XbaI
qtl11 AATAAA
...
..-Nde I Hindm Sal I Bam H I
o 0.2 kb
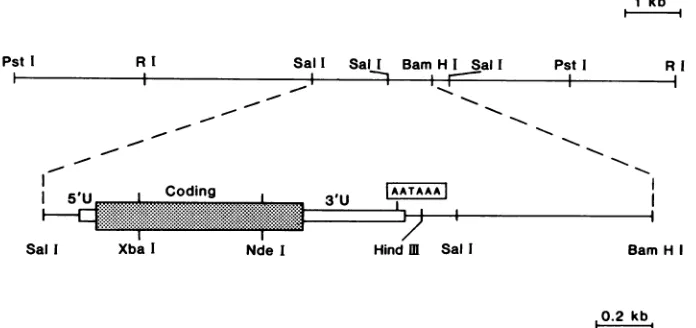
FIG. 2. Cloned portions ofBmNPV. The9.6-kbEcoRl and10-kbPstlfragments ofBmNPVcloned inplasmidpUC9arerepresented by the top line. Some of the enzymes relevant to the sequences of the polyhedrin gene are also presented. In the lower line an expanded version ofthe SalI-BamHI fragment containing the polyhedrin gene is presented togetherwith thecleavage sites usedfor generating end-labeled fragments for nucleotide sequence determination in the polyhedrin gene region. The designations 5'U and 3'U symbolize the 5' and 3' untranslated regionsofthe polyhedrin mRNA, and the end of the mRNA has been tentatively placed 30nucleotidesdownstreamfromthefirst AATAAApolyadenylation signal(seealsothe text).
100,uCi ofa-[32P]dCTP (ca. 3,000Ci/mmol),10UofKlenow
fragment,and 5 to 20pug ofthe DNA digest. Sites usedfor5' or3' end labelingare listedinorderfromleft torightin the
lower part of Fig. 2, and their locations are identified by referencetothe numbered DNA sequence shown inFig. 3 as
follows: Sallat -192,XbaIat 147,NdeI at595, and HindIll
at 1177. In all cases sequencing was performed in both directions.
RESULTS
To identify the part of the genome of BmNPV that contains the polyhedrin gene, viral DNA was isolated and
restricted with a large number of restriction enzymes with hexanucleotide recognition sites. After electrophoresis of the restrictedDNA,the resolvedfragmentsweretransferred
to amembrane filterandsubsequently hybridizedto aDNA
clonecontainingalargeportion ofthecodingsequencesand some 3' noncoding sequences of the polyhedrin gene of AcNPV (HindIII-V; see above for details). Typical results
fromsuch hybridizations are shownin Fig. 1. Through the
hybridizations ofthe BmNPV digests to the AcNPVprobe
(left hybridization panels ofFig. 1), the sizes of the
restric-tion fragments that included the BmNPV polyhedrin gene and the flanking sequences that are homologous to the
hybridization probe weredetermined. Thus, several
restric-tionenzymes were identified thatproduced single hybridiz-ing fragments whoselengthswerewithin the size range that may be cloned relatively easily in E. coli and also allowed
the prediction to be made that the entire polyhedrin gene may be contained withinthem.
Todetermineunequivocallytheposition of the polyhedrin
gene-containing fragmentswhen bandswere closely spaced and to visualize the smaller restriction fragments of the BmNPV genome that might have escaped detection in the ethidium bromide-stainedgels, the same immobilized digests weresubsequently hybridized to radioactive BmNPV DNA. Through such hybridizations (right hybridization panels of
Fig. 1), allrestrictionfragments of a size equal to or greater than 500 base pairs were detected. Based on this
informa-tion, we wereableto deducealength of125 ± 4kbforthe genome of BmNPV.
The EcoRI and PstI fragments of BmNPV DNA
hybrid-izing to theAcNPV polyhedringene probe (approximately
9.6 and 10kb,respectively; Fig. 1)werecloned andmapped
in detail. Partofthismapping information isshown inFig.2. ThecombinedEcoRIand PstI clonedfragmentswerefound to spanatotallengthof 11.8 kb of the viralgenome.On the basis ofthe complete restriction mapping information de-rived from these clones as well as from additional Southern
hybridizations (datanot shown), we were abletodetermine the approximate location of the polyhedrin gene in the middleofthe 11.8 kbof the cloned viral DNA andtoinitiate
its sequencing.
Thecompletenucleotidesequenceof the polyhedringene
withsomeof its5'and 3'flankingsequencesis showninFig.
3. Upon inspection ofthe 2,060-nucleotide-long sequenced
part ofthe cloned DNA, an open readingframewas identi-fied inoneofthe DNA strands thatresultedin thetranslation ofa 244-amino acid-long polypeptide that, with few excep-tions (seebelow),wasidentifiedasthepolyhedrin ofBmNPV based on comparisons with the published sequence ofthe authentic protein (26, 39)and with the predictedaminoacid
sequence of the AcNPV polyhedrin (23). The exact locali-zation ofthepolyhedringeneanditstranscriptional
orienta-tion in the cloned DNA,asdeduced fromthedetermination oftheprimarystructureofthe gene,are shown in the lower part of Fig. 2 along with the restriction sites used as the
major starting points forthenucleotide sequenceanalysis.
ThesequencedDNAwasalsoscannedforthe presenceof
other open reading frames.Two moreputativeopenreading
frameswere detected at thebeginningand at the endofthe sequenced DNA. The first one, at the beginning of the sequence, is read in the sameorientation asthe polyhedrin
gene, comprises 82 amino acids, and terminates at nucleo-tide -260. The second one, atthe end of the sequence, is read in the opposite orientation (from the complementary
strand).This openframe translates into 239 amino acids and terminatesatnucleotide 768 of the sequence shown inFig.3. The consequences of the possible presence ofone or two VOL. 54, 1985
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.128.476.77.241.2]440 IATROU ET AL.
