0022-538X/09/$08.00⫹0 doi:10.1128/JVI.01587-08
Copyright © 2009, American Society for Microbiology. All Rights Reserved.
The Amount of Hepatocyte Turnover That Occurred during Resolution
of Transient Hepadnavirus Infections Was Lower When Virus
Replication Was Inhibited with Entecavir
䌤
†
William S. Mason,
1* Chunxiao Xu,
1‡ Huey Chi Low,
2,3Jeffry Saputelli,
1Carol E. Aldrich,
1Catherine Scougall,
2,3Arend Grosse,
2,3Richard Colonno,
4§ Sam Litwin,
1and Allison R. Jilbert
2,3*
Fox Chase Cancer Center, 333 Cottman Avenue, Philadelphia, Pennsylvania 191111; Infectious Diseases Laboratories, SA Pathology,
Adelaide SA 5000, Australia2; School of Molecular and Biomedical Science, University of Adelaide, Adelaide SA 5005,
Australia3; and Bristol-Myers Squibb Pharmaceutical Research Institute, Wallingford, Connecticut 064924
Received 25 July 2008/Accepted 27 November 2008
Transient hepadnavirus infections can involve spread of virus to the entire hepatocyte population. In this situation hepatocytes present following recovery are derived from infected hepatocytes. During virus clearance antiviral cytokines are thought to block virus replication and formation of new covalently closed circular DNA (cccDNA), the viral transcriptional template. It remains unclear if existing cccDNA is eliminated noncytolyti-cally or if hepatocyte death and proliferation, to compensate for killing of some of the infected hepatocytes, are needed to remove cccDNA from surviving infected hepatocytes. Interpreting the relationship between hepato-cyte death and cccDNA elimination requires knowing both the amount of hepatohepato-cyte turnover and whether cccDNA synthesis is effectively blocked during the period of immune destruction of infected hepatocytes. We have addressed these questions by asking if treatment of woodchucks with the nucleoside analog inhibitor of viral DNA synthesis entecavir (ETV) reduced hepatocyte turnover during clearance of transient woodchuck hepatitis virus (WHV) infections. To estimate hepatocyte turnover, complexity analysis was carried out on virus-cell DNA junctions created by integration of WHV and present following recovery in the livers of WHV-infected control or ETV-treated woodchucks. We estimated that, on average, 2.2 to 4.8 times less hepatocyte turnover occurred during immune clearance in the ETV-treated woodchucks. Computer modeling of the complexity data suggests that mechanisms in addition to hepatocyte death were responsible for elimi-nation of cccDNA during recovery from transient infections.
Hepadnavirus infection can spread to the entire hepatocyte population of the liver and result in a period of high-titer virus production of 4 to 6 weeks or more. A vigorous immune re-sponse then initiates the immune clearance phase with hepa-tocyte destruction and suppression of virus replication (11, 12, 19, 23, 24). Previous work indicated that uninfected hepato-cytes in the recovered liver were derived either directly from or as progeny of hepatocytes that had been infected (12, 21). However, it remains to be determined how uninfected hepa-tocytes arise from infected hepahepa-tocytes, a process that requires elimination of virus replicative intermediate (RI) DNA from the cytoplasm and, more importantly, covalently closed circu-lar DNA (cccDNA) from the nucleus of the infected hepato-cytes or from their progeny.
Three models have been proposed to explain how transient hepadnavirus infections are cleared from the liver. In all three models hepatocytes present at recovery are derived from previ-ously infected hepatocytes. In model 1, cytokines released during the immune response to the infection inhibit virus replication, prevent synthesis of viral RI DNA and cccDNA, and eliminate both RI DNA and cccDNA from infected hepatocytes. Immune-mediated attack on infected hepatocytes may be considered cru-cial to the process that causes cytokine release, but the resulting hepatocyte death and compensatory proliferation per se have no essential role in the production of uninfected hepatocytes (7). In models 2 and 3, cytokines again inhibit virus replication and replenishment of cccDNA but do not act on preexisting cccDNA; infected hepatocytes are targeted for killing, provoking prolifer-ation of surviving infected and uninfected hepatocytes. In model 2, cccDNA does not survive mitosis and is therefore lost when an infected hepatocyte divides. Virus clearance in this model could be achieved with as little as 0.7 liver equivalents of cumulative hepatocyte turnover (i.e., death and compensatory proliferation to restore liver cell mass) (14, 18).
In model 3, cccDNA loss occurs only via hepatocyte death. cccDNA is proposed to survive mitosis and to be distributed in a binomial fashion to progeny hepatocytes. Repeated rounds of hepatocyte mitosis in response to destruction of other infected hepatocytes eventually result in the formation of lineages of un-infected hepatocytes. Starting with an average of 30 copies of cccDNA per hepatocyte, model 3 requires⬃2.6 liver equivalents of hepatocyte death to achieve virus clearance (14, 18). * Corresponding author. Mailing address for William S. Mason: Fox
Chase Cancer Center, 333 Cottman Avenue, Philadelphia, PA 19111. Phone: (215) 728-2462. Fax: (215) 728-2412. E-mail: ws_mason@fccc .edu. Mailing address for Allison R. Jilbert: School of Molecular and Biomedical Science, University of Adelaide, Adelaide SA 5005, Australia. Phone: 61-8-8303 5399. Fax: 61-8-8303 7532. E-mail: allison .jilbert@adelaide.edu.au.
‡ Present address: Drexel Institute for Biotechnology and Virology Research of Drexel University College of Medicine, Pennsylvania Bio-technology Center, 3805 Old Easton Road, Doylestown, PA 18902.
§ Present address: Presidio Pharmaceuticals, Inc., 1700 Owens Street, Suite 585, San Francisco, CA 94158.
† Supplemental material for this article may be found at http://jvi .asm.org/.
䌤Published ahead of print on 10 December 2008.
1778
on November 8, 2019 by guest
http://jvi.asm.org/
A key assumption in all three models is that cytokines inhibit virus replication and prevent new cccDNA formation, via in-tracellular pathways and infection from without (3), during the immune clearance phase. If this assumption were incorrect in some individuals and cytokines did not completely block new cccDNA formation concomitant with the onset of hepatocyte destruction, hepatocyte turnover during virus clearance might vastly exceed the amount required by these models, irrespec-tive of how cccDNA loss actually occurs.
To test the validity of this assumption, we examined whether the presumed inhibition of intracellular replication attributed to cytokines could be potentiated by the nucleoside analog entecavir (ETV) when it was used to treat woodchucks during transient woodchuck hepatitis virus (WHV) infection. ETV is
a potent inhibitor of virus DNA synthesis (5, 9) and has been approved by the FDA for the treatment of patients with chronic hepatitis B virus infections. Hepatocyte turnover was estimated by complexity analysis of virus-cell DNA junctions created by integration of WHV and present following recovery in the livers of WHV-infected control or ETV-treated wood-chucks. Since our expectation had been that cytokine-mediated inhibition of cccDNA formation and killing of infected hepa-tocytes were highly coordinated in time and that cytokines completely blocked cccDNA formation during the immune clearance phase, we assumed that ETV treatment would not reduce the amount of hepatocyte turnover during resolution of a transient infection.
