0022-538X/94/$04.00+0
Copyright© 1994, AmericanSociety forMicrobiology
Isolation of Epstein-Barr Virus
(EBV)-Negative
Cell Clones
from
the
EBV-Positive Burkitt's Lymphoma (BL) Line Akata:
Malignant
Phenotypes of
BL
Cells
Are
Dependent
on
EBV
NORIO SHIMIZU,AKIKOTANABE-TOCHIKURA, YASUYUKI KUROIWA,ANDKENZOTAKADA* DepartmentofVirology and Parasitology, Yamaguchi University School ofMedicine,
Kogushi,
Ube,
Yamaguchi
755,
Japan
Received 29 March 1994/Accepted 16June 1994
During cultivation of the Epstein-Barr virus (EBV)-positive Burkitt's lymphoma (BL) line Akata, it was noted that EBVDNA is lost from some of the cells. Isolation of EBV-positive and EBV-negative clones with the same originmade it possibletoexamine the effects ofEBV in BLcells.The results indicate that malignant
phenotypesof BL, such as growth in low serum, anchorage-independent growth in soft agar, andtumorigenicity
innude mice, are dependent on the presence of EBV genomes and underline theoncogenicfunctionofEBV in human cancer.
Epstein-Barr virus (EBV), a human herpesvirus, is associ-atedwith morethan 90% of Burkitt's lymphoma (BL) in the African regions of endemicity and less frequently (15to20%) with the sporadicBLoccurring worldwide.The mostconsistent finding inBL,whetherEBV-infected or not,isachromosome translocation involving immunoglobulin (Ig) and myc genes. The translocation results in deregulation ofc-myc expression andis regardedastheimportant stepin the genesis ofBL(2, 11). On the other hand, the role ofEBV is stillobscure. EBV infects primary B lymphocytes in vitro and transforms them into blasts that can proliferate indefinitely. Such EBV-trans-formed lymphoblastoid cells maintain the entire viralgenome mostly in a plasmid form and express a limited number of virus-encoded proteins, including six nuclear antigens
(EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNALP) and three membrane proteins(LMP1,LMP2A,and LMP2B), whichmustplayroles ininitiation and maintenance of transformation(7). However, inBLcellsthere is repression of the latent genes, exceptEBNA1 (17). These observations indicate that the functions of EBV that are required for transformation ofprimary lymphocytes are not necessary in the maintenance and propagation ofBLcells, although these functionsmaybeimportantduringBLgenesis.Itisnotknown whetherEBV contributes tothe growth of the EBV-carrying BLthatonlyexpressesEBNA1.
The Akata cell line is derived from an EBV-positive BL from aJapanesepatient, has aBL-type chromosome translo-cation,t(8;14), andexpressessurfaceIg of theG(K) class(21). TheAkata line isnowcommonlyusedtostudy reactivation of latent EBV because of its rapid and efficient response to cross-linking of cell surface Ig, using antibodyto Ig (anti-Ig)
(20, 23). The Akata line retainsthe tumorphenotype, which is characterizedby selective expression ofEBNA1,invitro (17). During cultivation of Akata cells, we have noted that EBV DNAislostfrom some of thecells. Isolation ofEBV-positive andEBV-negative Akata cell clones with thesameoriginmade it possible to examine the effects of EBV in BL cells. The results indicate that malignant phenotypes of BL, such as
*Correspondingauthor.Department ofVirology andParasitology,
YamaguchiUniversity School of Medicine,Kogushi, Ube, Yamaguchi 755,Japan. Phone: 836-22-2341. Fax: 836-22-2237.
growth in low serum, anchorage-independent growth in soft agar, andtumorigenicity in nude mice, are dependent onthe presenceofEBVgenomesandunderline theoncogenic func-tionofEBVin humancancer.