-l:. -55z -542 -S32 -522 -512-5i
CCGCCCACTA TTAATGAAATTAAAAATACC AATTTTAAAAAACGCAGCAAAAGAAACATT TGTATGAAAG
-492 -442 -472 -462 -452 -442 -472
AATGCGTAGAAG&AAAAAAT AATGTCATCG ACATGCTGAA CAGCAAGATC AATATGCCTCCGTGCATACA
-422 -412 -402 -392 -302 -772 -76:
AAAAATATTl GGCGATTTGA AAAAGAACAA TGCAGCGCGG CGGTATGTACAGGAAGAGGTTTATACTAAA
-352 -3,42 -@3 _-22 -312 -302 -292
CTGTTACATT GCAAACGTGGTTTCGTGTACCAAATGTGAAAACCGATGTTTAATCAAGGCTCTGACACAT
-208 -272 -26: -252 -242 -232 -222
TTTTACAATT ACGACTCCAAGTGTGTGGGT GAAGTCATGC ATCTTTTAATCAAATCCCAAGATGTGSATA
-212 -202 -192 -192 -172 -162 -152
AACCACCAAA CTGCCAAAAA ATGAAAACTGTCGACAAGCTCTGTTCGTTTGCTGGCAACT GCAAAGGTCT
-142 -132 -122 -112 -12 -92 -42
CAATCCTATCTGTAATTATTGAATAATAAA ACAATTATAAATGTCAAATTTGTTTTTTAT TAACGATACA
-72 -62 -52 -42 -32 -22 -1I
AATGGAAATA ATAACCATCT CGCAAATAAA TAAGTATTTTACTGTTTTCGTAACAGTTTT GTAATAAAAA
AACCTATAAA T
15 30 45 63
ATG CCGAATTAT TCATACAACCCC ACCATC GGG CGTACTTAC GTGTAC GAC AATAAATAT MetPro Asn Tyr SerTyrAs% ProThr lIe Gly ArgThrTyrVal Tyr AspAsnLys Tyr
75 90 105 1
TACAAAAAC TTG GGCGGT CTC ATCAAAAAC GCC AAG CGCAAG AAGCACCTAATC GAACAT
TyrLys A-nLeo GlyGlyLouIleLys Asn Al1 LysArg Lys Lys His LouII1GluHis
135 150 165
GAA 66A GA6 GAG AAGCAATGG GATCTTCTAGACAACTAC AT6 GTT GCC GAA GATCCC TTT Glu Lys GluGlu LysGinTrp Asp Leu Leu AspAsn TyrMet Val Al& GluAspProPhe
195 210 225 240
TTA GGACCG GGC AAAAAC CAA AAACTTACCCTTTTT AAAGAG 6TT CGC AAT0TG AAACCC
Leo GlyProGlyLys Asn Oln LysLouThrLouPh- LysGluVal ArgAsn Val LysPro
255 270 295 300
GATACCATGAAGTTAATCGTCAACTGG AGC GGC AAA GAGTTTCTGCOT G66 ACT TGG ACC Asp Th7r Ht Lye LouII Val AsnTrp Ser GlyLye GluPh-LouArgGluThrTrpThr
315 330 345 360
CGTTTT GTTGAG GAC AGCTTCCCC ATT GTAAACGACCAAGAG GTG ATGGAC GTG TAC CTC ArgPh- Vol Glu Asp S6r Phe ProIleVal AsnAspOlnOlu ValMetAspVal TyrLeu
375 390 405 420
GTC GCC AACCTCAAACCCACACGCCCCAACAGG TGCTAC AAGTTCCTC GCTCAACACGCT
Vol Al6 A-n LouLysProThr ArgPro Asn Aro Cy- Tyr Lys Ph- L-o AlaG61 HisAl&
435 450 465 480
CTTAGG T7GGACGAA GACTAC GTGCCCCAC GAAGTAATC AGA ATT ATG GAGCCATCCTAC
LouArgTrpAsp Glo Asp TyrVal ProHisGluVal leArg IleMetGI.Pro6-r Tyr
495 510 525 540
GT6 GGC ATG AACAAC GAATACAGA ATT AGT CTG GCT AAAAAG GGC GGC GGC TGCCCAATC
Val GlymetAsnAsnGlu Tyr Arg Ile Sor Lou A61 Lye LysGly Gly Gly CysPro
11-555 570 5ss 600
ATG AACATCCACAGC GAG TAC ACCAAC TCGTTCGAGTCG TTT GTG AAC CGCGTCATATGG MetAen lie HisSerGlu Tyr ThrAsn SerPh- Glu SOr Pho Val AsnArg Val Ile Trp
615 630 645 660
GAG AACTTC TACAAACCCATCGTTTACATCGGCACASAC TCTGCC GAA GAA GAG GAAATC
Glu Asn Ph- Tyr LysPro lieVal Tyr IleSlyThr Asp S-rAl&GluGlu Glu Glu lie
675 690 705 720
CTAATTGAG GTTTCTCTCGTTTTC AAA ATA AAGGAG TTT GCACCAGAC GCGCCT CTG TTC Lou11-GluVal SOr LouVal Ph- LysIle Lys Glu PhC Al6 ProAspAl&ProLeou
Ph-75
ACT GGT CCG GCA TAT TAA Thr GlyPro Al&Tyr Tor
749 758 768 779 798 798e 9o
AACACTATACATT0TTATTA GTACATTTATTAAGCGTTAGATTCTOTACGTTGTTGATTT ACAGACAATT
1e 92 93 94 95 96 97
GTTGTACGTA TTTTAATAAC TCATTAAATT TATAATCTTT AGGGTGGTAT GTTAGAGCGA AAATCAAATG
9ee e99 909 919 929 939 949
ATTTTCAGCGTCTTTGTATC TG7 7TTTAAATATTAAATCC TCAATAGATT TGTAAAATAGGTTTCGATTG
959 968 978 999 999 1009 0l1e
GTTTCAAACA AGGGTTGTTTTTOCAAACCGATGOCTGGACTATCTAATG7 ATTTTCGCTC AACACCACAC
1028 1439 1048 1059 1069 1078 1098 GACTTGCCAAATCTTGTAGC AGCAATCTAGCTTTGTCGATATTCGTTTGT GTTTT6 TTTT GTAATAAAGA
1099 l1o1 ill 1129 1139 1149 1159
TTCGACOTCG TTCAAAATAT TATGCGCTTT T7TATTTTTTTCATCACTOT CGTTA0 TGTA CAATTGACTC
116e 1179 l1es 1199 1209 1219 1229
GAC0TAAACACGTTAAATAAAGCTTGGACATATTTAACAT CGGGC0CGTTAG6CGCATTATT7CC0CC0T
1239 1249 12s3 1268 1279 1299 1299
CGTCCCAACCCTCOTCOTTAGAAGTTGCTT CCGAAOACGA TTTTGCCATA OCCACACGAC GCCTATTAAT
1309 1319 1329 1339 1349 1359 1369
TGTGTCGACTAACACOTCCOCGATCAAATT TTTAGTTGTT GA0CTTTTCG 6AATTATTTCTGATTGCGGA
1379 1389 1399 1409 1418 1429 1438
CGTTTTTG0 CGG0TTTCAATCTAACTGTG CCCGATTTTA ATTCAGACAA TAC7TTAGAA AGC6ATGGTG
1449 1458 1408 1479 1499
CAGGCG0 TG7 T7ACATTTCAACC60C06ATCTACTATG TGGCTGTAATG
FIG. 3. Nucleotide sequence of the polyhedrin gene and
sur-rounding region. The first nucleotideof the initiatorATG codonhas
beendesignatednumber 1. Forassignmentsofcap site, polyadenyla-tion signals,and otherconsensussequencesin thepromoterregion,
seethe text.
structuralgenes in the immediatevicinity ofthepolyhedrin gene are discussed below.