[image:2.585.52.539.80.462.2]We found instead that the amount of hepatocyte turnover TABLE 1. Time course of transient WHV infection
Woodchuck and treatment group
(age关yr兴)
No. of wks postinfection
cccDNA (copies/ hepatocyte)a
RI DNA (copies/ hepatocyte)a
% Core antigen-positive
hepatocytesb
% RI DNA-positive hepatocytesb
% PCNA-positive hepatocytesc
% Intralobular CD3-positive
cellsc
% Apoptotic hepatocytesd
% Ceroid-containing Kupffer cellse
Untreated animals
wc530 (1) 0 ⬍0.004 0 ⬍0.003 ⬍0.004 0.4 12,1 0.97 1.9
4 14.5 760 ⬎95 ⬎95 0.01 11.2 0.22 0.7
9 0.02 8 0.4 ⬍0.003 1.9 38.4 0.75 5.0
13 0.04 7 ⬍0.002 ⬍0.003 0.4 17.2 0.49 4.6
16 0.04 0.2 ⬍0.001 ⬍0.003 0.1 3.6 0.34 3.0
wc535 (2) 0 ⬍0.004 0 ⬍0.003 ⬍0.004 0.01 5.7 0.72 1.7
4 22.3 1,500 ⬎95 ⬎95 ⬍0.01 2.1 0.37 2.0
9 15.5 760 ⬎95 ⬎95 2.6 66.5 9.17 11.5
13 0.31 21 0.003 0.70 0.8 13.6 2.85 10.8
16 0.4 1.1 ⬍0.001 ⬍0.007 0.3 5.1 0.90 11.5
wc558 (2) 0 ⬍0.004 0.02 ⬍0.004 ⬍0.008 ⬍0.01 4.2 0.40 1.9
5 21.6 1,600 ⬎95 ⬎95 ⬍001 4 0.54 1.6
10 35.8 2,200 ⬎95 ⬎95 0.3 13.1 1.53 27.3
14 0.03 33 1.2 0.18 0.5 31.4 3.48 21.3
19 0.016 0.2 ⬍0.0015 ⬍0003 0.4 9.2 0.51 5.9
wc559 (1) 0 ⬍0.004 0 ⬍0.0028 ⬍0.006 0.01 9.4 1.45 0.6
5 27.8 1,700 ⬎95 ⬎95 0.02 10.6 0.58 2.3
10 45.5 1,500 ⬎95 ⬎95 0.07 36.3 2.00 11.5
14 6.2 320 20 11 0.9 53.2 1.52 2.9
19 0.05 110 ⬍0.001 ⬍0.002 0.03 15.1 0.34 7.1
ETV-treated animals
wc522 (3) 0 ⬍0.004 0 ⬍0.003 ⬍0.003 0.06 2.4 0.33 3.3
7 28.1 1,100 ⬎95 ⬎95 0.3 12.2 1.01 2.2
12 ⬍0.004 0.2 ⬍0.004 ⬍0.003 0.06 13.5 0.80 9.7
16 ⬍0.004 0.02 ⬍0.001 ⬍0.003 0.08 6.3 0.79 6.0
wc531 (3) 0 ⬍0.004 0 ⬍0.004 ⬍0.006 0.06 10.1 1.10 4.0
4 11.2 1,300 ⬎95 ⬎95 0.08 7.7 0.69 0.5
9 0.01 2 ⬍0.004 ⬍0.005 0.07 17.3 0.43 4.9
13 0.02 0.3 ⬍0.003 ⬍0.004 0.3 28.3 0.95 1.9
16 0.03 0.004 ⬍0.001 ⬍0.002 0.1 8.4 0.61 3.9
wc533 (1) 0 ⬍0.004 0 ⬍0.003 ⬍0.004 0.07 3.8 0.58 2.2
4 32.5 2,300 ⬎95 ⬎95 ⬍0.01 4.5 0.65 2.9
9 13 115 80 4.5 1.8 30.2 2.85 24.0
13 9.8 15 1.6 0.04 0.4 9.1 1.46 8.0
16 0.15 0.3 ⬍0.001 ⬍0.002 1.6 12.9 1.28 8.3
acccDNA and RI DNA copies per hepatocyte were determined by qPCR as described in Materials and Methods.
bThe percentages of WHV core antigen- and WHV DNA-positive hepatocytes were determined by immunostaining and in situ hybridization, respectively, as described in Materials and Methods.
cThe percentage of PCNA-positive hepatocytes was determined using a microscope eyepiece grid and scanning⬃100 fields at a magnification of⫻400. The percentage of CD3-positive liver cells was determined by scanning between 20 and 50 fields. The number of CD3-positive liver cells is expressed as a ratio per 100 hepatocytes.
dTerminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assays were performed on formalin-fixed liver sections as described in Materials and Methods.
eCeroid-containing Kupffer cells were detected using PAS-D staining, as described in Materials and Methods, and are expressed as a percentage of total liver cells using a microscope eyepiece grid.
VOL. 83, 2009 TRANSIENT HEPADNAVIRUS INFECTIONS 1779
on November 8, 2019 by guest
http://jvi.asm.org/
that occurred during resolution of a transient WHV infection was lower in the ETV-treated than untreated woodchucks. Analyses of these data suggest that a significant portion of the hepatocyte turnover observed in some woodchucks is due to incomplete suppression of virus DNA synthesis by antiviral cytokines and, therefore, a failure to completely prevent for-mation of new cccDNA during periods of active hepatocyte destruction by the cellular immune response. The effect of ETV on hepatocyte turnover approached statistical signifi-cance (P⫽0.057), but a larger study would be needed to prove that the effects are due to ETV and not to woodchuck-to-woodchuck variation during immune clearance of the virus.
Finally, using computer modeling of the infected liver, we were unable to reconcile the wide variation in hepatocyte turn-over between individual woodchucks or between woodchucks in the ETV-treated and untreated groups to cccDNA clear-ance by model 3. This suggests that model 1, model 2, or a combination of models 1 and 2, but not model 3, plays a major role in recovery from transient hepadnavirus infections.
MATERIALS AND METHODS
Infection of woodchucks with WHV.Adult woodchucks were trapped in south-east Pennsylvania and housed in the Laboratory Animal Facility of the Fox Chase Cancer Center. All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the Fox Chase Cancer Center. Animals ranged in age from 1 to 3 years at the time of WHV infection (Table 1). Infection was initiated by inoculation of 1 ml of serum from a chron-ically WHV-infected woodchuck (virus titer of⬃109per ml) into the saphenous
vein. Wedge liver biopsies were carried out as described previously (2, 11, 12). Liver tissues were fixed in 10% formalin in phosphate-buffered saline or
ethanol-acetic acid (3:1) and dehydrated and embedded in paraffin or quick-frozen and stored at⫺80°C for subsequent nucleic acid extraction. Woodchucks were bled each week following infection to detect levels of viremia using a hybridization spot test (15). Treatment with ETV was initiated when virus became detectable by this assay (⬎108WHV DNA genomes per ml). ETV, which was generously
supplied by Bristol-Myers Squibb, Wallingford, CT, was dissolved in water at a concentration of 1 mg per ml and administered orally (16) at a daily dose of 0.5 mg per kg of body weight until autopsy.