Initially, Akata cellswerevirtually 100%positive for EBNA (21).However,afterserialpassagefor about 2years, wefound that49% of the cellswerenegative forEBNAasindicatedby anticomplement immunofluorescence (21) with a polyvalent
human antiserum. Therefore, we intended to isolate EBNA-positive and EBNA-negative clones of Akata cells. Akata cells wereplated into 96-well platesat0.5cellperwell and 0.2 ml of conditioned mediumperwell. The conditioned mediumwas an equal mixture of freshRPMI1640mediumsupplemented with 20% fetal calfserum (FCS) (HyClone) and culture superna-tantof Akata cells thatweresuspendedinRPMI1640medium with 10% FCS at aconcentration of 5 x
105
cellspermland incubated for 3 days. Cellswere fed every4 dayswith condi-tioned medium. Threetofour weeks later, cell clonesemerged in 18% of the wells. Forty-two cloneswereexpanded infresh RPMI 1640mediumsupplemented with 10% FCS and assayed for EBNA expression. As a result, 18 clones were virtually 100%positive for EBNA,16cloneswerecompletelynegative,and 8 clones were a mixture of EBNA-positive and EBNA-negative cells.
Representative EBNA-positive and EBNA-negative clones werefurther examined for the expressionofEBNA
polypep-tidesby immunoblotanalysis (22).Cellpelletsweremixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis
loading buffer. After the pelletswere boiled for 5 min, equal
amounts of protein (corresponding to 2.5 x 105 cells) were
separated in polyacrylamide gels (gradient, 4 to 20%) and transferred tonitrocellulose. The blotswere blocked with5% milkin phosphate-buffered saline. After immunostaining, the blots were developed by the enhanced chemiluminescence
(ECL) method (Amersham) accordingto the manufacturer's instructions. The resultsareshown inFig. 1Aand B.Parental Akata cells and EBNA-positive clones were positive for EBNA1 polypeptide,while EBNA-negative cloneswere not.
Further examination, by Southern blot and PCR analysis,
wascarried out to determine whetherEBNA-negative clones contain EBVDNA. For Southern blot
analysis
(22),
cellular DNAs (10pLg
each)weredigestedwith BamHIendonuclease,
electrophoresed on 0.8% horizontal agarose
gels,
and trans-6069on November 9, 2019 by guest
http://jvi.asm.org/
A
1 2 3 4 5 6 7 8 9 10 11
N. 97
-m _ _ < E2
* E1
66
-B
1 2 3 4 5 6 7 8 9 10 11
97
-66
-* E2
- L
45
-C
TABLE 1. Characteristics of Akata cellclones
Growth in No. ofcolonies No.ofmice
Celltype low se- inagarose/ with tumor
rum' 104cells" (n =4)'
ParentalAkata cells + 97 ± 10 1
EBV-positive clones
19-16 + 158 + 18 1
19-19 + 136 + 11 2
19-33 + 172 + 20 1
19-88 + 144+ 19 0
EBV-negative clones
19-6 - 0 0
19-24 - 0 0
19-27 - 0 0
19-47 - 0 0
"Cells weresuspendedinRPMI1640 medium with0.1I%FCS at 3x 105per
mland incubated for 6 days.
'Tenthousand cells were seeded in 3 ml of mediumcontaining0.33% agarose
over3 ml of mediumcontaining0.66% agarose in a 60-mmpetridish. After3
weeks, colonies thatcontained >100 cells were counted. The results are means + standard errors of four dishes.
CNudeBALB/c mice wereinjected subcutaneously with 2 x 107cells persite. Animals were observed up to 8 weeks for tumors.
