0022-538X/87/051416-05$02.00/0
CopyrightX 1987, American SocietyforMicrobiology
Identification of gp350
as
the Viral
Glycoprotein Mediating
Attachment of Epstein-Barr
Virus
(EBV)
to
the
EBV/C3d
Receptor
of B Cells:
Sequence
Homology
of
gp350
and
C3
Complement Fragment
C3dt
GLEN R. NEMEROW,* CAROLYN MOLD,t VIRGINIA KEIVENS SCHWEND, VANESSATOLLEFSON,
AND NEIL R. COOPER
Department ofImmunology, Scripps Clinicand Research Foundation, LaJolla, California 92037 Received 2September 1986/Accepted 5 January 1987
The majorEpstein-Barrvirus(EBV) envelope glycoprotein, gp350,waspurifiedfrom the B95-8 cell lineand analyzed for its ability to mediate virus attachment to the isolated EBV/C3d receptor (CR2) of human B lymphocytes.Purifiedgp350andEBV,but notcytomegalovirus,exhibiteddose-dependent bindingtopurified CR2 in dot blotimmunoassays. Bindingwasinhibitedbycertain monoclonal antibodies to CR2 and togp350. Liposomes bearing incorporated gp350 bound to CR2-positive B-cell lines but not to CR2-negative lines. Liposome binding was also inhibited by the OKB7 anti-CR2 monoclonal antibody. A computer-generated comparison of the deduced gp350 amino acid sequence with that of the human C3d complement fragment revealedtworegionsofsignificant primarysequencehomology,afindingwhichsuggeststhata commonregion onthesetwounrelatedproteinsmay be involved in CR2binding.
Epstein-Barr virus (EBV) binds selectively to a 145-kilodaltonB-lymphocytemembraneglycoproteinwhich also
serves asthe receptor for thecomplementC3dfragment(7, 8, 12, 17, 18). Binding of EBV to this receptoron normal peripheral blood B cells is followed by viral endocytosis, whilebindingtothesamereceptoronBlymphoblastoidcells leadstoviral fusion withthe cell membrane(15).Wellsetal. found thatthe dualpresenceofamajor envelope proteinof
EBV, designated gp350/300 (gp350), and a related protein,
gp220/200 (gp220), is required for artificial lipid vesicles containingEBVantigenstobind to B cells(23). There have beennoother direct studiesof EBV attachmentprotein(s).It hasbeen found, however, thatsomemonoclonalantibodies (MAbs) to gp350 block EBV infection in vitro (21), and immunization of nonhuman primates with liposomes
con-taining gp350 protects against the development of fatal lymphoma (5). Inthe studiespresented here, we employed
purified gp350, EBV, and the EBV/C3d receptor (CR2) to identify gp350as the EBV attachment protein and to char-acterize certain aspects of the binding of gp350to CR2. In addition, two regions of amino acid sequence homology
werefound in the gp350 and C3d codingsequencesandmay
representtheCR2 bindingareasof thesetwoproteins. MATERIALS ANDMETHODS
EBVand CMVisolationand cell lines.EBVand the AD169 strainofcytomegalovirus (CMV)wereisolatedaspreviously described from phorbol ester-induced B95-8 cells (14) and foreskin fibroblasts (3), respectively. The Ramos cell line
wasobtained fromD. A. Thorley-Lawson,Tufts University School ofMedicine, Boston, Mass. As determined by the
*Corresponding author.
tPublication4512-IMM ofScrippsClinic andResearch Founda-tion.
tPresent address: Department ofMedicine, University of New Mexico, Albuquerque, NM 87131.
presenceof Epstein-Barrnuclearantigen, this line wasnot
infected with the B95-8 strain of EBV, nor did it possess
EBVreceptors,afact ascertained byfluorescence analyses
with anti-CR2 MAbs. Another Ramos cell line, designated Ramos-EF,wasobtained from ElizabethFowler, University
ofFlorida,Gainesville. The cell linewas infected withEBV and stainedwith anti-CR2 MAbs. Allcelllines were
main-tainedat 37°C inan atmosphere of5% CO2in RPMI 1640 containing10% heat-inactivatedfetal calfserum(Flow Lab-oratories, Inc., McLean, Va.), 50 jig ofgentamicin perml, 200 mM glutamine, and 0.2 jig of amphotericin B (Fungizone; GIBCOLaboratories, GrandIsland, N.Y.) per
ml.