Determination of virus titers by qPCR. To extract virion DNA, 25l of woodchuck serum was mixed with 400l of 0.1 M NaCl, 0.05 M Tris-HCl, pH 7.8, 0.01 M EDTA, 0.2% sodium dodecyl sulfate, and 0.1 mg/ml pronase and incubated for 2 h at 37°C. The mixture was extracted once with an equal volume of phenol. Forty micrograms of dextran carrier was added to the aqueous phase, and nucleic acids were collected by ethanol precipitation. Quantitative PCR (qPCR) was carried out as previously described (28) using the following primers specific to the 5⬘end region of WHV minus-strand DNA: P1, 5⬘-TCAGACGA GTCGGATCTCCCTT-3⬘(nucleotides [nt] 1644 to 1665); P2, 5⬘-AAGTCGCA TGCATTTATGCCTACA-3⬘(nt 1922 to 1899). All WHV primers were num-bered according to Cohen et al. (4) (WHV sequence with accession no. M18752).
Extraction and analysis of liver DNA.Total liver nucleic acids for quantitation of virus RI DNA and a Hirt supernatant fraction for quantitation of cccDNA were extracted from⬃20 to 50 mg of liver (11). The concentration of DNA in total nucleic acid extracts was determined as described by Labarca and Paigen (13).
WHV RI DNA was quantified by qPCR with the primers described above for serum DNA using 20-ng samples of liver DNA, equivalent to 4,000 diploid woodchuck liver cells. After digestion with EcoRI (to linearize cccDNA), the equivalent proportion of each Hirt supernatant fraction was assayed by qPCR for cccDNA using primers that spanned the cohesive overlap: P1, 5⬘-TCAGACGA GTCGGATCTCCCTT-3⬘(nt 1644 to 1665); and P3, 5⬘-AGCAATGTTCCCTA CCTGTTA-3⬘(nt 2165 to 2145).
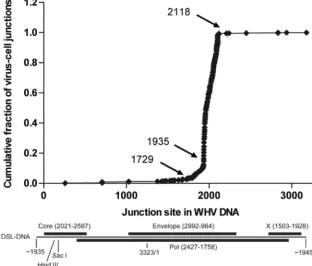
[image:3.585.135.447.72.338.2]Inverse PCR for detection of integrated WHV DNA.Inverse PCR for detection of integrated WHV DNA was carried out on DNA extracted from⬃1 to 3 mg of liver tissue, as previously described (17, 21). In brief, the DNA was digested with SacI, which cleaves WHV DNA at nt 2240 and cell DNA at an unknown position. FIG. 1. Map of virus-cell junction sites on the left-hand end of WHV DSL DNA. The genome position on WHV DNA of virus-cell junctions identified by inverse PCR in autopsy liver samples from all seven woodchucks is shown. The virus-cell junction is defined as the last nucleotide identical to WHV DNA. The putative left-hand end of DSL DNA at nt 1935 and the upstream end of the cohesive overlap of the relaxed circular DNA genome at nt 1729 are also shown. Of the 1,202 virus-cell junctions, 1,036 mapped between nt 1935 and nt 2118. The relationship between DSL DNA and the WHV open reading frames is shown at the bottom. The SacI and HindIII sites used during inverse PCR are also noted.
on November 8, 2019 by guest
http://jvi.asm.org/
The digested DNA was then incubated with T4 DNA ligase to circularize the restriction fragments and then linearized by digestion with HindIII, which cleaves WHV DNA at nt 2190, to create a fragment in which cellular DNA, including the virus-cell junction, is flanked by virus DNA. The DNA was then diluted into 96-well trays so that each well contained no more than one virus-cell junction, and the diluted DNA was subjected to nested PCR, as previously described (21). To prevent amplification products from being generated from WHV cccDNA, AflII, which cleaves WHV DNA at nt 2303, was included in the first PCR mix, and the trays were preincubated for 30 min at 37°C. The outer primers were P4 (5⬘-TTAGTATGCTGGGATGAATTAA-3⬘; nt 2195 to 2216) and P5 (5⬘-AATAGCTGTATGGTGCGGA-3⬘; nt 2185 to 2167); the inner prim-ers were P3 (5⬘-AGCAATGTTCCCTACCTGTTA-3⬘; nt 2165 to 2145) and P6 (5⬘-CTAAATTGATAGCTTGGATGA-3⬘; nt 2217 to 2237).
The inverse PCR detects virus-cell DNA junctions that occur near the left-hand end of double-stranded linear (DSL) DNA. Automated sequencing of virus-cell junctions was carried out using the WHV primer P3 (5⬘-AGCAATG TTCCCTACCTGTTA-3⬘; nt 2165 to 2145). Sequence alignments using the GCG program FASTA were used to locate virus-cell junctions. Cellular se-quences adjacent to virus DNA were screened using Sequencher (Gene Codes Corporation) to identify and align virus-cell junctions from hepatocytes that had undergone clonal expansion. A graphic summary of the virus-cell junctions de-tected in liver fragments collected at autopsy from seven woodchucks is shown in Fig. 1. Of the 1,202 virus-cell junctions shown in Fig. 1, 1,036 mapped between nt 1935, the putative left-hand end of WHV DSL DNA, and nt 2118.
Immunostaining and in situ hybridization of sections of woodchuck liver.
Formalin-fixed liver tissue was sectioned at 6m, deparaffinized, rehydrated, and used for hematoxylin and eosin staining, periodic acid Schiff-diastase(PAS-D), and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end label-ing for apoptotic cells, uslabel-ing a DeadEnd Colorimetric TUNEL System (Pro-mega). Immunostaining of WHV core antigen, proliferating cell nuclear antigen (PCNA) (Dako), and CD3 (Dako) were carried out using ethanol-acetic acid-fixed liver tissue sections without antigen retrieval, using normal rabbit serum as
a negative control, as previously described (8, 12). WHV DNA was detected by in situ hybridization using sections of ethanol-acetic acid-fixed liver tissue. Full-length WHV DNA and control (pBluebac4.5; Invitrogen) probes were labeled with digoxigenin-UTP by nick translation (Roche). Sections were prepared for in situ hybridization essentially as previously described (10, 17) incorporating a prehybridization step in 50% deionized formamide and 0.5 mg/ml carrier DNA without labeled probe for 60 min at 37°C. Sections were stained with hematoxylin.
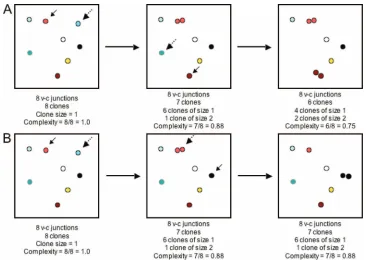
Computer modeling of the relationships between the complexity of virus-cell junctions and hepatocyte turnover.The ratio of the number of unique virus-cell integration junctions to total integration junctions detected defines the complex-ity of a population of virus-cell junctions present in a liver fragment. Thus, if all junctions are unique, occurring only once, the ratio will be 1.0, but if some hepatocytes are killed and others proliferate, then the overall complexity will be
⬍1.0 (see Results) (Fig. 2). In a liver of fixed size, the relationship between complexity and hepatocyte turnover can vary depending upon which hepatocytes are targeted. For instance, if all hepatocytes in a fully infected liver have an equal probability of being killed and of contributing to hepatocyte proliferation irre-spective of whether they are still infected, then the relationship between com-plexity and cumulative hepatocyte turnover will be different than if all hepato-cytes can proliferate, but infected hepatohepato-cytes are much more likely to be killed than uninfected hepatocytes. In addition, any losses of host DNA during exper-imental manipulations may increase the observed complexity because, for exam-ple, one or more members of a set of integrants are lost before the sample is subjected to nested PCR.