1 2 3 4 5 6 7 8 9 10 11 12
BamHlK -* s -
-D
1 2 3 4 5 6 7 8 9
E1
-____
239 bp
1 2 3 4 5 6 7 8 9
E2 _____
203 bp
1 2 3 4 5 6 7 8 9
L go___
434bp
FIG. 1. Detection of EBV latentproteinsandgenomesin EBNA-positiveandEBNA-negativeclones of Akata cells.(AandB) Immu-noblotanalysis (22)of proteinextractsfromanEBV-negative B cell line,BJAB(8) (lane 1),its B95-8 EBV-converted cellline BJAB-B95-8 (lane 2),theparental Akata cell line (lane3),EBNA-positiveAkata cell clones(lanes4to7),andEBNA-negativeAkatacell clones(lanes 8 to 11). For immunostaining, blot A was treatedwith a standard EBNA-positive human serumandperoxidase-labeled protein A. Blot Bwastreatedwithanti-EBNA2 and anti-LMP1 monoclonalantibodies (16, 26)andperoxidase-labeled anti-mouseantibody. Size markers in kilodaltonsareindicatedontheleft.EBNA1(El),EBNA2(E2), and LMP1 (L) bandsare indicatedbyarrows.(C) Southern blot analysis
(22) ofEBV genomes probed with theBamHI Kfragment of EBV
DNA. Lanes: 1, parental Akataline; 2to5,EBNA-positive Akata cell clones; 6to9,EBNA-negative Akata cellclones; 10, 2.22 pg of Raji cell(13)DNA,correspondingto10 EBVcopiespercell; 11,1.11 ,ugof Rajicell DNA,correspondingto5 EBVcopiespercell; 12, 222 ngof Raji cellDNA, correspondingto 1 EBVcopy percell. The Raji cell
line, an African BL line, maintains a stable copy number of EBV
genomes(approximately45copiespercell[18])inalatentstate.(D)
PCRanalysisof EBVgenomes.Lanes:1,parentalAkata cells;2 to 5,
EBNA-positive Akata cell clones; 6to9, EBNA-negative Akatacell clones. The controlexperiments indicated that this PCRsystemcould detect 1 to5 EBVcopies in the reaction mixture.
ferred to nitrocellulose. EBV DNAs were visualized by the ECL direct nucleic acidlabeling and detectionsystem (Amer-sham) accordingto themanufacturer's instructions. For PCR analysis, purified cellular DNAs (200ng, correspondingto3.2 x 105 cells) were subjected to PCR, with primer pairs specific forEBNA1 (5'GACGAGGGGCCAGGTACAGG3' and 5'G
CAGCCAATGCAACTTGGACGTllTTTGG3'), EBNA2 (5' ATAGTCCAGAACCACGGTCC3' and 5'TCTGGCCTTGA GTCTTAGAGG3'),and LMP1 (5'ACGCACTTTlCTCCTCT TTCCCC3' and5'GAGGGAGTCATCGTGGTGTGT3'). A 50-,lI portionofreaction mixture consisted of 50 mMKCl, 10 mM Tris hydrochloride (pH 8.3), 1.5 mM MgCl2, 50 p.M (each) dATP, dCTP,dGTP, and dTTP, 1 pFM (each) primer, and 2.5UofTaqpolymerase (Cetus).Reaction mixtureswere subjected to30cycles ofdenaturation (95°C,45 s),annealing (56°C,45s),andextension(72°C,75s). The PCR products (10 pL1ofeach)weresizeseparatedon2%agarosegels, transferred tonitrocellulose, and visualized by the ECL 3' oligonucleotide labeling and detection system (Amersham) with antisense oligonucleotides (5'GATGTGTCTGCCTTCTCTCCTA3' for EBNA1, 5'TAATGGCATAGGTGGAATGTTAT3' forEBNA2, and 5'CCTGCTCATCGCTCTCTGGAAT3' for LMP1) as probes. The results are shown inFig. IC and D. Analysis of Southern blots probedwith the BamHI Kfragment of EBV DNA and PCR analysis withprimerpairs specific for EBNA1, EBNA2, andLMP1 confirmed thatEBNA-negative clonesdo not contain EBV DNA.