DISCUSSION
Aprerequisitetothe successful genetic manipulationofa particularsite inagenomeis the detailed characterizationof the target site.Wearepredicting that,inamannersimilar to that showntooccurin AcNPV(8, 11, 27, 34, 40, 41), direct
or indirect mutational alteration, inactivation, and even removal ofthe polyhedrin gene from the BmNPV genome should bear no genetic consequences in terms ofviability and virulence of the nonoccluded form of the virus in
laboratoryanimals(silkmoths)and tissueculture cells.
There-fore, as the first step toward introducing foreign gene
se-quencesinto thegenomeofBmNPV,we identified, isolated
by molecular cloning, and characterized by sequence
anal-ysis thegene encodingthe viralprotein polyhedrin.
Genome sizeof BmNPV andpolyhedringenecloning.Inthe course of ourpreliminarycharacterization of thegenomeof BmNPV by restriction analysis and hybridizationaimed at the identification of restriction fragments containing the
polyhedringene, we hadtheopportunity tofirmly establish the size of theviral DNA. Over the past 30years, various
values have been published on the size of the circular
double-stranded BmNPVgenome, rangingfrom 3 to 180kb
(see reference 19 for a review). Through our restriction
digestion analysis (Fig. 1 and other data not shown), we calculatedalengthof 125 ±4 kb for the viralgenomea size very similar to that established for the genome ofAcNPV (32, 49). No restriction pattern polymorphisms (restriction
fragments appearing in submolar quantities) were detected when BmNPV DNA that had been serially passaged five
-5 IC 1S5
20-ProAsn TyrS-r Tyr Asn Pro Thr IleGly Arg ThrTyr Val Tyr AspAsn Lys TyrTyr
25 S.) ZX5 40
Lys Asn LeuGlyGly Leu Ile Lys Asn Ala LysArgLys LysHis Leou Ie GuHis Glu
Glu His
45 50 55 60o
|LysGluGluLysGln Trp Asp Lou Lou AspAsn TyrMWetVal Ala GluAspProPho Lou
65 70 75
GlyPro Gly LysAsnGin Lys LeuThr LeuPhe LysGluVal ArgAsnVal LysProAsp
85 90 95 100
Thr MetLysLeouIle Val AsnTrp SerGlyLysGlu Fhe LeouArg GluThrTrp ThrArg
105 -110 115 120
PheVal GluAspSer PhePro IleVal AsnAsp GinGluValMetAsp Val TyrLeuVal
125 1l0 135 140
AlaAsn Leu LysProThr ArgPro Asn ArgCysTyrLysPheLeuAlaGinHis AlaLeu
145 lS0 l5S 160
Arg TrpAspGluAsp TyrVal Pro HisG1u Val II Arg Ie*M1et Glu ProSer TyrVal
GinAsn
165 170 175 1_0
GlyMet AsnAen Glu Tyr Arg IleSer LouAla LysLysGlyGly Gly Cys Pro IieMet
185 190 195 200
Asn IlieHis Ser GluTyr.ThrAsn Scr PheGluSer Phe Val AsnArg Val IleTrpPlu
205 210 215 220
AsnPhe TyrLys Pro IleVal Tyr IleGly Thr AspISGr AlaGlu GluGIlo Gluo
Ala Seri . . IGInL
225 230 235 240
IleGluVal
~~~~~.
Ser LeuVal PheLysIle Lys GluPhe AlaProAsp AlaProLeuPheThr. . ...*.**.**.*.**. ..
G1lyProA1l& Tyr1
FIG. 4. Comparisonof two BmNPVpolyhedrinsequences.The
polyhedrinsequence predictedfromthegene sequence(upperline)
is compared to that previously reported for the purified protein
(lowerline;[26,39]).Dots in the lowerline indicatethe same amino
acidsas in thetopone and blocksof identicalsequencesareboxed.
Noticethechangeinreadingframeoccurringat aminoacid114and
itsrestoration at residue145.
J.
VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.612.320.559.289.658.2]BmNPV POLYHEDRIN GENE 441
Bm Ac Bm Ac
Bm
Ac Bm Ac Bm Ac Bm Ac Bm Ac Bm Ac Bm Ac Bm Ac
rF Asn Tyr Ser Tyr Asn Pro Thr Ile Gly Arg Thr Tyr Val Tyr Asp Asn Lys Tyr Tyr
I* Asp -5 Arg. . . .,)7,5. . .
40:
Lys Asn Leu G1y G1y Leu Ile Lys Asn Ala Lys Arg Lys Lys His Leu I 1e Giu His G1u
Ala Val .. . . Phe Ala
45 5 0 6
LysGluT Glu Lys Gln Trp[sp| Leu Leu Asp Asn Ty Met Vai Ala Glu Asp Pro Phe Leu
Ile| Ala Thr Leu Pro 0 Leu e ;
65 7
6uW--G1ly Pro G1ly Lys Asn G1ln Lys Leu Thr Leu Phe Lys G1lu ValI Arg Asn Val Lys Pro Asp
0 . .
~~~
. . . Ile . .85 90 95 1(C)
Thr MietLys Leu|IleVal sn r Ser Gly Lys Glu Phe Leu Arg Glu Thr Trp Thr Arg
* * * * Val Gly Lys Tyr[r* * *
105 1T1F 1151i5
e1ValIGiu Asp Ser Phe Pro le Vai Asn Asp Gin Giu Val Met Asp Val Tyr Leu Val
Met Phe
I-5 1 'M0 17i 140
Ala n Leu Lys Pro Thr Arg Pro Asn Arg Cys Tyr Lys Phe Leu Aia Gln His Ala Leu
Val I 18etM Arg . . . .
145 15iC 155 160e
rg Trp Asp Glu Asp Tyr Val Pro His Glu VaIle Arg Ile Met Giu Pro Ser Tyr Val
* Cys . Pro . Asp . Val .Trp
-. 165 170; 175 1E30
bMet Asn Asn Glu Tyr Arg Ile Ser Leu Ala Lye Lys Gly Gly Gly Cys Pro Ile Met
* Serj Gi. l T T A S Phe G . [ . . . .
sn Ile|His Ser Glu Tyr Thr Asn Ser Phe Glu |Ser |he jVal Asn Arg Val Ile Trp Glu
*ILeul * Glnl *Ile Asp
Bm Asn Phe Tyr Lys Pro Ile Val Tyr Ile Gly Thr Asp Ser Ala Giu Giu Glu Glu Ile Leu
Ac * * * * * * * * * * * * * * . . . 0
225 r=*>;230 2735 244:
Bm Ile Glu Vai Ser Leu Val Phe Lye Ile Lys Glu Phe Ala Pro Asp Ala Pro Leu Phe Thr
Ac Leu * * .Val . * . * . . . .
