Vol.41,No. 1 JOURNALOFVIROLOGY, Jan. 1982,p.250-257
0022-538X/82/010250-08$02.00/0
Avian
Myeloblastosis
Virus
Transforming
Gene Is Related
to
Unique Chicken DNA Regions Separated by
at
Least One
Intervening
Sequence
BERNARD PERBAL AND MARCEL A. BALUDA*
UniversityofCalifornia atLosAngelesSchoolofMedicine andMolecular BiologyInstitute, Los Angeles,
California90024
Received 30July 1981/Accepted 6 October 1981
Identification of several additional restriction endonuclease sites within the
cellular substitution (amv) inserted into the avian myeloblastosis virus proviral
genome haspermittedus toisolate differentregionsof theamvsequence. These
subsets ofthe avianmyeloblastosis virustransforming gene have been cloned in
theplasmidpBR322 and usedashybridization probestoinvestigatethetopology ofhomologous (proto-amv)normal chicken DNAsequences. The results showed
that thecellular proto-amv sequences in C/O chicken DNAareinterrupted by at
least oneinterveningsequence. Apartialarrangement of the proto-amv sequences
ispresented.
Avian myeloblastosisvirus(AMV)contains a
sequence of approximately 1,000 nucleotides
which appears to be responsible foracute
my-eloblastic leukemia in chickens (25, 26). This
sequence, designated amv, is homologous to
cellular DNA sequences(proto-amv) present in
all vertebrate species from amphibians to
hu-mans (5, 24, 27). This AMV leukemogenic
se-quenceis not present in thehelper
myeloblasto-sis-associated virus types 1 and 2 (MAV-1 and
MAV-2) orin Rous-associated virus type 0, the
endogenous virus produced by some chicken
strains (24-26). Previous endonucleasemapping
of the XCharon4Arecombinantclone(XllA1-1)
carrying the AMV provirus has revealed that the
amv sequence lies between the KpnI and
3'-proximalXbaIsites and contains one EcoRI site
but no BamHI or HindIII site (Fig. 1). The
hybridization results obtained earlier with the
cloned AMV HindIII fragment of 3.9 kilobase
pairs (kb)(5) and with other probes (26) (AMV
internal EcoRI-EcoRI and EcoRI-3' HindIII
fragments) indicated that three chicken DNA
fragments generated byEcoRI digestion contain
genetic information homologous to the amv
se-quences. Two of them are adjacent,the 5.4-kb
fragment and either the 2.2- or the8.7-kb
frag-ment, whereas the third one is farther apart or
present at adifferent homologous genetic locus.
The first alternative suggests that the normal
cellularproto-amv DNA may contain an
inter-vening sequence (intron). Mosteucaryotic genes
contain introns, up to approximately 50 in the a-collagen gene (32), whereas some genes have
none, e.g., histone (13) and interferon (15, 18).
The cellular analogs of viral transforming
se-quences analyzed so far also seem to fall into
two categories with respect to introns. Some have introns, e.g., the cellular sequences
ho-mologous to thetransformingregions of Abelson
murineleukemiavirus,felinesarcomavirus,and
Rous sarcoma virus (11, 22), whereassome do
not, e.g., the Moloney-murine sarcoma
virus-related sequences (19). The chicken cellular
DNA sequences homologoustothesrcgene of
thePragueC strainofRoussarcomavirus have
recently been shown to contain fiveorsix
inter-vening sequences (22).
The probes used in the earlier studies
con-tained some viral sequences common to AMV
and MAVinaddition toamv.Therefore,itcould
not be ruled out that some EcoRI or HindIII
chicken DNA fragments which hybridized to
theseprobes containedendogenousproviral
se-quences or that some cellular proto-amv
frag-ments had the same size as the endogenous
proviralfragments. To carry out a more detailed
and accurate analysis of the cellular proto-amv
sequences,weisolated and purified by
subclon-ing inpBR322 amv-containing DNA fragments
with fewer or no proviral sequences and used
themashybridization probes with C/O chicken
DNAdigested with three restriction enzymes.