Since theparentalAkata cells werealmost100%positivefor EBNA(21), it ismostprobable thatthe EBVplasmidwaslost from some of the Akata cells during cultivation. To further confirm this possibility, one of the EBV-positive Akata cell clones, clone 19,was maintained in culture. After 6 months, the EBNApositivity decreased from the initial 100% to83%. Clone 19 was plated into 96-well plates at 0.5 cell perwell. After 3 to4weeks ofincubation, cell clones emerged in 21% of the wells. Ninety clones were expanded and assayed for EBNAexpression. Of these, 63cloneswere 100%positive, 10 cloneswerecompletelynegative,and 17cloneswereamixture of EBNA-positive and EBNA-negative cells. PCR analysis confirmed that EBNA-negative clones do not contain EBV
on November 9, 2019 by guest
http://jvi.asm.org/
[image:2.612.320.561.84.237.2]EBV -positive clones EBV-negative clones
c)
C)
x
Days in culture
B
EBV
-positive clone
Days in culture
EBV
-negative
clone
C 1 2 3 4 5 6
Iw
.mqs_
qg<4- E2 .<- El
_- E2
[image:3.612.132.481.83.645.2]_ <~~~~~~L
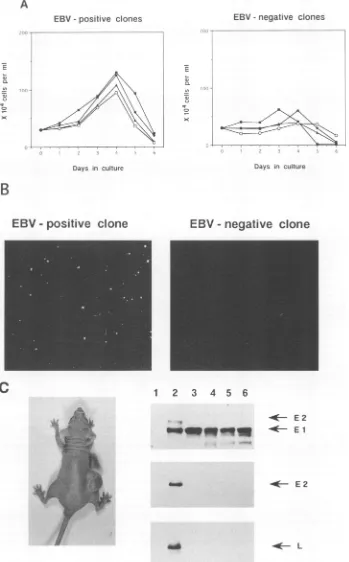
FIG. 2. Growthcharacteristics ofEBV-positive and EBV-negative Akata cell clones. (A) Growthcurvesinmedium with 0.1%FCS. (B)Growth
insoftagarofanEBV-positive clone (left) andanEBV-negative clone (right). (C) A nudemouseinjected with EBV-positive Akata cellclone
19-19(left). Themouse wasinjected 4 weeks previously with 2x 107cells. Immunoblot analysis shows that nudemousetumors(from Akata cell
clones19-16,19-19, and 19-33) consist of EBNA1-positive cells (right). Astandard EBNA-positive humanserumand monoclonal antibodies for
EBNA2 andLMP1wereusedforimmunostaining. EBNA1(El), EBNA2 (E2), and LMP1 (L) bandsareindicatedbyarrows.Lanes:1, BJABcells;
2, BJAB-B95-8 cells; 3, EBV-positive Akata cell clone 19-19; 4to6, nudemouse tumorcells.
A
c)
ci
st
la
x)
j ----7 ,.
on November 9, 2019 by guest
http://jvi.asm.org/
DNA. Both EBV-positive and EBV-negative clones were virtually 100% positive for surface Ig of the G(K) class and possessed chromosome markerscharacteristic ofthe parental Akata cells (21), and so they were clearly derived from the Akata cells and not fromcontaminated unrelatedcells. These results indicate that EBV DNA was lost from some of the Akata cellsduring cultivation.
Isolation ofEBV-positive and EBV-negative clones with the sameorigin makes itpossible toexamine the effects ofEBVin BL cells. Growth characteristics of EBV-positive and EBV-negative clones were compared (Table 1). Both clones grew wellinmedium with 10%FCS and reached themaximum cell density at around 2 x 106 cells per ml. In 0.1% FCS, EBV-positive clones grew at a slightly reduced rate and at 4 days reached the maximum cell density ofaround 1.3 x 106 cellsperml,while there was nogrowthofEBV-negativeclones (Fig. 2A). Anchorage dependenceofcell growth wasassayed in0.33% agarose (SeaPlaque; FMC). In EBV-positive clones, 1 to 1.7% ofthe cellsgrew tovisible colonies. In contrast,in EBV-negative clones no colony was macroscopically visible (Fig. 2B).Thesegrowth differences betweenEBV-positive and EBV-negative clonesinlowserumandsoftagarosehave been stably retained over 6 months of passages in culture. We further assayed tumorigenicity of EBV-positive and EBV-negative clones in nude mice. Of four EBV-positive clones, three produced tumors atthe site of inoculation. By4weeks,
the tumors ranged in size from 0.7 to 2 cm. Tumor cells consisted of EBNA1-positive cells and were negative for EBNA2 and LMPI (Fig. 2C). On the other hand, EBV-negative clones were not tumorigenic in nude mice. These results indicate that malignant phenotypes of EBV-carrying BL, such as growth in low serum, anchorage-independent growth in soft agar, and tumorigenicity in nude mice, are dependentonthe presenceofEBVgenomesand underline the
oncogenic function ofEBVin humancancer.