Bm Gly Pro Ala Tyr
Ac *
FIG. 5. Comparisonof thepolyhedrinsequencesofBmNPVand AcNPV. The BmNPV
polyhedrin
sequencepredicted
from thesequence of thegeneis comparedwith thatpredictedfromthecorresponding
geneofAcNPV(23). Dotsin thesequenceof AcNPV indicate thesame amino acids asthosein the BmNPV sequence,andblocksofidentical sequencesareboxed.times through Bm-5 cells and restricted with enzymes that
produce reasonably small numbers ofwell-separated DNA
fragments wasanalyzed onagarosegels.
After hybridizations of Southern transferred restriction
digests of BmNPV DNA to the probe (937 base pairs) containing part of the AcNPV polyhedrin gene, single re-strictionfragmentswerefound tohybridizeto theprobeina number of different digests (Fig. 1). These fragments pre-sumablycontainthecorrespondinggeneof BmNPV. Two of
them, a 9.6-kb EcoRI and a 10-kb PstI fragment (Fig. 2), werecloned,and theapproximate locations ofthesequences hybridizing to the AcNPV probe were determined. After nucleotide sequence determination (Fig. 3), the hybridizing portion of BmNPV DNA was identified as the polyhedrin
gene. This identification was based on the amino acid sequence ofthe polypeptide that resultedfrom the concep-tual translation of the corresponding RNA sequence after
comparisons with the published sequence of the authentic
BmNPV polyhedrin (26, 39) and with that derived from the
polyhedrin gene sequences of AcNPV (23).
Nucleotide sequences of the polyhedrin gene. The
se-quencesdeterminedfor thepolyhedringene of BmNPV and
its vicinity (Fig. 3) include 571 nucleotides offlanking and
mRNA noncoding sequences to the 5' end of the ATG
initiator codon (nucleotide 1 inFig. 3),acodingsequenceof
738 nucleotides(includinginitiationand terminationcodons), and 751 nucleotides of mRNA noncoding and flanking re-gions downstream from the 3' end of the TAA terminator
(nucleotide 738 in Fig. 3). Since no information is yet availableonthe BmNPVpolyhedrin mRNA sequences, the cap addition site has been inferred at position -57 by homologytothecapsitereported recentlyforthepolyhedrin
gene of AcNPV (23, 42). DNA sequences similar to the
consensus TATAand CCAAT boxes that have been shown to
represent
important
elementsofeucaryotic
genepromot-ers
(6)
werefound in the 5'flanking region
ofthe BmNPVpolyhedrin
gene 28 and 63 nucleotidesupstream
from theputative
capsite,
respectively (Fig.
3,
positions
-85 and-120).
As with the AcNPVpolyhedrin
gene(23),
theob-served TATAbox deviates from the established canonical
sequence
(TACAAA
forTATAAA).
Deviations of thistype,
however,
are notunusual,
particularly
in genes of viralorigin
(2).
Anextra set ofTATAand CAATsequences wasalsoobservedat
positions
-116and-151,
respectively (Fig.
3).
Of course, the contributions ofany ofthesignals
men-tioned above to
polyhedrin
genepromoter
function arespeculative
and mayonly
be deducedthrough
transcrip-tionalstudies.
The
coding
portion
ofthepolyhedrin
gene of BmNPV isnot
interrupted
by intervening
sequences. This is also truewith the
polyhedrin
genesofAcNPV(23, 42)
and ofthe NPVof
Orgyia
pseudotsugata
(OpNPV [36]),
whose N-terminalpolyhedrin
sequencewasfound tobe verysimilartothatofBmNPV
polyhedrin
(37).
In termsofcodonusage,theTACcodon for
tyrosine
was found to be used at afrequency
of13/16,
whereas the AAC codonforasparagine
occurredatafrequency
of 15/18(the
frequencies
of thecorresponding
codons in the AcNPV
polyhedrin
gene are 12/15 and12/14,
respectively
[23]).
Except
for these two cases, no otherstrong codon usage
preferences
were noted.The
polyhedrin
mRNAs of AcNPV andOpNPV
havebeen shown to be
polyadenylated
(36, 50),
and mostlikely
the same is true for thepolyhedrin
mRNA of BmNPV. Because thepublished
sequenceofthe AcNPVpolyhedrin
genedoesnotextendtothepoint
ofthe mRNA polyadenyl-ateadditionsite,
this sitecannotbe inferredonthegeneforVOL.54,1985
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.612.124.496.71.346.2]-18. -172 -162 -152 -142 -132 -122 Sm TCGACAAGsI C1,TTCTTT G rE&C GrE&AGGTCT CAATCCTATC TGIAATIATI IT4AAp
Ac iiGTCTGCGAGG A AGCA TTGTAATGAGACGCACAAAC TAAXAAC
-11 -102 -92 -82 -72
Bm ACAATTA A ATGT AATT TGTTTTTTAT TAAATc5CA --
-Ac
AWT664A
ATGTC*A;CA ATATATAGTT A-67 -57 -47 -37 -27 -17 -7
Sm AAATAATAAC CATCACGCAA ATAAATAAGT ATTTTACTGT TT'TCGTAACA GTTTTGTAAT AAAAAAACCT
Ac AAATGATAAC CATCTCGCAA ATAAATAAGT ATTTTACTGT TTTCGTAACA GTTTTGTAAT AAAAAAACCT
Bm ATAAAT.