Purification of gp350 and CR2. gp350 was isolated from B95-8 cells aspreviously described(19), except that either
high-pressure liquidchromatography (HPLC) gelfiltrationor
immunoaffinity chromatography was also used to remove
low-molecular-weightcontaminants.For HPLCpurification, gp350 obtained from chromatography on ricin-agarose
col-umns (19) was chromatographed on an HPLC TSK-250
column (Bio-Rad Laboratories, Richmond, Calif.) which
was equilibrated with 10 mM phosphate-buffered saline (PBS; pH 7.4; 0.15 M NaCl) containing 0.01% sodium deoxycholate(Calbiochem-Behring, San Diego, Calif.) (Fig. 1). Thecolumnwas runataflowrateof 0.5ml/h. Fractions containing gp350 wereidentified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (10) on
5%resolving gels which weresilverstained(13)and bydot
blot assays with anti-gp350 MAbs. The column was
precalibrated withgelfiltration standards (Bio-Rad). Forpreparationofliposomes containinggp350 (seebelow)
aswellasfor dotblotassays, immunoaffinity
chromatogra-phy with MAbs specific for gp350 was used as the final
purification step. Anti-gp350 MAbs were cross-linked to proteinA-agarose (17).
CR2, which was isolated from Raji lymphoblastoid cells by HB5 MAb immunoaffinity chromatography, was more
1416
on November 10, 2019 by guest
http://jvi.asm.org/
MW
103
Daltons
-200
-116
-92
-67 -45
-dye
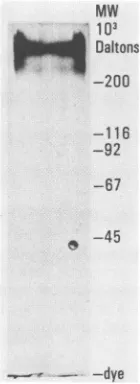
FIG. 1. Affinity purification of gp350. Partially purified gp350
waschromatographed on an anti-gp350 column. Forty microliters
(approximately 100 ng) ofgp350waselectrophoresedon a5to 15% gradient SDS gel. The gel was silver stained. MW, Molecular
weight.
than 90% homogeneous asjudged by SDS-PAGE, as
pub-lished previously (12, 17).
MAbstogp350 and CR2.MAbstogp350wereraised inthe
following manner. B95-8 cells were treated with 20 ng of phorbol myristate acetate (tumor-promoting agent [TPA])
per ml for 4 to 6 days, and the intact cells were injected
intravenously into 4-to6-week-old BALB/cmice. Themice werereinjected every 7to 10days and at 3 days before the spleenswereremovedfor fusion withthe Fox NYmyeloma cell line (Hyclone Laboratories, Inc., Ogden, Utah). Wells containing hybridomas were screenedby an EBV
enzyme-linked immunosorbent assay (ELISA) as previously
de-scribed (16). Briefly, approximately
105
EBV particles perwell were coated onto the plastic wells of ELISA plates (Immulon II; Dynatech Industries, Inc., Alexandria, Va.), reacted with culture supernatants, and incubated with biotinylated anti-murine immunoglobulin, streptavidin-horseradish peroxidase (Amersham Corp., Arlington Heights, Ill.), and substrate. A secondary screen for the
identification of clones recognizing EBV envelope antigens
was carried out by membrane fluorescence assay of
TPA-treated B95-8 cellsaspreviouslydescribed (18).Hybridomas
whichwerepositivein the EBV ELISA and in the
TPA-B95-8fluorescenceassayweresubclonedatleastthreetimesand
then grownin ascites. Four oftheseantibodies, designated
BOS-1, 18A7, 5B11,andliB5, wereof theimmunoglobulin
G2a(K) isotype and were purified on protein A-agarose (6).