EBNA1 plays an integral role in the replication of viral plasmids in EBV-infected cells (24, 25). EBNA1 binds to specific sequences within the plasmid origin of replication
(oriP), localized in the BamHI Cfragment ofEBV DNA (5,
14). It alsoregulates the expression of the EBV latent genes (15, 19). It is not known whether or not EBNA1 affects the replication of any cellular origins of DNA synthesis or the expression ofanycellulargenes. Itseemslikely, however, that if there are sequences homologous to the EBNA1-binding
sequence within cellularDNA, EBNA1 will bindtothem and affect DNA replication and/or RNAsynthesis. In addition to EBNA1, two small, nonpolyadenylated RNAs known as EBER1 and EBER2 (4, 10) and the transcripts from the BamHIAregion of thegenome(1,7)arecommonly expressed in BLand EBV-transformedlymphoblastoid cells. These prod-uctsand/or additional, presently unidentifiedviral gene prod-uctsmaycontribute to malignant phenotypes ofBLcells.
MostEBV-negative BLlinesare susceptible toEBV infec-tion, leadingto stable EBV-positive lines (3, 9). This process has permitted the analysisofbiologic roles ofEBV in clonal cell populations. However, in EBV-negative BL cells, EBV
infection, like infection ofprimary B lymphocytes, invariably results inexpressionof afullsetof latent genes(12). There has beennoinfection system such as that whichproduces BL-type geneexpression,which is characterizedbyselectiveexpression of EBNA1. The EBV-positive and EBV-negative Akata cell clones, for the first time, make it possible to study the phenotypic andbiochemical roles of EBV in BL cells.
This is thefirstreport of lossofEBV DNAfrom BLcells. StudiesofwhyEBV plasmid DNAis not maintainedin some Akatacells are currently beingcarried out.
We thank B. Sugden (McArdle Laboratory for Cancer Research, Madison, Wis.)forhelpfulcomments on themanuscript.
This work was partly supported by grants from the Japanese Ministries of Education, Science, and Culture and of Health and Welfare.
REFERENCES
1. Brooks,L.A.,A. L.Lear,L.S.Young, andA. B.Rickinson. 1993.
Transcriptsfrom the Epstein-Barrvirus BamHI Afragment are
detectable in all three forms of virus latency. J. Virol. 67:3182-3190.
2. Epstein,M. A.,and B. G.Achong. 1979. The relationshipof the virus to Burkitt's lymphoma, p. 321-337. In M. A. Epstein and B.G.Achong(ed.),TheEpstein-Barrvirus.Springer-Verlag,New York.
3. Fresen,K.O.,and H.zurHausen. 1976.Establishmentof
EBNA-expressing cell lines by infection of Epstein-Barrvirus (EBV)-genome-negative human lymphoma cells with different EBV strains.Int. J.Cancer 17:161-166.
4. Howe, J.G.,andJ.A. Steitz. 1986. Localization ofEpstein-Barr virus encoded small RNAs by in situ hybridization. Proc. Natl. Acad. Sci. USA 83:9006-9010.
5. Jones,C.H.,S.D.Hayward,and D. R.Rawlins. 1989.Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1with itsDNA-bindingsites. J.Virol. 63:101-110. 6. Karran, L.,Y.Gao,P. R.Smith,and B. E.Griffin. 1992.
Expres-sionofa familyofcomplementary-strand transcriptsin Epstein-Barrvirus-infected cells. Proc. Natl. Acad. Sci. USA 89:8058-8062. 7. Kieff, E., and D. Liebowitz. 1990. Epstein-Barr virus and its replication, p. 1889-1920. In B. N. Fieldset al. (ed.), Virology. RavenPress,New York.
8. Klein, G., T. Lindahl, M. Jondal, W. Leibold, J. Menezes, K.
Nilsson,andC.Sundstrom.1974.Continuouslymphoidcell lines with characteristics of B cells(bone marrow-derived), lackingthe Epstein-Barrvirusgenomes andderived from three human lym-phomas.Proc. Natl.Acad. Sci. USA 71:3283-3286.