Ac ATAAATA
15 30 45 60
Bm ATG CCG AAT TAT TCA TAC AO CCC ACC ATC GGGCGT ACITAC GTG TAC GAC AAI AAATAI
Ac ATG CCG SAT TAT TCA TAC CGT CCC ACC ATC6G6 CGT ACCTAC GTGTAC GAC AACAAGTAC
75 90 105 120
Bm TAC AAA AAC TTG GGC GGT CTC ATC AAA AACGCSAAG CGC AAG AAG CAC TA ASICGAA CAT
Ac TAC AAA AAT TTASSTG CCGTT ATC AAGAAC GCT AAG CGC AAGAAG CAC TTC GCC GAA CAT
135 150 165 180
Bm GAA AAA GAGGAGAAG CAA TGG GAT CII CTA GAC AAC TACeTG GTGCCTGAAG GAT CCC TTI
Ac GAG ACGAA GAG GCTACC CTCGACCCC CTA GAC AAC TAC CTA GTG GCT GAG GAT CCT TTC
195 210 225 24i
Sm ITA GGA CCG GGCAAA AAC CAA AAA CTT ACC CTTTTI AAA GAG GTT CGC AAT GTGAAA CCC
Ac CTGGSACCCGGC AAS AAC CAA AAA CTCACTCTCTTC AAGGAA ATCCGT AAT GTt AAA CCC
255 270 285 300
Bm GAT ACC ATGAAG TTA ATC GTCAA, TGGA,G GG, AAA GAS TTI CI ,GST GAA ACT TGG ACC
Ac SAC6AC8 ATGAAGCT;tTC GTt GGbTGGAAA GGA AAA GAG TTCtA A GG GAA ACT TGG ACC
31S 330 345 360C
Bm CGTTTT STT GAG GAC AGC TTC CCC ATT GTA AAC GAC CAA GAG GTG ATG GACGTG TAC CTC
Ac CGC TTC ATG GAA GAC AGC TTC CCC ATT GTT AAC SAC CAA GAA GTG ATG GAT GTt TTCCTT
375 390 405 42C)
Bm GTC GCC AAC CTC AAACCC ACA CGC CCC AAC AGGTGC TAC AAG TTC CT, GCT CAA CAC GCT
Ac GTT G6CAAC ATGS6TCCC ACT AGA CCC AAC CGT TGTTAC AAA TTC CTG GCCCAA CAC GCT
435 450 465 480
Bm CTTAGGTGGSAC SAA GAC TAC GTGCCCCAC GAO GT ATC AGAAT! ATGGAGCCA TCC TA
Ac CTG CGT TGCGAC CCC GAC TAT GTACCTCAT GAC GTG ATT AGG ATC GTCGAGCCt TCA TGG
49S 510 525 540
Bm GTG GGC ATGAAC AAC GAA TAC AGA AT! AGS CTG GCTAAA AAG GGC GGC GGC TGC CCA AT;
Ac GTG GGC AGCAAC AAC GAG TAC CGCATCAGC CTG GCTAAGAAG GGC GGC GGC TGC CCA ATA
555 570 585 600
Bm ATG AAC ATCCAC AGCGAG TAC ACC AAC TCGTTC GAGICG TT GTG A,AA CGS GTCATA TGG
Ac ATGAAC CTT CAC TCT GAGTACACC AAC TCGTTC GAA CAGTTCATC A CG GTC ATCTGG
61S 630 645 66L0
Bm GAG AAC TTC TAC AAACCC ATC GTT TAC ATC GGC ACAGAC TCT GC GAA GAAGAG GAA AT,
Ac GAGAAC TTC TAC AAGCCC ATC GTT TAC ATC GGt ACC GACTCT GCTGAA GAG GAG GAA AT?
675 690 705 720
Bm CTA ATT GAG GTT TCI CTC GTTTTC AAA ATA AAGGAGTTT GCA CCA GAC GCG CCT CTGTTC
Ac CTC CTT GAA GTT TCCCTGGTGTTC AAA GTA AAG GAGTTT GCA CCA GAC GCA CCTCTGTTC
735 Bm ACT GGT CCG GCA TAT TAA
Ac ACTGGT CCG GCGTAT TAA
748 758 768 778 788 798 B80
Bm AACACTATAC ATTGTTATTA STACATTTAT TAAGCGTTAG ATTCTGTACGTTGTTGATTTACAGACAATT
Ac AACAC6ATAC ATTGTTATTA GTACATTTAT TAAGCGCTAGATTCTGTGCG TTGTTGATTTACAGACAATT
18B 828 838 848 858 868 878
Bm GTTGTACGTA TTTTAATAAC TCATTAAATT TATAATCTTT AGGGTGGTAT GTTAGAGCGA AAATCAAATG Ac GTTGTACGTA TTTTAATAAt TCATTAAATT TATAATCTTT AGGGTGGTAT GTTAGAGCGA AAATCAAATG
s88 898 908 918 928 938
Bm ATTTTCAGCGTCTTTQTATC TGAATTTAAA TATTAAATCC TCAATAGATT TGTAAAATAG GTTTCGA Ac ATTTTCAGCG TCTTTATATC TGAATTTAAATATTAAATCC TCAATAGATT TGTAAAATAGGTTTCGA
FIG. 6.
Sequence comparison
betweenthe BmNPVand AcNPVpolyhedrin
genesandtheirsurrounding
sequences.(A)The 5'flanking sequencesof thetwogeneswhich have diversifiedconsiderably
arecompared
aftergapswereintroducedtomaximizehomologies.
Blocks oftwoor moreidentical nucleotidesareboxed,anddotsindicate nucleotide differences.(B)Thecomparison begins
atnucleotide -76of the sequenceofthe BmNPV gene andcontinuestothe lastknown nucleotideofthe AcNPVpolyhedrin
gene sequence(23).Except
forablank introduced after nucleotide-1ofthe BmNPVpolyhedrin
gene sequencetoaccommodateanextraAresiduepresentin thegeneof AcNPVpolyhedrin,
no sequencerearrangementswerenecessaryfor maximumhomology.
Nucleotide differencesareindicatedby
dots.BmNPV polyhedrin by homology. However, the sequence AATAAA, showntobe presentin the 3'noncodingregion of
almostallpolyadenylatedeucaryoticmRNAs approximately 25to30nucleotidesupstreamfrom the site of polyadenylate addition (35), occursatposition 1081ofourgene(Fig. 3). It should be noted thatasecond AATAAA sequence ispresent
approximately 90nucleotides downstream from the firstone
(position 1174).Anunambiguousanswer astothe location of thepolyadenylationsite mayonlybederivedbySinuclease
protection experiments (5). Protection experiments ofthis kindto determine the length ofAcNPV polyhedrin mRNA have indicated a cytoplasmic mRNAsize of approximately
1,200 nucleotides, excluding the polyadenylate tail (42). Assuminga similar mRNAsizefor BmNPVpolyhedrin, that value wouldplace theend of the BmNPV mRNA at around 60 nucleotides downstream from the first polyadenylation signal.
Amino acid sequence ofpolyhedrin. When the amino acid sequencepredictedfrom the geneprimary structure (Fig. 3) was compared to that of the authentic protein (26, 39), a small number ofdiscrepancies were noted whichprevented
the absolutematchingof thetwosequences.The differences are shown in Fig. 4 and are summarized as follows (the numbers inFig. 4 and those mentioned below correspond to
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.612.149.461.73.528.2]BmNPV POLYHEDRIN GENE 443
oursequence and areexclusive ofthe initiating methionine residue): (i) amino acid substitutions in dipeptides at posi-tions 39-40 (His-Glu for Glu-His) and 213-214 (Ser-Ala for
Ala-Ser); (ii) an amino acid substitution at position 218 (Glu for Gln); (iii) the presence of a single valine residue at
position 114instead oftwo,resulting in ashift oftheamino acidreading frame by one residue; (iv)the presence ofan extraamino acid residueat position 144(Glu) missing from
thepublished polyhedrinsequence (26, 39),whichresults in therestoration ofthecolinearamino acid reading frame;and (v) amino acid substitutions at positions 142 (Trpfor Gln)
and 143 (Aspfor Asn) within theregion in whichthereading frameshift has occurred.