Thespecificity of the MAbs was confirmedby
immunopre-cipitationof
125I-labeled
B95-8 cellsfollowed bySDS-PAGE performedaspreviously described (18).MAbsto CR2 usedin this study were OKB7, generously
provided by Ortho Diagnostics, Inc., Raritan, N.J.; AB-1, kindly donated by Barry Wilson, Hybritech, San Diego, Calif.;and HB5, purchasedfrom theAmericanTypeCulture Collection, Rockville, Md. These antibodies were purified by protein A-agarose chromatography as previously de-scribed (6).
Dot blot immunoassays. Detection of EBV and gp350 bindingtoisolatedCR2wascarriedoutbydotblotassaysas
previously described (17). The specificity ofligand binding
wasassessedby the ability ofthe CR2 MAb OKB7 toblock receptor
binding
toEBVandtogp350.TheOKB7antibodyhas previously been shown to directly inhibit both C3d and EBVbinding to intact B cells (18). Another anti-CR2 MAb,
designatedAB-1,whichrecognizes adifferent epitope on the receptor(20), wasusedas acontrol in the inhibition studies.
Purifiedreceptor(40 ng) was preincubated with 6,ugofeach MAb for 60minat
4°C.
This mixture was then reacted with immobilized EBV, CMV, or gp350 for an additional 60minat
22°C.
Detection of receptor binding was determined byincubation with biotinylated HB5 MAb, streptavidin-horseradish peroxidase, and chromogenic substrate as
pre-viously described (17).
In parallel studies, MAbs specific for gp350 were used to
inhibit receptor binding to immobilized gp350. In these experiments, 100 ng of gp350 was coated onto nitrocellulose,
and after nonspecific binding sites were blocked with 2% bovine serum albumin (BSA), the gp350 blots were reacted with various amounts of purified MAbs to gp350 or irrelevant MAbs.Afterfurther incubation for 60 min at22°C, the blots were reacted with purified CR2 and developed as described above.
Preparation of liposomes containing gp350. Liposomes
containing gp350 were prepared by detergent dialysis as previously described (12), except that cholesterol was added tothe phosphatidylcholine in preparation of themembranes. The composition of the liposome mixtures was 50 ,ug of gp350 per mlin 10 mMTris-150 mM NaCl (pH 8.0) contain-ing 0.5% deoxycholate, 6 mM egg yolkphosphatidylcholine (Supelco Inc., Bellefonte, Pa.), 0.75 mM cholesterol (Supelco), and 0.5
,uCi
of[14C]dipalmitoyl
phosphatidyl-choline (New England Nuclear Corp., Boston, Mass.).Con-trol liposomes were prepared with 10 mM Tris-150 mM NaCl (pH
8.0)-0.5%
deoxycholate lackinggp350. Liposomes were separated from unincorporated protein by ultracen-trifugation on discontinuous sucrose gradients (12). gp350was detected in liposomes by dot blot assays with purified
MAbs to gp350 as described above. Control liposomes
prepared in the absence of gp350 did notreact inthis assay. gp350 liposome-binding studies. Forcell-binding assays, 10
,ul of liposomes containing 2,500 cpm of
14C-lipid
was incubated for 60 min at4°C with 5 x10'
B lymphoblastoidcells in 250
RI
of PBS containing 0.1% BSA. Cells were pelleted for 30 s at 2,000 x g in a Microfuge (Beckman Instruments, Inc., Fullerton, Calif.) andwashed three times in PBS-BSA. The supernatants and pellets were counted for14C.
To assess thespecificity of theliposome binding,5 x 106 lymphoblastoid cells were incubated with 5 ,ug of HB5 or OKB7 anti-CR2 MAbfor 30 min on ice. Cells werepelleted,and supernatants were removed before the liposomes were added forbinding.
Quantitation of CR2 sites on B lymphoblastoid cells.