9. Klein, G.,B.Sugden,W.Leibold,andJ.Menezes.1974. Infection of EBV-genome negative and positive human lymphoblastoid lines withbiologicallydifferentpreparationsof EBV.Intervirology 3:232-244.
10. Lerner,M. R., N. C.Andrews, G.Miller,andJ. A. Steitz. 1981. Twosmall RNAs encoded by Epstein-Barrvirus andcomplexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus.Proc. Natl. Acad.Sci. USA 78:805-809.
11. Miller, G. 1990. Epstein-Barr virus. Biology, pathogenesis, and medicalaspects, p.1921-1958.In B. N.Fieldsetal.(ed.), Virology. RavenPress,NewYork.
12. Murray,R.J.,L.S.Young,A.Calender,C.D.Gregory,M.Rowe, G. M. Lenoir, andA. B. Rickinson. 1988. Different patternsof Epstein-BarrvirusgeneexpressionandofcytotoxicT-cell recog-nition in B-cell lines infected with transforming (B95.8) or
non-transforming(P3HR1)virus strains. J.Virol. 62:894-901. 13. Pulvertaft,R.J.V.1965. AstudyofmalignanttumorsinNigeria by
shorttermtissue culture.J. Clin. Pathol. 18:261-273.
14. Rawlins,D.R.,G.Milman, S.D. Hayward,andG. S. Hayward. 1985. Sequence specificDNA binding of the Epstein-Barrvirus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region.Cell 42:859-868.
15. Reisman, D.,andB.Sugden. 1986.transactivation ofan
Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclearantigen1. Mol. Cell. Biol. 6:3838-3846.
16. Rowe, M., H. Evans, L. Young, K. Hennessy, E. Kieff, and A. Rickinson. 1986.Monoclonal antibodiestothe latent membrane proteinofEpstein-Barrvirusrevealheterogeneity of theprotein andinducibleexpressioninvirus-transformed cells. J. Gen. Virol. 68:1575-1586.
17. Rowe, M.,D. T.Rowe,C. D.Gregory,L.S.Young,P.J.Farrell,H.
Rupani,andA. B. Rickinson. 1987. Differencesin Bcellgrowth phenotypereflect novelpatternsofEpstein-Barrvirus latentgene
expression in Burkitt'slymphomacells. EMBOJ. 6:2743-2751. 18. Sternas, L., T. Middleton, and B. Sugden. 1990. The average
number of molecules ofEpstein-Barr nuclearantigen 1 per cell
on November 9, 2019 by guest
http://jvi.asm.org/
doesnotcorrelate with theaveragenumber of Epstein-Barr virus
(EBV) DNA moleculespercell amongdifferent clones of
EBV-immortalized cells. J. Virol. 64:2407-2410.
19. Sugden, B., and N. Warren. 1989. Apromoter ofEpstein-Barr virus that can function during latent infection can be transacti-vated byEBNA-1,aviralproteinrequiredfor viralDNA replica-tion duringlatent infection. J. Virol. 63:2644-2649.
20. Takada, K. 1984. Cross-linking of cell surface immunoglobulins
induces Epstein-Barr virus in Burkitt lymphoma lines. Int. J. Cancer 33:27-32.
21. Takada, K.,K. Horinouchi, Y.Ono, T. Aya,T.Osato, M. Taka-hashi, and S. Hayasaka. 1991. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. VirusGenes 5:147-156.
22. Takada, K., Z. Ji, S. Fujiwara, N. Shimizu, and A. Tanabe-Tochikura. 1992. Partial elimination of Epstein-Barr virus
plas-mids from Burkitt's lymphoma cells bytransfecting the BZLF1
gene.J. Virol. 66:5590-5593.
23. Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol.63:445-449.
24. Yates, J. L., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viralgenome that
per-mits stable replication of recombinant plasmids in latentlyinfected cells. Proc. Natl.Acad. Sci. USA 81:3806-3810.
25. Yates, J. L., N.Warren, and B. Sugden. 1985. Stable replication of plasmidsderived from Epstein-Barrvirus invarious mammalian cells. Nature(London) 313:812-815.
26. Young, L. S., C.Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson,J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, and J.I.
Cohen. 1989. N.Engl. J. Med. 321:1080-1085.