Ofthe observed differences, those described in (i) above are positional. Ofthe remaining amino acid substitutions, those at positions 143 and 218 may be explained by the
accumulation of single point-mutational events. The amino
acid substitution at position 142, however, requires the
accumulation ofatleasttwo point mutations on the
partic-ular codon. More complex mechanisms should, ofcourse,
be used toexplain the deletion and insertion ofthe codons fortheamino acid residuesat positions 114and 144. These
differences areobviously morethan whatonewouldexpect from simple variants of the same virus. Since we have
confirmed our nucleotide sequence analysis and since the
sequenced moiety of the cloned
fragment
was the only portion of the 3mNPV genome that hybridized to the AcNPVpolyhedringeneprobe,wefeel thatareexamination of the polyhedrinprotein sequencing data may beappropri-ate.
Polyhedrins of BmNPV and AcNPV. The sequence of
BmNPV polyhedrin predicted by the primary structure of
the gene was also compared with that predicted from the
correspondingsequences ofAcNPV
(Fig.
5).No rearrange-mentswere requiredtoalignthetwoamino acidsequences,which areclearly highly homologous. The degree of
diver-gence between the two sequences is 13.9% (34 amino acid substitutions inatotalof244
residues).
The collectiveresultofthe amino acid substitutionsappears tobe
neutral,
sincethe net change in charge effected by them amounts to one and the overall degree of
hydrophobicity
remainsun-changed. In
addition,
computerized predictions
ofthesec-ondary structures of BmNPVand AcNPV
polyhedrins (15,
28) reveal that although some regional differences may occur, the overalldistributions
of a-helical andp-pleated
sheet structures within the two molecules were identical (datanot shown). Inthis respectitis also worthmentioning
that BmNPV polyhedrin has 11 arginine
residues,
all of which arealso present in the polyhedrin ofAcNPV, whichhas atotal of13 arginine residues. Ofthecodonsfor the 11
arginine residues sharedbythe twosequences, 5 have been altered by double point mutations resulting in codons again specifying arginine residues. Thus, itappears that thechanges
in thepolyhedrin molecule during evolution occurred under considerable selectivepressure. This isfurther corroborated by the results obtained when the N-terminal amino acid
sequences of the
polyhedrins
from two types ofOpNPV
werecompared with those of the polyhedrin of BmNPV (37). Asequenceidentityof
88.9%
wasfoundinthecomparisons involving the first 36 residues of a multicapsid OpNPV,whereas the sequence homology for 34 residues of a
uni-capsid OpNPVwas85.3% (thecorrespondingvaluesfor the
comparisonbetween BmNPV and AcNPVpolyhedrinswere 88.2and 86.8%for the first 34 and 36residues, respectively
[Fig.
5]).
Sequence homologybetweenthepolyhedringenesofBmNPV
and AcNPV. The nucleotide sequences
determined
for thepolyhedrin
gene ofBmNPV werecompared
with those of thepolyhedrin
geneof AcNPV. Thiscomparison
indicatedthatmajor portionsofthetwosequences may becompletely correlated
(Fig.
6).Forthe protein-encoding
regions
ofthe genes (shown to havediverged by
13.9% at the amino acid level[Fig. 5]),
a22.2%
divergence
was determined(164
nucleotide substitu-tions in a total of738).
Of the observedchanges,
110(two-thirds) represented
silent substitutionsresulting
inthe appearance ofsynonymouscodons,
whereas 54(one-third)
were replacement substitutions
leading
to amino acidchanges.
From these results it is apparent that alarge
number of silent substitutionshavebeenfixed inthe
coding
portion
of the genesduring
evolution and that the twonucleotide sequences have
diverged
somewhat more thanthe
corresponding
aminoacid
sequences. Thisindicates,
inturn,
that theprotein
itselfhas been undermorestringent
selectivepressure than thecorresponding
DNA sequence.Incontrast tothe
divergence
found in thecoding regions
ofthetwopolyhedrin
genes, thenucleotidesequencesof the5'
noncoding regions
andthose ofasmallportion
of the5'flanking
regions
show a remarkable conservation. Infact,
exceptforan extra
nucleotide
that is presentin the58-nu-cleotide-long
5'noncoding
region
ofthe AcNPV genejust
before the ATGinitiation codon(Fig. 6),
thetwononcoding
sequences are identical. Even this difference may not be
significant,
since in another isolate of AcNPV the extranucleotide was not observed
(E.
B.Carstens, unpublished
data).
This remarkablelength
and nucleotidesequencecon-servationin the 5'
noncoding regions strongly
suggests thatcapping
ofthetwomRNAsmayoccur atthe sameposition
(-57 in the sequences shown in
Fig. 6).
The sequencehomology
continues fartherupstreamfor another20 nucleo-tides(19
identical residuesoutof20)
and stopsatnucleotide-76
just
beforethebeginning of
thepresumed
TATAboxes. Asimilarly
striking homology
wasalsoobserved in the 3'noncoding regions
ofthegenes.Only
5nucleotidesubstitu-tions
wereobserved in the 207 nucleotidesofthe sequences that werecompared
(2.4%
sequencedivergence).
Unfortu-nately,
because more sequence information on the AcNPVpolyhedrin
genewasnotavailable,
wecould notextendthecomparison
to cover the entire 3'noncoding region
andflanking
gene sequences.Taking
intoconsideration,
how-ever, that mutations
probably
occurreduniformly
through-outthetwogenomes
(including
thepolyhedrin genes),
it is obviousthat,
in contrast to thepoint
mutations
that have beenfixed inthecoding
regions, only
averysmall numberof substitutions were allowed to be fixed in the 5' and 3'noncoding regions
and inthe immediate5'flanking
regions
of the genes
during
evolution. This of course should beindicative of the
degree
of selective pressure which thedifferent
portions
of the genes areunder
because of thefunctions
they
havetoaccomplish.
No
significant homology
canberecognized
atfirstglance
in the 5'
flanking
gene sequences upstreamfromnucleotide-76
(Fig.
6,
upperpanel).
With the aid of a dot matrixprogram,wescanned thetwosequences for the presence of
homologies
thatmayoccurwithout respecttoposition.
Asa result of this search we were able to construct different versionsof sequencealignments.
Oneofthesealignments
ispresented
inFig.
6(upper
panel).