1II-HB5,
which was generously donatedby JohnBohnsack,Scripps Clinic and Research Foundation, La Jolla, Calif.,
was trace labeled with 1251andlodo-Beads (PierceChemical Co., Rockford, Ill.) according to the instructions of the manufacturer to a specific activity of 250,000 cpm/,g. B lymphoblastoid cells (5 x 106) were incubated with 2,ug of
125I-HB5
in 500RI
ofPBS-0.1%BSA for 30 min at4°C. The cells were then washed three times, and pellets were counted. Background binding was determinedbyincubatingcells with 200,ug of unlabeled HB5 for 15 min before the labeled antibody wasadded and represented less than 0.5% of the total counts perminute.
gp350 and C3d sequencecomparison. The deduced amino
acid sequences of gp350 (1, 2) and human C3d
(4)
wereon November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.146.216.70.262.2]TABLE 1. Antigenicpropertiesof MAbs
Membrane
EBV
gp350MAba fluorescence ELISA dot blot
(%positive)b
reactivity
reactivity(4405)c
ectvtBOS-1 13.2 1.690 +
18A7 19.7 2.031 +
11B5 17.2 1.915 +
SB11 20.2 1.299 +
AB-1 2.3 0.158
-Medium (control) 1.0 0.029
aBOS-1, 18A7, 11B5, and5B11areMAbsgenerated byimmunizationwith B95-8cells. AB-1 is MAbtoCR2 includedas acontrol.
b B95-8 cells weretreated for4days with20 ng ofTPAper mlbefore fluorescence analysis with each MAb and fluorescein isothiocyanate anti-mouseimmunoglobulin.
IPurified EBV, approximately 105 particles perwell, was coated onto plastic wells of ELISA platesandreacted with undilutedculture supernatants
containingantibody and then withbiotinylatedanti-murineimmunoglobulin,
streptavidin-horseradish peroxidase, and substrate.
comparedbyanalignmentprogram,
ALIGN,
of theNationalBiomedical Research Foundation, Georgetown University MedicalCenter,
Washington,
D.C.RESULTS
Isolation of gp350. Affinity-purified gp350 was devoid of
low-molecular-weight contaminants (Fig. 1). The purified protein also contained detectable amounts of the related gp220,onlysmall amountsof whicharepresentin theB95-8
cell linefromwhich the gp350waspurified. gp350 isolated by HPLC gel filtration lacked the gp220 but contained minor
amounts
(c5%)
ofa host cell 160- to170-kilodaltonprotein (datanotshown).Characterization of anti-gp350 MAbs. Four MAbs were
produced which reacted with the membranes ofa subpop-ulation of TPA-treated B95-8 cells, indicating that they probably reacted withastructural EBVglycoprotein (Table
MW BOS 1
10'
Dakoans
2 HB5
350- 200-
116-
92-
67-
45--CMV
*** * * * * *~~~~' -EBV
* * * * *
-gp350
2.3 1.15 0.57 0.28 0.14 0.07 0.03 0.015
Micrograms
FIG. 3. Dot blot immunoassay for CR2 binding. Various amountsofCMV, EBV,orgp350 wereimmobilizedon nitrocellu-lose. Theblotswereblocked with 5% nonfat milkandreactedwith purified CR2 and then with biotinylated anti-CR2, streptavidin-horseradishperoxidase, and substrate.
1). TheseMAbsalso reacted stronglywithpurifiedEBV in anELISA and withpurified gp350inadot blot assay.
AB-1,
animmunoglobulinG2a CR2 MAbincludedas acontrol,did not react with B95-8 cells, EBV, or gp350. The BOS-1 antibody-stainedTPA inducedB95-8 cells but didnotreact with untreated orphosphonoformate-treated B95-8 cells as
determined by fluorescence-activated cell sorter analyses (datanotshown).To confirm the specificity of the MAbs for
gp350, immunoprecipitation analyseswere carried out with
radiolabeled, TPA-treatedB95-8 cells. BOS-1, 18A7,
11B5,
and5B11 all reacted stronglywitha350-kilodalton
protein
and reacted weakly with a 200-kilodalton protein, asshown
for BOS-1 inFig. 2. A controlMAb of the sameisotypebut
directed againstCR2 (HB5) didnot immunoprecipitateany
proteinsfrom B95-8 cells.