[image:4.585.109.475.66.326.2]To examine how hepatocyte turnover might be related to complexity and clonal expansion of hepatocytes during virus clearance, a computer simulation of an infected liver, comp10, was constructed. The model relates hepatocyte turn-over occurring during resolution of a transient hepadnavirus infection to changes in the complexity of the population of virus-cell junctions. We suppose that at time zero the liver is fully infected and contains a fraction of hepatocytes in which virus genomes are integrated into random, distinguishable sites in cellular DNA FIG. 2. The relationship between complexity of virus-cell junctions and cumulative hepatocyte turnover. The relationship between complexity and cumulative hepatocyte turnover is shown for a hypothetical liver of eight hepatocytes. (A) An example of hepatocyte turnover reducing complexity. In the upper-left panel, all the hepatocytes are uniquely identified and occur only once. Thus, the complexity of the population is 1.0 (unique/total hepatocytes). The hepatocyte marked with a dashed arrow will be killed and be replaced by proliferation of the hepatocyte marked by the solid arrow, as shown in the middle panel. The complexity is now 7/8 or 0.88. The process is repeated in the middle panel, reducing the complexity to 6/8 or 0.75 (right panel). (B) An example of hepatocyte turnover that does not reduce complexity. In the bottom left-hand panel, a hepatocyte marked with a dashed arrow is killed and is replaced by the hepatocyte marked with a solid arrow, reducing the complexity to 7/8 or 0.88. The process is repeated in the bottom middle panel, but this time one of the hepatocytes that has just divided will die and another will divide. Since there were two copies of the hepatocyte that died, death and proliferation do not change the complexity of the population, which remains at 7/8 or 0.88.
VOL. 83, 2009 TRANSIENT HEPADNAVIRUS INFECTIONS 1781
on November 8, 2019 by guest
http://jvi.asm.org/
to create specific virus-cell junctions. The nucleus of each infected hepatocyte contains a random number of cccDNA molecules, a number which is uniformly distributed or follows Poisson distribution but truncated to be limited. In the current study we assumed a uniform distribution of between 10 and 50 cccDNA molecules per nucleus, with an average of 30.
For each cycle in a simulated liver, a fraction of infected hepatocytes is killed, and a smaller fraction of uninfected hepatocytes also dies. In the next time cycle hepatocytes are selected at random to proliferate to restore liver cell mass, and others are selected to be killed since, to our knowledge, there is no evidence that a significant, sustained loss of liver cell mass typically occurs during recovery from transient infections.
It should be noted that the rate of killing, at least in the range we looked at (up to 35% per cycle [see Fig. S7 in the supplemental material]), did not appreciably change the relationship between complexity and cumulative hepatocyte turnover needed to clear an infection. This means that a lag of up to 35% between death and compensatory proliferation would not significantly alter the complexity/liver turnover predictions, provided the liver returns to full size after every cycle.
One percent killing of infected hepatocytes per cycle was used in the simula-tions illustrated in Tables 2, 3, and 4; killing was assumed to be zero order (i.e., if 1% of hepatocytes were killed during cycle 1, then the same number of hepatocytes would be killed on all subsequent cycles), and the liver was assumed to replicate back to full size after every cycle. During this phase of infection, virus replication is blocked (the model was also programmed to measure the conse-quences of random hepatocyte turnover during active virus replication or during a period of active virus replication that is followed by a period in which virus replication is blocked, as illustrated for some of the simulations in Table 4). cccDNA is distributed to progeny hepatocytes at user-defined efficiencies of 0 to 1 until it is all lost due to hepatocyte death and/or proliferation, signifying resolution of the infection. Hepatocyte death may thus cause loss of some hepatocytes with integrated virus DNA and proliferation of other hepatocytes with integrated virus DNA. In this way some virus-cell junctions are duplicated while others are lost (Fig. 2). A Monte-Carlo simulation of the liver comprised of 1 million hepatocytes was used to estimate the variation of complexity and hepatocyte clone size (of hepatocytes containing specific virus-cell junctions) over time and as a function of the amount of hepatocyte turnover, that is, to estimate how much hepatocyte turnover may be necessary to remove all cccDNA and cure the infection or reduce it to a low but defined level. The model estimates the average cccDNA copy number variation over time as well as the distribution of copy number on the final day simulated.
The model was also designed to predict how delayed replication of hepatocytes during the recovery phase would affect the predicted relationships between
complexity and hepatocyte turnover, based on the hypothesis that significant losses of hepatocytes might occur before liver regeneration was initiated. In the example shown in Fig. S8 in the supplemental material, we have calculated this relationship for different rates of liver regeneration, assuming that liver regen-eration did not initiate until there was a 50% drop in the number of hepatocytes. As shown, significant changes due to the lag in liver regeneration occur only if the rate of liver regeneration is either very rapid or else very close to, though larger, than the rate of hepatocyte death. A comparison of the simulations described in Table 4 is discussed in the legend to Fig. S8 in the supplemental material.
To determine the consequences of incomplete sample recovery during DNA isolation and experimental manipulations, integrants were selected at random for deletion at the end of each simulation, as illustrated in Table 4.
The Fortran code for comp10 and the supporting file, comp10.in, are available in the supplemental material. A copy of the program is available upon request.
RESULTS
[image:5.585.44.542.80.254.2]Basis for the analysis of the complexity of virus-cell junc-tions and hepatocyte turnover. Integration of hepadnavirus DNA at random sites in host DNA occurs at a low frequency during infection, largely from DSL DNA, which is formed as TABLE 2. Virus-cell junctions and complexity in untreated and ETV-treated woodchucks infected with WHV
Woodchuck and treatment group
Sample date (wk p.i.)a
Total no. of unique virus-cell junctionsb
Total no. of virus-cell junctionsb Complexity of virus-cell junctionsc Minimum cumulative hepatocyte turnoverd Cumulative hepatocyte turnover (random)e Untreated animals
wc530 16 98 151 0.65 0.35 0.53
wc535 16 146 312 0.47 0.53 1.1
wc558 19 84 320 0.26 0.74 2.8
wc559 19 57 236 0.24 0.76 3.1
Avg 0.41f 0.59 1.88
ETV-treated animals
wc522 16 121 182 0.66 0.34 0.51
wc531 16 39 58 0.67 0.33 0.49
wc533 16 118 138 0.86 0.14 0.16
Avg 0.73f 0.27 0.39
ap.i., postinfection.
bExtracted liver tissue was analyzed using inverse PCR followed by sequencing for the detection of unique and total virus-cell junctions as described in Materials and Methods.
cThe complexity of virus-cell junctions was calculated as the ratio of unique to total virus-cell junctions detected. dThe minimum cumulative hepatocyte turnover was determined as shown in Fig. 2A as 1.0 minus complexity.
eThe cumulative hepatocyte turnover due to random killing and compensatory proliferation needed to reach the measured complexity was calculated using the program comp10 described in Materials and Methods. In this simulation infected hepatocytes were assumed to be killed at 20 times the rate of uninfected hepatocytes (1% versus 0.05% per cycle, respectively); virtually identical values were calculated when the death rate for uninfected hepatocytes was set at zero.