MATERIALS AND METHODS
Cloning. Escherichia coli HB101 (pro leu thilac-4 hsdR endA recA rpsL20 ara-14 galK2 xyl-5 mtl-l
svpE44) was the recipient in transformation
experi-ments(8). The vector used was pBR322. The transfor-mation procedure of Villa-Komaroff et al. (30) was used with minor modifications (S. Horowitz, personal communication). Bacteria were grown in 300 ml of
250
on November 10, 2019 by guest
http://jvi.asm.org/
INTRON IN proto-amv DNA SEQUENCES
EcoRI 2.6 (4 kb)
5' 7 T zIF ,T ,IY 3'
Hindl2.6 (4 kb)
AMV 5 7 T I wW I T Y
+~~~~~~~~~~~-~
1
s
3'I
c
C
0.-pBR 322/KX162
(1.3 kb)
r
I.
E 0
co
pBR322/HAX4
(1.0 kb)
pBR322/EX II
(0.8 kb)
Sx1
(0.45 kb)
ES2
(0.35 kb)
-~~ Iu
t
I
t
t
t
I t 1(r)0E(c> Tr LU (.')
Q-HaeIl
E I,_
GD
i
i
It
_
0
x
0I
0
CD
-XTC
_n:
-O
X
in
0
Eal O
[image:2.504.70.429.67.507.2]I~t4~~~U g a I
~~~~~Sol
I Sal I (pBR322) EtM LLJ
a BarmHI linkers
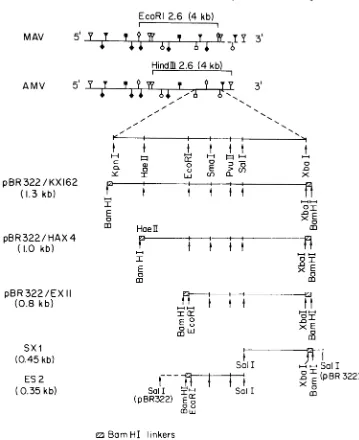
FIG. 1. Derivation of thehybridization probes. The restriction endonuclease sitesin MAVand AMVare
mappedaspreviously described (26). Symbols: V,HindlIl; *, BamHI;V,XhoI; *, BgII,O,EcoRI;0,XbaI;
0,KpnI.Themappingof restrictionsiteswithintheKpnI-XbaI fragmentwascarriedoutbydigestionof the
32P-labeledpurified fragment.DNAfragmentsbearingBamHIlinkersatboth endsweresubcloned afterinsertionat
theBamHI site of pBR322. Arrows indicateknown restriction sites still available aftersubcloning.
tryptone-yeast extract medium (10 g of tryptone [Difco]perliter,5gofyeastextractperliter,and 5 g of NaCl per liter) until the optical density at 600 nm
reached0.3, spundown, suspended in 30ml of cold buffer(25mM sodiumacetatebuffer[pH5.6],70mM
MnCI2, 30 mM CaCl2), andkeptfor 20 min at4°C. Competentcellswerespunat4°Candresuspendedin 10 ml of thesamebuffer,and 200,ulofthesuspension
wasmixed with 25IlIof theplasmidsolution.After 1 h of incubation at4°C, 0.1-ml samples were platedon tryptone-yeast extractagarplates containing100 pLgof
ampicillinperml.Screeningofindividual transformed clones for their plasmid content was performed as
previouslydescribed(6, 14).
DNApurification.Purification ofplasmidDNAwas
performed as described by Curtiss et al. (10) after
amplification in thepresenceofchloramphenicol (9). High-molecular-weightcellular DNA(45kborlarger)
was purified from C/O H&N chicken embryos as
described earlier(3).