Dot blot assays for gp350 binding to CR2. Dot blot im-munoassays wereused to assessthe ability of purifiedgp350
tobind toisolatedCR2. EBV, as well as purified gp350(Fig. 3), showed dose-dependent binding to CR2. Binding ofas little as 70 ng of gp350 or 15 ng of EBV to CR2 was
detectable. No binding of CMV to CR2 in doses as highas
2,300ng wasdetectable (Fig. 2).
Effect of anti-CR2 MAbs on gp350 and EBV binding to CR2. Further experiments were done to document the
specificityofgp350 bindingtoCR2 in the dot blot assay. The substitution of BSA for CR2 abrogated reactivity (Fig. 4,
upper right), indicating that gp350 and EBV were not
di-gp350 EBV CMV
CR2
[image:3.612.322.565.71.186.2]CR2 MoAb OKB7
FIG. 2. SDS-PAGE analysis of immunoprecipitates of 1251I labeledTPA-treated B95-8 cells. 125I-labeled B95-8 cell membrane lysates were immunoprecipitated with 5 ,ug of protein A-linked
BOS-1orwith HB5,an anti-CR2 MAb,as acontrol. The
precipi-tateswereeluted with 2% SDS (and 5%BME) andelectrophoresed on5 to15%SDS-PAGEgradient gels.MW, Molecular weight.
gp350 EBV CMV
0
IBSA
[image:3.612.65.304.81.190.2]CR2 MoAb , AB- 1 FIG. 4. Specificity of CR2 bindingto gp350. Purified CR2 was reacteddirectlywithEBV,CMV,or gp350 (upper left); with 6 ,ug of MAb OKB7 (lower left); or with 6 ,ug of MAb Ab-1 (lower right) in solution beforeincubationwithimmobilized gp350,EBV, or CMV. BSAwas substituted forCR2(upperright)in the blotting experi-ments. Thereactionsweredevelopedas described in the legend to Fig. 3.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.134.234.476.662.2] [image:3.612.320.561.540.656.2]AB-1- 0 0
5Bll- *
11B5- i -X 4 * * * * 0
18A7-* * 0 * 0 0
BOS 1- 0*
*
20 6.6 2.2 0.74 0.24 0.08 0.02 0.009 Micrograms
FIG. 5. Effect of anti-gp350 MAbs on CR2 binding to gp350. Variousamountsofanti-gp350 MAbs 5B11,llB5,18A7,and BOS-1
were reacted for 60 min with immobilized gp350. After further incubation with purified CR2, theblotsweredevelopedasdescribed
inthe legendtoFig. 3. AB-1,ananti-CR2 MAb, wasalsoincluded
as acontrol. The four wells atthe upper rightwere coatedwith
albumininstead ofgp350 andreactedwithCR2.
rectlyinteracting with the immunologicreagents. The prein-cubation of CR2 with OKB7 MAb, which blocks EBV binding to B cells and to CR2, completely abrogated CR2 binding toboth gp350 and EBV(Fig. 4, cf. lower left with
upperleft). Another anti-CR2 MAb, AB-1, which does not directly block CR2 ligandbindingtoBcells,didnotinterfere with the binding of purified CR2 to EBV orgp350 (Fig. 4,
lowerright). Thesefindings document the specificity of the dotblotreceptor assayfor CR2-EBVligand interactions.