Although
the overall sequenceidentity
isonly
38.8%,
several blocksofcommon sequences may bediscerned(boxes
inFig. 6). Except
for thelarge
gap of ninenucleotidesintroducedatthe 3' endof the BmNPV sequence,only
threesingle
nucleotide gaps haveVOL.54,1985
on November 10, 2019 by guest
http://jvi.asm.org/
beenintroduced inthe sequence of AcNPVtoallow forthe alignment. We would like to emphasize not the nucleotide sequencesassuch but thefact that thedistancesamong all of theobservedhomology blocks are thesame(±1nucleotide) in both flanking sequences. At this point we can only speculateonthesignificance of these blocks.Transcriptional
experiments would be needed to determine whether they participate in the regulation of polyhedrin gene activity. Despite our uncertainties, sequences of this type should be
considered, particularly when molecular engineering in the
vicinity of thepolyhedringene isto beundertaken. Is the polyhedrin gene immediately flanked by other viral genes? Since we may need to insert a fragment offoreign
genetic material such as B. mori genomic DNA in the
vicinity ofthepolyhedringenewhile preservingtheabilityof the virus to function properly (including the formation of
polyhedra),itisimportantnotonlytoknow the exact limits of the polyhedrin gene but also what lies next to it. In this respect we have noticed the two openreadingframesat the twoendsof thesequenced portionof theclonedDNA. More extensive studies are needed to determine whether the observed open reading frames are parts ofadjacent struc-turalgenes. Such studies are now in progress.
ACKNOWLEDGMENTS
We thankJ. L. Vaughn and R. Stone ofthe Insect Pathology Laboratoryfor thegifts ofBm-5cellsandinitial inocula ofBmNPV as wellasfor theiradvice for the establishment ofthecellsin our laboratory, E.B. Carstens for supplying us with the cloned HindIII-Vfragment of the AcNPV genome and forcommunicating to ushis sequences of the AcNPVpolyhedrin gene before publica-tion;G. Chaconas of theCancer ResearchLaboratory, University of WesternOntario,formakingavailableto usthecomputer program
onprotein secondary structurepredictions, J.C. States of the De-partmentofMedicalBiochemistry, UniversityofCalgary,for hisgift ofpUC9 and host cells, D. McKay of the Department of Medical Biochemistry, University of Calgary, for providinguswithsomeof the computer software used for the analysis ofour nucleic acid sequences, B. Pinder and R. Haselden for photography, Anne Vipondfor technicalassistance, and Susan Carlson for secretarial assistance.
This work was supported by grants from the Cancer Grants Program of the Alberta Heritage Savings Trust Fund (Alberta CancerBoard)andthe Medical Research Council of Canada (toK. Iatrou)and byapostdoctoralfellowshipfromthe AlbertaHeritage Foundation for Medical Research (to H.W.).
LITERATURE CITED
1. Adang,M.J.,and L. K. Miller.1982.MolecularcloningofDNA complementarytomRNAofthe baculovirusAutographa cali-fornicanuclearpolyhedrosisvirus: locationandgeneproducts ofRNAtranscriptsfound late in infection.J.Virol.44:782-793. 2. Baker, C. C., J. Herisse, G. Courtois,F. Galibert,and E. Ziff. 1979. MessengerRNAfor the Ad2 DNAbinding protein: DNA sequences encoding the first leader and heterogenity at the mRNA 5'end. Cell 18:569-580.
3. Bergold,G. H. 1947. DieIsolierungdesPolyeder-Virus unddie Natur derPolyeder. Z. Naturforsch.26:122-143.
4. Bergold,G. H. 1953. Insectviruses. Adv. Virus Res. 1:91-139. 5. Berk, A. J., and P. A. Sharp. 1978. Spliced early mRNA of simianvirus 40. Proc. Natl.Acad. Sci. U.S.A.75:1274-1278. 6. Breathnach, R., and P. Chambon. 1981. Organization and
ex-pression ofeukaryotic split genes coding for proteins. Annu. Rev.Biochem. 50:349-383.
7. Brown, M.,P.Faulkner,M. A.Cohran,and K. L.Chung.1980. Characterization of two morphology mutants ofAutographa californicanuclearpolyhedrosisviruswithlargecuboidal inclu-sionbodies.J.Gen. Virol. 50:309-316.
8. Carstens,E. B. 1982. Mappingthe mutation site of an Auto-graphacalifornicanuclearpolyhedrosis virus polyhedron
mor-phologymutant.J. Virol.43:809-818.
9. Couch,J.A.,S.M.Martin, G. Tompkins, and J. Kinney. 1984. Asimplesystemforthepreliminaryevaluation ofinfectivityand pathogenesisofinsect virus inanontargetestuarineshrimp.J. Invertebr.Pathol. 43:351-357.
10. Dulbecco,R.,and M.Vogt. 1954.Plaque formation and isolation of pure lines with polyomyelitis viruses. J. Exp. Med. 99: 167-182.
11. Duncan,R.,K.L.Chung,and P. Faulkner.1983.Analysisofa
mutant ofAutographa californica nuclear polyhedrosis virus with a defect in the morphogenesis of the occlusion body macromolecular lattice. J. Gen. Virol. 64:1531-1542.
12. Duncan, R., and P. Faulkner. 1982. Bromodeoxyuridine-in-ducedmutantsofAutographacalifornicanuclearpolyhedrosis virusdefective in occlusion body formation. J. Gen. Virol. 62: 369-373.
13. Faulkner, P. 1981. Baculovirus, p. 3-37. In E.W. Davidson (ed.), Pathogenesis of Invertebrate Microbial Diseases. Allan-held, Osmum &Co., Totowa, N.J.
14. Fraser,M.J.,and W. F.Hink. 1982. Theisolation and charac-terization of theMPand FPplaque variants of Galleria mellon-ellanuclearpolyhedrosis virus. Virology 117:366-378. 15. Garnier, J.,D.J. Osguthorpe,and B.Robson. 1978.Analysis of
the accuracy andimplicatiops of simple methods forpredicting the secondarystructureofglobular proteins. J. Mol. Biol. 120: 97-120.
16. Goldsmith, M. R., andF. C. Kafatos. 1984. Developmentally regulated genesin the silkmoths. Annu. Rev. Genet. 18:443-487. 17. Grace, T. D. C. 1967. Establishment ofalineof cells from the
silkwormBombyxmori. Nature(London) 216:613.
18. Groner, A.,R.R.Granados,andJ.P.Burand.1984. Interaction ofAutographa californica nuclear polyhedrosis virus withtwo
nonpermissivecell lines. Intervirology 21:203-209.
19. Harrap, K. A., and C. C.Payne. 1979. The structuralproperties andidentification of insect viruses. Adv. VirusRes.25:273-355. 20. Heckman, J. E.,andU. L.RajBhandary. 1979. Organization of
tRNA and rRNA genes in N. crassamitochondira: intervening sequences in thelarge rRNA gene and strand distribution of the RNA genes. Cell 17:583-595.
21. Himeno, H.,F.Sakai,K.Onodera, H. Nakai,T.Fukada, andY. Kawade. 1967. Formation of nuclear polyhedral bodies and nuclear polyhedrosis virus of silkworm in mammalian cells infected with viral DNA.Virology 33:507-512.