[image:5.585.299.543.609.676.2]fThe complexity measurements in individual untreated woodchucks were compared to those in the ETV-treated group using a Wilcoxon two-samples, two-sided nonparametric test (P⫽0.057), and approached statistical significance.
TABLE 3. Evidence in untreated and ETV-treated woodchucks infected with WHV that complexity did not increase in the
weeks following recovery
Woodchuck Treatment
Sample date (wk p.i.)a
Total no. of virus-cell junctionsb Total no. of unique virus-cell junctionsb Complexity of virus-cell junctionsc
wc530 None 9 192 134 0.70
wc522 ETV 12 82 51 0.62
ap.i., postinfection.
bExtracted liver tissue was analyzed using inverse PCR followed by sequenc-ing for the detection of unique and total virus-cell junctions as described in Materials and Methods.
cThe complexity of virus-cell junctions was calculated as the ratio of unique to total virus-cell junctions detected.
on November 8, 2019 by guest
http://jvi.asm.org/
an aberrant by-product of normal virus DNA synthesis as a result of in situ priming of the virus plus-strand DNA (20). DSL DNA represents⬃10 to 20% of virion DNA in chroni-cally infected woodchucks (25) and is the preferred substrate for integration into host DNA (1, 6, 27). When DSL DNA enters the nucleus to amplify cccDNA copy number, it can either form an aberrant cccDNA by illegitimate recombination (26) or integrate into host DNA, again by nonhomologous end joining (1, 6, 27).
Since integration occurs at random sites in cellular DNA, each integration event creates a unique virus-cell junction that can be used as a genetic marker of that hepatocyte to assess hepatocyte turnover. Sequencing of virus-cell junctions serves to identify individual integration events. Thus, the number of times a unique virus-cell junction appears in a liver fragment indicates how many times the hepatocyte containing that virus-cell junction had divided following integration to form a he-patocyte clone of defined size.
Measurement of the complexity of virus-cell junctions in a liver fragment from a recovered woodchuck provides an esti-mate of how much hepatocyte turnover (death and prolifera-tion) occurred during the infection (21). This is illustrated in Fig. 2A for a population of eight hepatocytes. Thus, if one hepatocyte dies and another divides, the complexity will be reduced to 7/8 or 0.88, and so on. Therefore, complexity allows a direct, albeit minimal estimate of the amount of hepatocyte death and proliferation that has taken place (i.e., 1⫺ com-plexity). The relationship between hepatocyte turnover and complexity can be more complicated (Fig. 2B). For instance, if a hepatocyte is killed that already belongs to a hepatocyte clone of size 2 and if any other hepatocyte proliferates to maintain liver mass, the complexity does not change because there has been no loss of genetic diversity (Fig. 2B). Thus, a complexity value of 7/8 or 0.88 predicts that at least one he-patocyte has died. In this example, two hehe-patocytes have ac-tually been killed. Using complexity analysis, we estimated in a previous study of transient WHV infection that, cumulatively, at least 1 liver equivalent (100%) of hepatocytes were killed
during a transient WHV infection, presumably during the im-mune clearance phase, based on an assumption of random death and proliferation of both uninfected and infected hepa-tocytes throughout the recovery phase (12, 21). However, as discussed below, the relationship between complexity and he-patocyte turnover is actually more complicated and is influ-enced by the mechanism of virus clearance. Thus, our previous estimates (12, 21) would provide a close approximation to virus clearance by model 3, in which uninfected hepatocytes accumulate in large numbers only toward the end of virus clearance but, as discussed later, would not apply to clear-ance via model 2.
Transient WHV infection in the presence and absence of ETV.To determine if incomplete inhibition of virus replication might contribute to the large amount of immune-mediated hepatocyte death during virus clearance, seven woodchucks were infected with WHV. Once⬎95% of hepatocytes were infected, as assessed by detection of a high-titer viremia (⬎108
per ml) (Fig. 3) and subsequently confirmed by immunostain-ing of liver sections for WHV core antigen and in situ hybrid-ization of WHV DNA (Table 1; Fig. 4 and 5), ETV treatment was initiated in three woodchucks to inhibit synthesis of RI DNA and cccDNA, with the remaining four serving as WHV-infected, untreated controls.
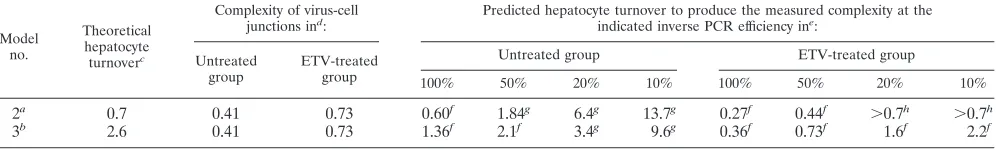
The four WHV-infected but untreated control animals showed the expected pattern of WHV infection, with spread of the virus to infect ⬎95% of hepatocytes by 4 or 5 weeks postinfection. With the exception of woodchuck 530 (wc530), in which infection cleared by 9 weeks postinfection, in the three other woodchucks widespread virus antigen expression and RI DNA persisted in ⬎95% of hepatocytes at 9 or 10 weeks postinfection, followed by rapid resolution of infection with complete clearance (wc535) at week 13 or partial clear-ance (wc558 and wc559) at week 14 postinfection. In all four woodchucks clearance of WHV-infected hepatocytes was mir-rored by decreases in cccDNA and RI DNA detected by qPCR and increased levels of PCNA expression, CD3 cell infiltration, TABLE 4. Efficiency of inverse PCR experiment: effect on estimates of predicted hepatocyte turnover
Model no.