Effectof anti-gp350 MAbs ongp350 binding toCR2. The ability of the variousanti-gp350 MAbstoalter gp350 binding toCR2 was examined. Pretreatment ofimmobilized gp350
with BOS-1 MAb strongly inhibited binding of CR2 in a
dose-dependent manner (Fig. 5). MAbs 5B11 and 11B5
inhibitedmodestly,while18A7waswithoutinhibitory
activ-ity. AB-1,ananti-CR2 MAb includedas acontrol,failedto inhibitbinding of CR2togp350. CR2 also didnot reactwith uncoatednitrocellulose.
gp350 liposome-binding studies. To confirm by another experimental approach the binding of gp350 to CR2, the ability of liposomes containing gp350 to bind to several B lymphoblastoid cell lines was assessed. gp350-containing
liposomes, but not control liposomes, bound to Raji and Ramos-EFcells, bothof which had approximately 2 x 104
CR2 sites per cell (Table 2). gp350-containing liposomes
failedtobindtoB95-8 and Ramoscells,both of which lacked
detectable CR2. Furthermore,binding of gp350 liposomesto Raji and Ramos-EF cells was abrogated by pretreatment withOKB7 MAb butnotwith HB5MAb,indicatingthatthe
liposome binding is mediatedthrough CR2.
DISCUSSION
In the studies presented here, purified ligands and
recep-torshavebeenusedtoshow thatgp350mediatesbindingof EBV to B-cell CR2. Affinity-purified gp350 employed in these studies was devoid of minor amounts of cellular contaminantsandcontainedaminoramountofgp220. gp220 is related to gp350 but differs in size because of altered mRNA splicing (2, 9). Isolated gp350boundto thereceptor inadose-dependent manner. This reactionwas specifically
blocked by OKB7 anti-CR2 MAb, which also blocks EBV and C3dbindingtoBcells (18),butwasunaffectedbyAB-1
and HB5anti-CR2 MAbs, which donotdirectly inhibitEBV
and C3d binding to B cells. Certain anti-gp350 MAbs also specifically blocked the interaction of purified gp350 with the
receptor. Finally, liposomescontainingpurifiedgp350 bound to B cells expressing CR2 but did not bind toCR2-negative cell lines. This bindingalso was blockedby OKB7 but was notblocked by HB5anti-CR2MAb. These studies indicate
that gp350 mediates adsorption of EBV to CR2. We are unable toexplain the gp220requirement previously reported
by Wells et al. with gp350- andgp220-containing liposomes
(23) since the HPLC-purified gp350, which lacked gp220,
also mediated CR2binding.
The nature of the ligand-binding site on CR2 to which
EBV and C3d bind is not known, and it is also unclear whether both ligands react with the same site or bind to
distinct sites. It is not possible to address this question directly by performing competitive binding studies with C3d,g andgp350because thebinding affinity ofmonomeric C3d,g for CR2is very low(22)andbecause isolated gp350 is
insoluble in the absence of detergent. The ability to
selec-tively inhibit EBV orC3d binding to B cells with different anti-CR2 MAbswhichwasrecently reported (R. Frade, M.
Barel,L.
Krikoria,
and C. Charriaut, 6th Int.Congr.Immu-nol.,p.189, 1986) doesnotprove that EBVandC3d bindto
different sites because the attachment of large ligands such
as EBV- and
C3d-coated
particles tocells mayinvolvenotonly the
primary
ligand-binding site but also secondary interactions. In an alternative approach to determining thebinding sites, we questioned whether gp350 and C3d, two
proteins of diverse
origin,
might exhibit regions ofprimary
sequence homology. A computer search done by using the ALIGN program identified two regions of homologous se-quence in the deduced gp350 coding sequence (1) and the humanC3dsequence(4). One oftheseregions(region1,Fig. 6) consisted of five identical amino acids.Thissequence was notfound inanyofthe other3,000proteinscontained inthe data base. In addition, a second region (region 2, Fig. 6) encompassing the region of C3d which has been reportedto
beinvolvedinCR2binding (11)wasalsoidentified. Whether
eitherofthese two sequences has any role in CR2binding remains to be determined, but their presence raises the
posibility
that acommondomain on gp350 andC3d isusedfor CR2 binding.