22. Hink, W. F., and E. Strauss. 1976. Replication and passage of alfalfa looper nuclear polyhedrosis virus plaque variants in cloned cell cultures and larval stages of four host species. J. Invertebr. Pathol. 27:49-55.
23. Hooft vanIddekinge, B. J. L., G. E. Smith, and M.D.Summers. 1983. Nucleotide sequence of the polyhedrin gene of Auto-grapha californica nuclear polyhedrosis virus. Virology 131: 561-565.
24. Iatrou, K., and S. G. Tsitilou. 1983. Coordinately expressed chorion genes of Bombyx mori: is developmental specificity determined by secondary structure recognition? EMBO J. 2:1431-1440.
25. Ignoffo, C.M.1973. Effects ofentomopathogensonvertebrates. Ann. N.Y. Acad. Sci. 217:141-172.
26. Kozlov, E. A., T. L. Levitina, N. M. Gusak, and S.B. Sere-bryani.1981. Comparison of the amino acid sequence of inclu-sion body proteins of nuclear polyhedrosis viruses Bombyx mori, Porthetriadispar and Galleria mellonella.Bioorg. Khim. 7:1008-1015.
27. Lee,H.H.,and L. K.Miller. 1979.Isolation, complementation, and initial characterization oftemperature-sensitive mutantsof the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol.31:240-252.
28. Lifson, S., and C. Sander. 1979. Antiparallel and parallel -strands differ inamino acid residue preferences. Nature (Lon-don)282:109-111.
29. Maxam, A. M., and W. Gilbert. 1977. A new method for sequencingDNA. Proc.Natl. Acad. Sci. U.S.A. 74:560-564. 30. Messing, J., B. Gronenborn, B. Muller-Hill, and P.H.
Hof-schneider. 1977. Filamentouscoliphage M13 as acloning
on November 10, 2019 by guest
http://jvi.asm.org/
BmNPV POLYHEDRIN GENE 445
cle: insertion ofaHindllfragment of the lac regulatory region in M13 replicative form in vitro. Proc. Natl. Acad. Sci. U.S.A. 74:3642-3646.
31. Miller, D. W., and L. K. Miller. 1982. Avirus mutant with an insertion ofacopia-liketransposable element.Nature (London) 299:562-564.
32. Miller, L. K., and K. P. Dawes. 1979.Physicalmapof the DNA genomeof Autographa californica nuclear polyhedrosis virus. J. Virol. 29:1044-1055.
33. Miller, L. K., A. J. Ling, and L. A. Bulla,Jr. 1983. Bacterial, viral andfungal insecticides. Science 219:715-721.
34. Pennock, G. D., C. Shoemaker, and L. K. Miller. 1984. Strong andregulated expression of Escherichia coli
P-galactosidase
in insect cells with a baculovirus vector. Mol. Cell. Biol. 4:399-406. 35. Proudfoot, N. J., and G. G. Brownlee. 1976. 3' non-coding sequencesineukaryoticmessenger RNA. Nature(London) 263: 211-214.36. Rohrmann, G. F., D. J. Leisy, K.-C. Chow, G. D. Pearson, and G.S. Beaudreau. 1982.Identification, cloning andR-loop map-ping of the polyhedrin gene from the multicapsid nuclear polyhedrosis virusof Orgyia pseudotsugata. Virology 121:51-60. 37. Rohrmann, G. F., M. N. Pearson, T.J. Bailey, R. R. Becker, and G. S. Beaudreau. 1981. N-terminal polyhedrin sequences andoccluded baculovirus evolution. J. Mol. Evol. 17:329-333. 38. Ruther, U. 1982. pUR250 allowsrapid chemical sequencing of
both DNA strands of its inserts. Nucleic Acids Res. 10: 5765-5772.
39. Serebryani, S. B., T. L. Levitina, M.L. Kautsman, Y. L. Radavski, N. M.Gusak, M. N. Ovander, N. V. Sucharenko, and E. A. Kozlov. 1977. The primary structure of the polyhedral proteinof nuclear polyhedrosis virus(NPV)of Bombyxmori.J. Invertebr. Pathol.30:442-443.
40. Smith,G. E., M. J. Fraser, and M. D. Summers. 1983. Molec-ular engineering of the Autographa californica nuclear poly-hedrosis virusgenome:deletionmutations withinthepolyhedrin gene. J.Virol. 46:584-593.
41. Smith, G. E., M. D.Summers,and M.J.Fraser. 1983. Produc-tion of human beta interferon in insect cells infected with a
baculovirus expressionvector. Mol. Cell. Biol. 3:2156-2165. 42. Smith, G. E., J. M. Vlak, and M. D. Summers. 1983. Physical
analysis of Autographa californica nuclear polyhedrosis virus transcripts for polyhedrin and 10,000-molecular-weightprotein. J.Virol. 45:215-225.
43. Smith, H. O., and M. L. Birnstiel. 1976. A simplemethod for DNArestriction site mapping. Nucleic AcidsRes.3:2387-2398. 44. Southern, E. M. 1975. Detection of specificsequences among DNAfragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517.
45. Tinsley, T. W. 1978. Use of insect pathogenic viruses as pesticidal agents, p. 199-210.In M. Pollard (ed.), Perspectives in Virology vol. 10. Raven Press, NewYork.
46. Tjia, S. T., G. M. Altenschildesche, and W. Doerfler. 1983. Autographa californica nuclear polyhedrosis virus (AcNPV) DNA does not persist in mass cultures of mammalian cells. Virology 125:107-117.
47. Trager, W. 1935. Cultivation ofthe virus ofgrasserie in silk-worm tissue cultures.J. Exp. Med. 61:501-517.
48. Vieira, J., and J. Messing.1982. ThepUC plasmids,an M13mp7-derivedsystemfor insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268.
49. VIak, J. M., and K. G. Odink. 1979. Characterization of Autographa californica nuclear polyhedrosis virus deoxyri-bonucleic acid.J. Gen. Virol. 44:333-347.
50. VIak,J. M.,G. E. Smith,andM. D. Summers. 1981. Hybridi-zation selection and in vitro translation ofAutographa califor-nicanuclearpolyhedrosis virusmRNA.J. Virol.40:762-771. 51. Wahl, G. M., M.Stern,and G. R. Stark.1979.Efficient transfer
of largeDNAfragments fromagarosegels to diazobenzyloxy-methyl-paper andrapid hybridization by usingdextransulfate. Proc. Natl. Acad. Sci. U.S.A. 76:3683-3687.
52. Weiss, S. A., G. C.Smith,S. S. Kalter, andJ. L.Vaughn. 1981. Improved method forthe production of insect cell cultures in largevolume. InVitro 17:495-502.
53. Wood, H. A. 1980. Isolation and replication of an occlusion body-deficient mutant of the Autographa californica nuclear polyhedrosis virus. Virology105:338-344.
VOL. 54,1985