Isolation of A DNA recombinant dones containing
proto-amvsequences.A total of 3 x 105 plaquesfroma
VOL. 41, 1982
MAV
251
on November 10, 2019 by guest
http://jvi.asm.org/
252 PERBAL AND BALUDA
ACharon 4Alibrary of leukemic chicken DNA partial-ly digestedwithEcoRJ(27)werescreened by using the in situ plaquehybridizationtechniqueof Benton and Davis (4). Nitrocellulose filters were duplicated and hybridized eithertotheKpn-Xbaprobeor tothe MAV EcoRI 2.6(3.9-kb) probe. Plaques which hybridized
with the Kpn-Xba DNA fragment but not with the MAV-1 DNAfragmentwereconsideredlikelyto con-tain proto-amv sequences. A total of 18plaqueswere picked, and each phage isolate wasreplaqued twice
more. Nineproto-amv-containing recombinantclones were grown tohigh-titer stocks in E. coli DP50 supF (7).The DNAwaspurifiedasdescribed(27).
Purification ofDNA fragments. Restriction enzyme digestions wereperformedin the buffers recommend-ed by Bethesda Research Laboratories, Rockville,
Md. A 10-fold-excess ratio of enzyme to DNA was generally used. Digestion of cellular DNA and gel
electrophoresisprocedures havebeen described(26).
The Kpn-Xbafragment wasisolated from meltedsea plaque gel (Seakemlow-temperature-meltingagarose; Marine Colloids, Rockland, Maine) (26). All other DNAfragments wereelectroeluted fromregular agar-ose(SigmaChemical Co.,St.Louis, Mo.)gelsat150 mA for at least 90 min in 5 ml of TAE buffer(40mM
Tris-hydrochloride [pH 7.6], 5 mM
Na-C2H302 *3H20,1 mMEDTA). DNAfragments were
thenextractedtwice withamixtureof
phenol-chloro-form-isoamyl alcohol(1:24:1),twicewith
chloroform-isoamyl.(24:1),and twice with ether before
precipita-tionwithethanol.
Ligatton ofBamHIlinkersto DNAfraginents. BamHI
linkers (17, 21)were phosphorylated in reaction vol-umes of 30 ,ul containing 0.8 nM BamHI linkers
(Collaborative Research,Inc.,Waltham, Mass.),0.07 MTris-hydrochloride buffer(pH7.4),0.01 MMgCl2,
0.01 M2-mercaptoethanol, 0.06 mM ATP, and 5 U of T4polynucleotidekinase(Bethesda Research Labora-tories). After 2 h of incubationat 37°C, the reaction wasstopped byincubationat70°Cfor 10 min. Restric-tion DNAfragmentswithcohesive ends were incubat-ed in the presence of DNA polymerase I (Bethesda Research Laboratories) to generateblunt ends (1, 2, 12). Inthe case of enzymes generating 3' protruding ends (KpnI, HaeII), DNA fragments (5 ,ug) were incubated in the presence of 60 mM Tris buffer (pH
7.5),10mMMgCl2,10 mM2-mercaptoethanol,and1
Uof E. coliDNApolymerase I for 10min at 12°C to
takeadvantageofthe 3' -+5'exonucleaseactivityof
polymeraseI.Thefour deoxynucleotide triphosphates were then added to a final concentration of 0.2mM,
and polymerization was allowed to proceed for 1 h at
12°C. In the case of enzymes generating5'protruding ends(EcoRI,XbaI), the restriction sites werefilledin by polymeraseI.After inactivation ofpolymerase I for 10minat70°C, ATP was added to a final concentra-tion of1mM,and1U of T4DNAligase(New England Biolabs, Beverly, Mass.) was added to the mixture. Ligation was performed at 12°C for 10 h.