Theidentification ofgp350astheEBVproteinresponsible forreceptor attachmentrepresents the firstidentification ofa
[image:4.612.57.306.72.215.2]distinct herpesvirus envelope
glycoprotein
whichisutilizedTABLE 2. Binding ofgp350-containingliposometoB lymphoblastoid cells
Liposome binding(cpm)c
CeH
lineCR2
MbCell line (molecules/cell)a MAbb With Control
gp350
Raji 20,500 200 256 31 48 + 4
OKB7 58 6 ND
HB5 281 7 ND
Ramos-EF 19,000 ± 1,300 141 ± 21 34 + 11
OKB7 46 8
Ramos 0 76± 12 67
B95-8 0 62 2 ND
aDeterminedbybindingof1251-labeledHB5.Resultsare mean+standard
error.
bCellswerepreincubatedwith 5
p.g
ofMAbtoCR2.cLiposome-bindingassaysweredoneinduplicate.Resultsofthree
exper-imentswerecombined andexpressedasmean± standarderrorofcountsper
minute inpellet.ND,Notdetermined.
L
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.322.563.567.677.2]REGION
#1 Position
372 378
gp350/300THRePRO-SERoGLYoCYS-GLUOASN
C3d THR*PRO*SER*GLY*CYS*GLY*GL U
1006 1012
REGION
#2 Position
21 28
gp350/300GLU*ASP*PROOGLY.---*- --*PHE*PHE*ASN*VAL*GLU
C3d GLU'ASPoPROoGLYeLYSoGLNoLEU*TYR*ASN-VAL.GLU
1221 1231
FIG. 6. Primary sequence homology between gp350 and C3d. Thededuced gp350 codingsequence andthatof C3dwerecompared for homologybyALIGN.
in virus adsorption to host cells. This system provides a unique modelfor the analysis of the structure and function of virus receptor-ligand interations. Since CR2, with which bothgp350 and C3d interact, mediates viralendocytosisor fusiondepending onthe cell type, this system also provides the means toaddress not only the nature of, and epitopes
involved in, ligand-receptor interactions but also the func-tionalconsequences of these reactions.
ACKNOWLEDGMENTS
We thank Bonnie Weier for assistance in preparation of the manuscript.Wealsogratefullyacknowledge the excellent technical assistance ofMaryEllen McNaughton.
Funding for this research projectwasprovided by Public Health ServicegrantsCA36204,A116082,A117354,CA35048, and CA14692 from the National Institutes of Health and by a PEW Scholars Award in Biomedical Sciences toGlenR.Nemerow.
LITERATURE CITED
1. Baer, R., A. T. Bankier, M. D. Biggin, P. L.Deininger, P.J. Farrell,T.J.Gibson,G.Hatfull,G. S.Hudson,S.C.Satchwell, C. Sequin, P.S. Tuffnell, and B.G. Barrell. 1984. DNA
se-quenceandexpression of the B95-8Epstein-Barr virusgenome. Nature(London) 310:207-211.
2. Biggin, M., P. J. Farrell, and B. G. Barrell. 1984.Transcription and DNA sequence of the Bam HI-Lfragment of B95-8 Ep-stein-Barr virus. EMBO J. 3:1083-1090.
3. Booth, J. C., G.Hannington,T. A.G.Aziz, and H. Stern. 1979. Comparison of enzyme-linked immunosorbent assay (ELISA) technique and complement-fixation test for estimation of cytomegalovirus IgG antibody.J. Clin. Pathol.32:122-127. 4. DeBrui,n,M.H.L., and G. H. Fey. 1985. Humancomplement
component C3: cDNA coding sequence and derived primary structure. Proc. Natl.Acad. Sci. USA 82:708-712.
5. Epstein, M. A., A. J. Morgan, S. Finerty, B. J. Randle, and J. K. Kirkwood. 1985. Protection ofcottontop tamarinsagainst Ep-stein-Barr virus-induced malignant lymphoma by a prototype subunitvaccine.Nature(London) 318:287-289.
6. Ey,P.L.,S.J.Prowse,and C. R.Jenkin. 1978.Isolationofpure IgGl, IgG2a and IgG2b immunoglobulin from mouse serum usingprotein-ASepharose. Immunochemistry 15:429-436. 7. Fingeroth, J. D., J. J. Weis, T. F. Tedder, J. L. Strominger,
P. A.Bird,and D. T. Fearon.1984.Epstein-Barrvirusreceptor ofhuman Blymphocytesis theC3d receptorCR2. Proc. Natl. Acad. Sci. USA81:4510-4516.