Theoretical hepatocyte turnoverc
Complexity of virus-cell junctions ind:
Predicted hepatocyte turnover to produce the measured complexity at the indicated inverse PCR efficiency ine:
Untreated group
ETV-treated group
Untreated group ETV-treated group
100% 50% 20% 10% 100% 50% 20% 10%
2a 0.7 0.41 0.73 0.60f 1.84g 6.4g 13.7g 0.27f 0.44f ⬎0.7h ⬎0.7h
3b 2.6 0.41 0.73 1.36f 2.1f 3.4g 9.6g 0.36f 0.73f 1.6f 2.2f
aIn model 2, cccDNA is lost during mitosis.
bIn model 3, cccDNA survives mitosis. Hepatocytes are assumed to contain an average of 30 copies of cccDNA prior to the initiation of clearance, and cccDNA is distributed in a binomial fashion to progeny hepatocytes during mitosis.
cThe theoretical level of hepatocyte turnover required to clear WHV infection from 100% of hepatocytes, each containing 30 copies of cccDNA, was previously defined to be 0.7 and 2.6 liver equivalents for models 2 and 3, respectively (14, 18).
dThe complexity of virus-cell junctions present in the liver of WHV-infected control and ETV-treated woodchucks was determined as shown in Table 2. eThe efficiency of the inverse PCR reaction is assumed to fall within the range of 10 to 100%. The predicted cumulative hepatocyte turnover required to clear WHV infection with model 2 (cccDNA loss) or model 3 (cccDNA survival) was calculated based on the measured complexity of virus-cell junctions using the program comp10, as described in Materials and Methods, allowing for the range of efficiencies of the inverse PCR reaction. The liver was assumed to regenerate to full size after each cycle of hepatocyte killing.
fThe predicted hepatocyte turnover based on measured complexity is less than the theoretical hepatocyte turnover for model 2 or model 3 and is not sufficient to eliminate the virus from the liver.
gThe predicted hepatocyte turnover based on complexity is greater than the theoretical hepatocyte turnover for model 2 or model 3 and suggests that a period of random hepatocyte death and proliferation, to maintain liver cell mass, occurs prior to the clearance phase of the infection.
hThe predicted hepatocyte turnover based on complexity was not calculated because virus replication is inhibited by ETV even prior to initiation of immune clearance.
VOL. 83, 2009 TRANSIENT HEPADNAVIRUS INFECTIONS 1783
on November 8, 2019 by guest
http://jvi.asm.org/
[image:6.585.44.542.83.158.2]apoptotic hepatocytes, and ceroid-positive Kupffer cells de-tected by PAS-D staining (Table 1).
In contrast, treatment of woodchucks with ETV, as expected (5, 9), caused a rapid drop in viremia (Fig. 3) but did not appear to accelerate the clearance of WHV from the hepatocyte popu-lation (Table 1; Fig. 4 and 5) or to substantially alter the levels of PCNA expression, CD3 infiltration, apoptotic hepatocytes, or Kupffer cells compared to untreated woodchucks. While there was a wide range of differences in time to clearance between animals in both groups, this did not fall outside the ranges seen in previous studies of transient WHV infection of woodchucks (8, 12, 21). These markers, although providing supporting evidence for hepatocyte turnover in both control and ETV-treated animals, do not provide quantitative estimates. As described below, we therefore used inverse PCR and sequencing to determine the
complexity of virus-cell junctions present in one or more⬃1- to 3-mg fragments of liver collected from all control and ETV-treated woodchucks at autopsy and used these measurements to estimate levels of cumulative hepatocyte turnover.
[image:7.585.110.477.68.509.2]Complexity measurements of virus-cell junctions in autopsy liver samples.Complexity analysis was carried out on autopsy liver samples collected following recovery from infection. In brief, total DNA was purified from⬃1- to 3-mg fragments of liver and subjected to inverse PCR, essentially as described previously (21). Following nested PCR in 96-well plates, PCR products were resolved by gel electrophoresis, extracted, and sequenced to detect virus-cell junctions located near the left-hand end of DSL DNA (Fig. 1). The complexity of these collections of virus-cell junctions is summarized in Table 2, and the hepatocyte clone sizes observed are shown in Fig. 6. FIG. 3. Time course of viremia in WHV-infected untreated and ETV-treated woodchucks. Virus titers (WHV genomes per ml of serum) were determined by qPCR as described in Materials and Methods. The start of ETV (St. ETV) therapy (0.5 mg/kg/day) is also indicated for wc522, wc531, and wc533.
on November 8, 2019 by guest
http://jvi.asm.org/
FIG. 4. Time course of infection in the liver of a WHV-infected and untreated woodchuck. Liver tissue sections from wc559 (Table 1) were subjected to immunostaining to detect WHV core antigen (left panels) and in situ hybridization to detect WHV RI DNA (right panels), as described in Materials and Methods. More than 95% of hepatocytes were scored positive for WHV core antigen and RI DNA at 5 and 10 weeks (wk) postinfection. By 14 weeks, 20% remained positive for core antigen, and 11% were positive for RI DNA. Neither WHV core antigen nor WHV DNA was detectable at 19 weeks. Bar, 200 um.
1785
on November 8, 2019 by guest
http://jvi.asm.org/
FIG. 5. Time course of infection in the liver of a WHV-infected ETV-treated woodchuck. Liver tissue sections from wc533 (Table 1) were subjected to immunostaining to detect WHV core antigen (left panels) and in situ hybridization to detect WHV RI DNA (right panels), as
on November 8, 2019 by guest
http://jvi.asm.org/
There was a considerable amount of variation among indi-viduals in both the untreated and ETV-treated groups, sug-gesting the presence of unknown factors that influence the results, independent of the assignment of animals to the un-treated or ETV-un-treated groups. Not surprisingly, such factors might include qualitative, quantitative, or kinetic differences in the immune response to infection as well as variations in the efficiency of recovery of liver DNA during phenol extraction and subsequent manipulations. However, major sample-to-sample variation in complexity was not found when we ana-lyzed more than one fragment from the same woodchuck (e.g., for wc522, 0.59 and 0.72; wc530, 0.58, 0.62, and 0.73; wc533, 0.80, and 0.91; wc535, 0.43 and 0.51; wc559, 0.24 and 0.30), indicating that the differences in measured complexities reflect differences between woodchucks rather than variability of the assay.
One WHV-infected, untreated control woodchuck (wc530) had higher levels of complexity than the other three untreated controls, indicating that less hepatocyte turnover occurred in this woodchuck than in wc535, wc558, and wc559. This may be related to the observation that wc530 had a shorter course of WHV infection than the other three animals, with a shorter period of high-titer viremia (Fig. 2) and more rapid clearance of WHV-infected hepatocytes (before week 9 postinfection) than the other three untreated woodchucks that had WHV core antigen and RI DNA in⬎95% of hepatocytes at weeks 9 or 10 postinfection (Table 1).
Nevertheless, despite the higher level of complexity in wc530, a difference approaching statistical significance (P ⫽
0.057) exists in the average complexity measurements between the four untreated control woodchucks and the three ETV-treated animals. Thus, the average complexity values for the four untreated woodchucks (complexity of 0.41⫾ 0.19) was almost half that of the three ETV-treated animals (complexity of 0.73⫾0.11) after clearance. Moreover, samples from three out of four of the untreated woodchucks displayed clones of 10 or more hepatocytes, wc530 being the exception, whereas none of the clones in the ETV-treated animals were greater than 5 hepatocytes in size (Fig. 6).
It was possible that the difference in complexity between the two groups was due to a slightly longer lapse between resolu-tion of the infecresolu-tion and collecresolu-tion of samples for complexity analysis in the treated woodchucks; i.e., that the higher com-plexity reflected an unexpected loss of hepatocytes containing virus-cell junctions following recovery. To evaluate this possi-bility, we did a complexity analysis of virus-cell junctions in liver collected from one untreated and one ETV-treated wood-chuck (wc530 and wc522) when the infections first appeared to have resolved (Table 1 and Table 3). The results were similar to those collected at the later time point (Table 2). A similar stability of integrated WHV DNA was also observed when chronically infected woodchucks were treated with clevudine
(2⬘-fluoro-5-methyl--L-arabinofuranosyluracil), another in-hibitor of hepadnavirus DNA synthesis (22).