Ligation of DNA fragments to pBR322 vector. The pBR322 vector, cleaved with the appropriate restric-tion endonuclease, was treated with calf intestine alkaline phosphatase (Boehringer Mannheim Bio-chemicals, Indianapolis, Ind.) to reduce colony back-ground caused by recircularization of vector DNA (29). Six units of calf intestine alkaline phosphatase as ammonium sulfate precipitate were spun in an
Eppen-dorf microfuge tube and suspended with 5 p.g of cleaved pBR322. The reaction mixture (100 ,ul)was
adjustedto20mMTris-hydrochloride buffer(pH8.0) and incubated for 1 h at 37°C. The reaction was
stoppedby the addition of 10 mMTris-hydrochloride
buffer (pH 7.5)-i mM EDTA (pH 7.5)-0.2 M NaCI-0.5% sodiumdodecylsulfate. The treatedvector was thenextracted twice withphenol-chloroform-isoamyl
alcohol (1:24:1), twice withchloroform-isoamyl alco-hol(24:1),and twice with ether before ethanol
precipi-tation. Ligation of DNAfragmentstotreatedpBR322
was performed in 250 mM Tris-hydrochloride (pH
7.4)-S50mMMgCI2-5mMATP-50 mM
2-mercaptoeth-anolat aDNAconcentration of 200jig/mlwith 1
RI
of T4ligase (NewEngland Biolabs).Nick translation ofpurified DNA fragments. Nick translation(20) wasperformedin the presence of 200 ,uCi of [a-32P]dCTP (Amersham Corp., Arlington
Heights,Ill.), 30,uMeach ofdATP, dGTP,anddTTP,
5 ,ul of nick translation buffer (Bethesda Research Laboratories), 2 ,ul of DNApolymerase1(2U/,ul),and 2 ,ulof DNase I(WorthingtonDiagnostics, Freehold,
N.J.; DPFFgrade0.2
jig/ml).
DNaseincubationwas started 5 minbefore the additionof,DNApolymeraseI. The 32P-labeled fragments and the remaining [a-32P]dCTPwereseparated by chromatographyon
Se-phadex G-75 equilibrated in 0.1 x SSC plus 0.1% sodium dodecyl sulfate. The specific activity of the 32P-labeled probeswasapproximately2x108cpm/p.g.
Sizing of the 32P-labeled DNA fragments obtained afterdigestion of nick-translated probes with restriction enzymes.Thechainlengthoffragmentsobtained after
singleand doubledigestionsofDNAswasdetermined byelectrophoresis in5%acrylamidegelasdescribed by Maniatisetal.(16). Kodak X-ray filmswereplaced
onthe top of thegels,covered with SaranWrap,and
exposedfor 3 hat4°C.
Preparation and hybridization of DNA (Southern) blots.The DNAfragments separated by
electrophore-sis were blotted on nitrocellulose membrane filters (Millipore Corp., Bedford, Mass.) as described by Southern(23).Hybridizations were performed
accord-ingtoWahletal. (31).Filters were washed twice in 2x SSC plus0.1% sodiumdodecylsulfate atroom tem-peratureandtwo tothreetimesat68°C in 2xSSC plus 0.1% sodiumdodecyl sulfate,rinsedtwice in 2x SSC, dried,andexposed with Kodak X-ray films (XR5) and aDuPontintensifying screen at -700C.
Physical and biologicalcontainment.This work was carriedout atthe P2-EK2containment levels accord-ing to the revised guidelines of the National Institutes of Health.
RESULTS
Derivation of the amv-specific probes. The
preparation of specific amv probes started with
the isolation of the KpnI-XbaI (3'-proximal)
fragment from the AMV HindIII 2.6 fragment
(2.6x 106 daltons [4 kb]) (Fig. 2) already cloned
in pBR322 (5). The Kpn-Xba DNA fragment was
isolated by gel electrophoresis after double
di-gestion of AMV HindlIl 2.6/pBR322
recombi-nant DNA with KpnI and XbaI, ligated to
BamHI linkers, and cloned in pBR322
(pBR322/KX162). This probe still contains
ap-J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
INTRON IN proto-amv DNA SEQUENCES
O DC b C
2"'37-... _
--- _ _
t.-:
fti04Z O#k4-3
a
K>x 6 i_>P 4
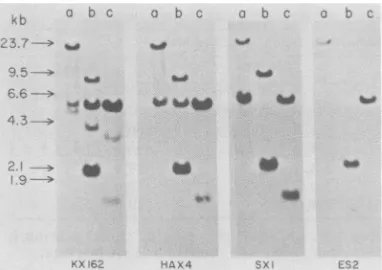
FIG. 2. Hybridization ofresti
ated DNAfragmentswithamv-c4
chicken DNA was digested wit
(b), orHindlll (c), electrophore
gel, and transferredtonitrocellu blots). The three different restri
were runin parllel inthesameg
four separate sets. A sample digested with Hindlll endonuw parallel lane wasused asmolec pressed in kilobase pairs. The shownin Fig. 1wereusedafter dCTPby the nick translationpr(
proximately 200 base pairs
located at the 5' terminus,
previous electron microsc
Therefore, a search was ma
enzyme which could remo'
pairsfrom the 5' end of the j
The 32P nick-translated
was treated with several re
AluI, MspI,Sau3A1, CfoI,
PvuII, Hinfl, HpaI, Sail,
Among the few enzymes w
cleavagein theKpn-Xbafral
at450 basepairs from theX
cleavedat290 basepairsfror
1).A HaeII(XbaI-BamHI)-J isolated afterdigestionof K) nuclease. AfterligationofB
HaeII sitesand digestionw
clease,aHaeII-Xbafragmei
ersatboth endswas cloned
One ofthe
resultinf
clonibridized with the 3P nic
EcoRI 2.6 fragment (also 2
kb]), to determine whethe
quencesremainedat the 5'ei
fragment.The results(datan
thathelperviral sequencesv
end oftheamvinsertinHA)
digestedwithSalltoestabli.