8. Frade,R.,M. Barel,B. Ehlin-Henriksson, andG. Klein. 1985. gpl40, the C3d receptorof human Blymphocytes, isalso the Epstein-Barr virus receptor. Proc. Natl. Acad. Sci. USA 82:1490-1493.
9. Hummel, M.,D. Thorley-Lawson, and E. Kieff. 1984. An Ep-stein-Barr virus DNAfragmentencodesmessages forthetwo major envelope glycoproteins (gp350/300 and gp220/200). J. Virol.49:413-417.
10. Laemmli,U. K. 1970.Cleavageofstructuralproteins duringthe assembly of the head ofbacteriophage T4. Nature (London) 227:680-685.
11. Lambris, J. D., V. S. Ganu, S. Hirani, and H. J. Muller-Eberhard.1985.Mappingof theC3d receptor (CR2) bindingsite andaneoantigenicsite in theC3ddomain of the third compo-nentofcomplement. Proc. Natl.Acad. Sci. USA82:4235-4239. 12. Mold, C.,N. R.Cooper,and G. R. Nemerow. 1986. Incorpora-tion of thepurified Epstein-Barr virus/C3dreceptor(CR2)into liposomes anddemonstration ofits dual ligand binding func-tions. J. Immunol. 136:4140-4145.
13. Morrisey, J.H. 1981. Silver stain forproteinsinpolyacrylamide gels: amodified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310.
14. Nemerow, G. R., and N. R. Cooper. 1981. Isolation of Ep-stein-Barr virus and studies of its neutralization byhumanIgG andcomplement. J. Immunol. 127:272-278.
15. Nemerow, G. R., and N. R. Cooper. 1984. Early events in infection of human B lymphocytes by Epstein-Barrvirus: the internalizationprocess. Virology132:186-197.
16. Nemerow, G. R., F. C. Jensen, and N. R. Cooper. 1982. Neutralization of Epstein-Barr virus by nonimmune human serum: role of crossreacting antibodytoherpes simplexvirus andcomplement. J.Clin. Invest.70:1081-1091.
17. Nemerow, G. R., M. F. E. Siaw, and N. R. Cooper. 1986. Purification of theEpstein-Barrvirus/C3dcomplement receptor ofhuman Blymphocytes: antigenicandfunctional propertiesof thepurified protein. J.Virol. 58:709-712.
18. Nemerow, G. R., R. Wolfert, M. E. McNaughton, and N. R. Cooper. 1985. Identification and characterization of the Ep-stein-Barr virus receptor on human B lymphocytes and its relationship to the C3dcomplementreceptor (CR2). J. Virol. 55:347-351.
19. Qualtiere, L. E., R. Chase, and G. R. Pearson.1982. Purification and biologic characterization ofa major Epstein-Barr virus-induced membraneglycoprotein. J. Immunol. 129:814-818. 20. Siaw, M. F. E., G. R. Nemerow, and N. R. Cooper. 1986.
Biochemical and antigenic analysis of the Epstein-Barr virus/C3dreceptor(CR2). J. Immunol. 136:4146-4151. 21. Thorley-Lawson, D. A., and K. Geilinger. 1980. Monoclonal
antibodies against the major glycoprotein (gp350/220) of Ep-stein-Barr virus neutralize infectivity. Proc. Natl. Acad. Sci. USA 77:5307-5311.
22. Vik, D. P., and D. T. Fearon. 1985. Neutrophils express a receptorforiC3b,C3d,g and C3d that is distinct from CR1, CR2 and CR3. J. Immunol. 134:2571-2579.
23. Wells, A., N. Koide, and G. Klein. 1982. Two large virion envelope glycoproteins mediate Epstein-Barrvirus binding to receptor-positive cells.J. Virol.41:286-297.