Complexity and cumulative hepatocyte turnover.The mini-mum cumulative hepatocyte turnover in the two groups (Table 2) (fraction of hepatocytes killed⫽1⫺complexity) indicates that hepatocyte turnover, on average, was 2.2 times higher in the group of untreated woodchucks. However, complexity measurements do not provide a direct indication of the amount of hepatocyte turnover. The value equal to 1⫺complexity is the minimum amount of cumulative hepatocyte turnover that could have occurred over time (Fig. 2), but this minimum requires that only hepatocytes in the original population were killed, as in model 2, for example, in which only undivided hepatocytes remain infected and therefore are targets for im-mune-mediated death. A much larger turnover for the same change in complexity would occur during a period of random death and proliferation, that is, when hepatocytes are selected at random for death or proliferation, as would occur in model 3, while most of the progeny hepatocytes are initially still infected and therefore remain targets for killing. The amount of cumulative hepatocyte turnover that would result from the indicated changes in complexity during a period of random death and proliferation was calculated using comp10 and is shown in Table 2. Assuming that a similar process of hepato-cyte turnover occurred in all woodchucks, this analysis suggests that there was on average 4.8 times more hepatocyte turnover in the untreated woodchucks.
DISCUSSION
This study was initiated to test the hypothesis that there is a strong suppression of virus replication by the host immune response throughout the immune resolution phase of a tran-sient hepadnavirus infection. Thus, it was expected that no difference in hepatocyte turnover (death and proliferation) would be seen between the WHV-infected, control, and ETV-treated woodchucks. Moreover, based on an earlier study (21), we expected to see rather consistent complexity measurements in all of the woodchucks. In the previous study, the complexity measurements were 0.5, 0.5, and 0.6, respectively, for the WHV-infected, control, and ETV-treated woodchucks. How-ever, this was not the case. In WHV-infected and untreated control woodchucks, there was a higher level of hepatocyte turnover than in the ETV-treated woodchucks (P ⫽ 0.057). This result, taken in conjunction with the wide range of differ-ences between individual woodchucks, suggests that significant hepatocyte turnover can occur when there is a failure or delay of the immune system in eliminating RI DNA and preventing formation of new cccDNA, i.e., in completely controlling virus replication.
Using comp10, we asked if the observed complexities, rang-ing from 0.24 to 0.86 between individual woodchucks and from
described in Materials and Methods. WHV core antigen and RI DNA were detected in⬎95% of hepatocytes at 4 weeks (wk) postinfection prior to the start of ETV therapy. Despite the fact that 80% of hepatocytes remained core antigen positive (9 wk; left), only a very weak signal from RI DNA was detected by in situ hybridization in 4.5% of hepatocytes at 9 weeks postinfection (right) or 5 weeks after initiation of ETV therapy. WHV core antigen was still detected in a small percentage of hepatocytes at 13 weeks postinfection but was no longer detectable at 16 weeks. Bar, 200 um.
VOL. 83, 2009 TRANSIENT HEPADNAVIRUS INFECTIONS 1787
on November 8, 2019 by guest
http://jvi.asm.org/
0.41 to 0.73 between the ETV-treated and untreated groups of woodchucks, were compatible with clearance by either model 2 or model 3. As shown in Fig. S7 in the supplemental material, model 2 predicts a complexity of⬃0.3, corresponding to⬃0.7 liver turnovers, while model 3 predicts a complexity of⬃0.15 corresponding to⬃2.6 liver turnovers during clearance, start-ing with WHV infection of 100% of hepatocytes, each contain-ing on average 30 copies of cccDNA (14, 18). However, this assumes 100% sample recovery during tissue extraction and processing.
Unfortunately, we cannot definitively know how closely the measured complexity values reflect the true complexities in the samples and therefore the amount of hepatocyte turnover they indicate according to each model. The complexity measure-ments depend on the efficiency of detection of all virus-cell junctions in the liver samples analyzed. The efficiencies of the inverse PCR assay are affected by the efficiency of each of the steps of DNA extractions, enzyme digestions, ligations, and PCR amplifications and are necessarily below 100%. While the restriction digestion and ligations appeared to go to comple-tion in test reaccomple-tions, the efficiency of DNA extraccomple-tion is un-certain but likely above 50%. As noted earlier, however, these sources of inefficiency, to the extent they exist, appear to be consistent as duplicate tissue samples from the same liver gave
essentially identical complexity values. We therefore used comp10 to predict the effect of assay efficiency on complexity measurements and hepatocyte turnover during immune clear-ance of an infection by either model 2 or model 3.
The calculated values for cumulative hepatocyte turnover at 100% efficiency and corrected values for 50, 20 and 10% effi-ciencies for both models 2 and 3, reflecting the hepatocyte turnover needed to achieve the experimentally measured com-plexities for untreated and ETV-treated woodchucks, are shown in Table 4. In some cases the hepatocyte turnover pre-dicted by model 2 or model 3 was not sufficient to clear the infection, whereas in others, much more hepatocyte turnover appeared to occur, based upon the observed complexity change, than was needed for clearance (i.e., 0.7 for model 2 and 2.6 for model 3). In this situation the total amount of hepatocyte turnover needed to reach the experimentally mea-sured complexity was calculated assuming that the immune clearance phase was preceded by a period of random death and proliferation in which virus replication was not inhibited. As shown in Table 4, the theoretical level of hepatocyte turnover predicted for model 2 of 0.7 liver equivalents is achieved in both the untreated and ETV-treated groups with an assumed assay efficiency of between 20 and 50%. The the-oretical level of hepatocyte turnover associated with model 3 of 2.6 liver equivalents would require an even lower efficiency to achieve clearance in the ETV-treated group (i.e., 5 to 10%). In that case, the predicted hepatocyte turnover in the WHV-infected control group would be extremely high (⬎9 liver equivalents). It should be noted that even larger amounts of hepatocyte turnover would be predicted if we attempted to reconcile differences between individual woodchucks (e.g., wc533 in the ETV-treated group and wc559 in the untreated group) to clearance by model 3.
Thus, the results by this analysis seem more consistent with the idea that virus clearance from the liver occurs via either model 1 or model 2 in which cccDNA does not survive mitosis since we are aware of no evidence for the consistently large amount of hepatocyte turnover predicted by model 3 (8, 12, 24). A similar conclusion was reached by a completely different approach involving a data-fitting analysis of the immune clear-ance phase of transient hepatitis B virus infections in chimpan-zees (19). The assumption that liver regeneration during immune clearance does not begin until at least 50% of hepa-tocytes have been killed did not significantly alter complexity and turnover predictions using model 3 (see Fig. S8 in the supplemental material) and therefore did not lend any unex-pected support to this model.
In addition, our data suggest that cytokine suppression of virus replication during the immune clearance phase of a hepadnavirus infection is often incomplete, resulting in 2.2 to 4.8 times or more hepatocyte turnover than would otherwise be needed to clear the virus.