the insertinpBR322. Clone,
XbaI DNAfragmentin the t
tionswithin theplasmidDN
a b c a b c The presence of a single EcoRI site in the
HaeII-XbaIfragment and in pBR322 DNA (28)
was exploited to generate an
EcoRI-XbaI
DNAfragment from pBR322/HAX4 after digestion
_ ~_ with EcoRI. This fragment was cloned into
pBR322 after treatment with DNAPoll,ligation
of BamHI linkers, and digestion with BamHI
endonuclease. The orientation of the
EcoRI-XbaI DNA
fragment
with respect toplasmid
a DNA in one
(pBR322/EXII)
of the clonesob-tained was determined by digestion with Sall and EcoRI endonucleases. This clone was then
used to isolate twofragments carrying the amv
sequences located on the 3' side of the EcoRI
riction enzyme-gener-
site:
anEcoRI-SalI
(ES2) and aSalI-XbaI
frag-ontaining probes.C/O ment (SX1).
'h BamHl (a), EcoRI
esed in 0.7% agarose The sizes and derivations of the different
ilose filters(Southern hybridization
probes
are shown inFig.
1.iction enzymedigests Hybridization of chicken DNA fragment to
gelbut were blotted in
amv-containing
probes. High-molecular-weightofeaseXnd
(0.5
ig)
(larger than 45 kb) DNA from C/O chickenularesizemarkners
ex-
embryos
wasdigested
with eitherEcoRI,
Hin-hybridization probes
dIll,
or BamHI restriction endonuclease,elec-labelingwith
[a-32p]-
trophoresed in agarose gels, and transferred toocedure. nitrocellulose. The blots were
hybridized
withthe
32P-labeled
probes described above (Fig. 2andTable 1).
(i) Hybridization with the KpnI-XbaI probe.
of viral sequences The Kpn-Xba DNA fragment obtained by
as estimated from BamHIdigestion of pBR322/KX162 was purified
opic
studies (27). byagarose
gel electrophoresis, electroeluted,Lde fora restriction and 2P nick translated. The chicken DNA
ve about 250 base EcoRIfragments hybridizing with the Kpn-Xba
Kpn-Xbafragment. probe are 8.7, 5.4, 3.9, and 2.2 kb. The low
Kpn-Xba fragment intensity of hybridization obtained for the EcoRI
.striction enzymes: 3.9-kbfragment suggests thatthis hybridization
WaeIII,
TaqI,HaeII, resultsfrom the 200 base pairs of viral sequencesEcoRl, and SmaI. present at the 5' end of the KX162 probe.
ihich gave a
single
However, this probe no longer detects thegment,
Sallcleaved EcoRI20.8-kbfragmentwhichhybridizedtothe'bal
site,
andHaeII HindIll2.6probe (5).Thechicken DNAHindIII ntheKpnIsite(Fig.
fragments detected are 5.4, 3.3, and 1.4 kb.ffaeII
fragment
was Hybridization was notdetected with the 2.6-kb(162byHaeII endo- fragmentwhich hybridizedwith theHindIII2.6
tamHIlinkers to the probe. The intensity of the HindIII3.3-kb
frag-rithBamHIendonu- mentrelativetothe othertwois
greatly
reduced,
nthavingBam link- suggesting that this may also be due to the intopBR322. presence of viral sequences
remaining
in thees
(HAX4)
washy-
probe. The detection of the 5.4-kbfragment
Sk-translated
MAV might be due either to the viral sequencesre-.6 x 106 daltons
[4
maining
in theKX162probe
or to the existencesr helper viral se- ofanamvfragmentwhichhasthesame
molecu-ndof theHaeII-Xba lar
weight
as theendogenous proviral
DNAiotshown)indicated
fragment
whichhybridized
to the MAV EcoRIvereabsentatthe5' 2.6
probe
described earlier(5).
The patternsK4.HAX4wasalso obtained with the C/O DNA BamHI digest
sh theorientationof showed that four
fragments hybridize
with thesbearingtheHaeII- AMV
Kpn-Xba
probe
(19.6,
12.4, 5.5, and 4.8 :wopossibleorienta- kb) and fourfragments
hybridized
with the{Awereidentified. MAV
probe
(12.4,
5.8, 4.8,
and 1.9kb).
There-253
VOL.41,1982
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.504.55.246.70.205.2]254 PERBAL AND BALUDA
TABLE 1. Hybridization of C/O chicken DNA fragments with different amv-containingprobesa
Cellularfragments(kb)generatedwith:
DNAprobes EcoRI HindIII BamHI
MAV-1(EcoRI2.6) 3.9 5.4, 3.3, 2.6, 1.9 12.4, 5.8, 4.8,1.9
AMV
HindIl 2.6 20.8, 8.7, 5.4, 3.9, 2.2 5.4, 3.3, 2.6, 1.4 NDb
KpnI-XbaI 8.7, 5.4, 3.9,2.2 5.4, 3.3, 1.4 19.6, 12.4, 5.5, 4.8
HaeII-XbaI 8.7, 5.4, 2.2 5.4, 1.4 19.6,5.5
EcoRI-SalI 2.2 5.0 19.6
SalI-XbaI 2.2, 8.7 5.4, 1.4 19.6,5.5
aThe MAV-1 (EcoRI 2.6) and AMV
(HindlIl
2.6) probes were obtained from pBR322 clonespreviously
described(5). The sizes of thefragmentsweredeterminedbycomparisonwith those ofXDNAmarkers. bND,Notdetermined.
fore, it appears that the 19.6- and 5.5-kb
frag-ments are amv specific and that the 12.4- and
4.8-kb fragmentsare detectedby KX162owing
to thepresenceofviral sequences in thisprobe.
(ii) Hybridization with the HaeHI-XbaI probe.
The HaeII-XbaI DNA fragment obtained by
BamHIdigestionofpBR322/HAX4wasisolated
by
adarose gel electrophoresis, electroeluted,and
3
P nick translated. Three bands wereob-tained after hybridization with C/O chicken
DNAdigested with EcoRI:8.7, 5.4, and 2.2 kb.
Thus, there is no EcoRI 3.9-kb bandcontaining
amv-related sequences. Hybridization of this
probe toC/O chicken DNA digested with
Hin-dIIIrevealed the presenceof onlytwofragments
complementary to the amv sequences, the
5.4-kb band being much darker than the 1.4-kb
band. Therefore, the detection of the HindIII
3.3-kb fragment by the Kpn-Xbaprobe resulted
from the
presence.
ofendogenous proviralse-quences.Hybridization of theHaeII-XbaIprobe
tochicken DNA digested with BamHI confirms
theconclusions reached above from the
compar-isonof the bandpatternsdetected withthe MAV
EcoRI 3.9-kb and the AMV Kpn-Xba fragments.
Only two amv-related BamHI bands were
de-tected:19.6 and 5.5 kb, the latter being relatively
darker than the 19.6-kb band.