ACKNOWLEDGMENTS
[image:11.585.55.272.66.359.2]We are grateful to Jesse Summers (University of New Mexico), John Taylor, and Christoph Seeger for helpful discussions during the course of this work and to Glenn Rall and Jesse Summers for a critical reading of the manuscript. We are also grateful for technical assistance from the facilities for animal care, DNA sequencing, histopathology, and oligonucleotide synthesis in the completion of this work.
FIG. 6. Assay for clonal expansion of hepatocytes following reso-lution of transient WHV infections. The number of identical virus-cell junctions and, by inference, the extent of clonal expansion of hepato-cytes, is plotted for autopsy liver samples from all seven woodchucks. Hepatocyte clone sizes greater than 10 were found for all of the untreated woodchucks except wc530 (which had a shorter time course of WHV infection) while all hepatocyte clone sizes were⬍6 for all woodchucks treated with ETV.
on November 8, 2019 by guest
http://jvi.asm.org/
Our research was supported by grants from the U.S. National Insti-tutes of Health (grants 5R01AI018641 and CA06927) and the National Health and Medical Research Council (Australia) (grant 453505) and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
1.Bill, C. A., and J. Summers.2004. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc. Natl. Acad. Sci. USA101:
11135–11140.
2.Carp, N. Z., J. Saputelli, T. C. Halbherr, W. S. Mason, and A. R. Jilbert.
1991. A technique for liver biopsy performed in Pekin ducks using anesthesia with Telazol. Lab. Animal Sci.41:474–475.
3.Ciupe, S. M., R. M. Ribeiro, P. W. Nelson, G. Dusheiko, and A. S. Perelson.
2007. The role of cells refractory to productive infection in acute hepatitis B viral dynamics. Proc. Natl. Acad. Sci. USA104:5050–5055.
4.Cohen, J. I., R. H. Miller, B. Rosenblum, K. Denniston, J. L. Gerin, and R. H. Purcell. 1988. Sequence comparison of woodchuck hepatitis virus replicative forms shows conservation of the genome. Virology162:12–20. 5.Genovesi, E. V., L. Lamb, I. Medina, D. Taylor, M. Seifer, S. Innaimo, R. J.
Colonno, D. N. Standring, and J. M. Clark.1998. Efficacy of the carbocyclic 2⬘-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hep-atitis B virus infection. Antimicrob. Agents Chemother.42:3209–3217. 6.Gong, S. S., A. D. Jensen, C. J. Chang, and C. E. Rogler.1999.
Double-stranded linear duck hepatitis B virus (DHBV) stably integrates at a higher frequency than wild-type DHBV in LMH chicken hepatoma cells. J. Virol.
73:1492–1502.
7.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari.1999. Viral clearance without destruction of infected cells during acute HBV infection. Science284:825–829.
8.Guo, J.-T., H. Zhou, C. Liu, C. Aldrich, J. Saputelli, T. Whitaker, M. I. Barrasa, W. S. Mason, and C. Seeger.2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infection. J. Virol.
74:1495–1505.
9.Innaimo, S. F., M. Seifer, G. S. Bisacchi, D. N. Standring, R. Zahler, and R. J. Colonno.1997. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob. Agents Chemother.41:1444–1448. 10.Jilbert, A. R.2000. In situ hybridization protocols for detection of viral DNA using radioactive and nonradioactive DNA probes. Methods Mol. Biol.123:
177–193.
11.Jilbert, A. R., T. T. Wu, J. M. England, P. de la M. Hall, N. Z. Carp, A. P. O’Connell, and W. S. Mason.1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J. Virol.
66:1377–1388.
12.Kajino, K., A. R. Jilbert, J. Saputelli, C. E. Aldrich, J. Cullen, and W. S. Mason.1994. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J. Virol.
68:5792–5803.
13.Labarca, D., and K. Paigen.1980. A simple, rapid, and sensitive DNA assay procedure. Anal. Biochem.102:344–352.
14.Mason, W., and S. Litwin.2002. Pathogenesis of hepadnavirus infections, p. 99–113.InS. Locarnini and C. L. Lai (ed.), HBV human virus guide. Inter-national Medical Press, London, United Kingdom.
15.Mason, W. S., C. Aldrich, J. Summers, and J. M. Taylor.1982. Asymmetric replication of duck hepatitis B virus DNA in liver cells: free minus-strand DNA. Proc. Natl. Acad. Sci. USA79:3997–4001.
16.Mason, W. S., J. Cullen, G. Moraleda, J. Saputelli, C. E. Aldrich, D. S. Miller, B. Tennant, L. Frick, D. Averett, L. D. Condreay, and A. R. Jilbert.1998. Lamivudine therapy of WHV-infected woodchucks. Virol-ogy245:18–32.
17.Mason, W. S., A. R. Jilbert, and J. Summers.2005. Clonal expansion of hepatocytes during chronic woodchuck hepatitis virus infection. Proc. Natl. Sci. USA102:1139–1144.
18.Mason, W. S., S. Litwin, C. Xu, and A. R. Jilbert.2007. Hepatocyte turnover in transient and chronic hepadnavirus infections. J. Viral Hepatitis14(Suppl. 1):22–28.
19.Murray, J. M., S. F. Wieland, R. H. Purcell, and F. V. Chisari.2005. Dynamics of hepatitis B virus clearance in chimpanzees. Proc. Natl. Acad. Sci. USA102:17780–17785.
20.Staprans, S., D. D. Loeb, and D. Ganem.1991. Mutations affecting hepadna-virus plus-strand DNA synthesis dissociate primer cleavage from transloca-tion and reveal the origin of linear viral DNA. J. Virol.65:1255–1262. 21.Summers, J., A. R. Jilbert, W. Yang, C. E. Aldrich, J. Saputelli, S. Litwin, E.
Toll, and W. S. Mason.2003. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc. Natl. Acad. Sci. USA 100:11652– 11659.
22.Summers, J., and W. S. Mason.2004. Residual integrated viral DNA after hepadnavirus clearance by nucleoside analog therapy. Proc. Natl. Acad. Sci. USA101:638–640.
23.Wieland, S. F., L. G. Guidotti, and F. V. Chisari.2000. Intrahepatic induc-tion of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol.74:4165–4173.
24.Wieland, S. F., H. C. Spangenberg, R. Thimme, R. H. Purcell, and F. V. Chisari.2004. Expansion and contraction of the hepatitis B virus transcrip-tional template in infected chimpanzees. Proc. Natl. Acad. Sci. USA101:
2129–2134.
25.Yang, W., W. S. Mason, and J. Summers.1996. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J. Virol.70:4567–4575.
26.Yang, W., and J. Summers.1995. Illegitimate replication of linear hep-adnavirus DNA through nonhomologous recombination. J. Virol. 69:
4029–4036.
27.Yang, W., and J. Summers.1999. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J. Virol.73:9710–9717. 28.Zhu, Y., J. M. Cullen, C. E. Aldrich, J. Saputelli, D. Miller, C. Seeger, W. S.
Mason, and A. R. Jilbert.2004. Adenovirus based gene therapy during clevudine treatment of woodchucks chronically infected with woodchuck hepatitis virus. Virology327:26–40.
VOL. 83, 2009 TRANSIENT HEPADNAVIRUS INFECTIONS 1789