(iii) Hybridization with Sall-XbaI and
EcoRI-Sail probes. These probes were generated by
Sall digestion of pBR322/EX11, purified by
agarose gel electrophoresis, electroeluted, and
32P nick translated. The SalI-XbaI probe
con-tains amv sequences complementary to the
chicken DNAEcoRI8.7- and 2.2-kb fragments,
totheHindlIl5.4- and1.4-kb fragments, and to the BamHI 19.6- and 5.5-kb fragments. The
hybridizationintensityof theEcoRI8.7-kb, Hin-dIII 1.4-kb, and BamHI 5.5-kb bands was
great-ly increased when the
SalI-XbaI
probewas usedinstead of the HaeII-XbaI probe, reflecting the
increasedconcentrationofamvsequences in the
former probe. Only one chicken DNA EcoRI
fragment (2.2kb),oneHindIII fragment (5.4kb),
and one BamHIfragment (19.6 kb) were found
tocontainsequences homologoustothe
EcoRI-SalIprobe.
Analysis ofchicken-A DNA recombinants
car-rying proto-amv sequences. Nine DNA
recombi-nant clones carrying cellular proto-amv
se-quencesidentified as describedin Materials and
Methods were digested with EcoRI and
fraction-ated by electrophoresis. They fall into three
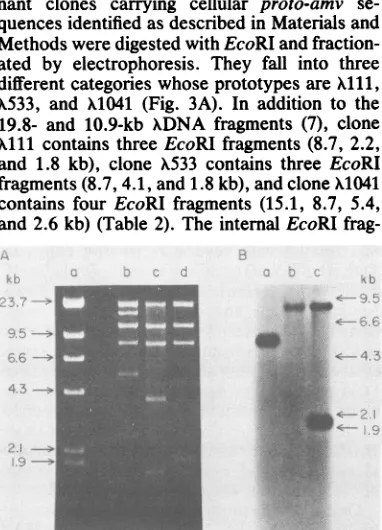
different categories whose prototypes areX111,
X533, and X1041 (Fig. 3A). In addition to the
19.8- and 10.9-kb XDNA fragments (7), clone
Xlll contains three EcoRIfragments (8.7, 2.2,
and 1.8 kb), clone X533 contains three EcoRI
fragments (8.7, 4.1, and 1.8 kb), and clone X1041
contains four EcoRI fragments (15.1, 8.7, 5.4,
and 2.6 kb) (Table 2). Theinternal EcoRI
frag-< -.
_ _
--* _
E. .g.. <... _ ....-. v,.'
8£
Y, dsx:.3n': ':'.:.se'.!: #':: ,...
FIG. 3. Analysis of chicken-A recombinant DNA clones containing proto-amv sequences. Chicken-A Charon 4A recombinants carrying proto-amv se-quences were isolated and their DNA was digested with EcoRI as described in the text. (A) a, X+; b,
X1041; c,X533; d, Xlii.Paralleldigests were blotted on nitrocelluloseand hybridized with the 32P-labeled Kpn-Xba probe. (B) a, X1041; b,X533; c, Alii. The marker A DNA fragments with sizes in kilobase pairs were run in the same gel.
J. VIROL.
"I.._
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.504.266.457.308.573.2]TABLE 2. Size of the DNAfragmentsgeneratedby
EcoRIdigestion of chicken-A DNA recombinants
IN
Recombinant clone Fragmentsize (kb) kil1 19.8,a 10.9,a8.7,b 2.2,b 1.8 X533 19.8 a10.9a87,b4.1, 1.8 X1041 19.8,9 15.1,10.9,a 8.6,5.4,b2.6
aACharon 4A EcoRI fragments.
bFragments which hybridize with the AMV
(KpnI-XbaIorHaeII-XbaI)probes.
mentsof A Charon 4A can be replaced by foreign
DNA inserts in the size range of 8.2 to 22.2 kb
(7). Also, the leukemic chickenDNAfragments
used to prepare our chicken-A library ranged in
A
I-I'-4 ~ 4 c
C 4) 0
Q. Oa
-Y rI L1J
i
j
0.221
[TRON IN proto-amv SEQUENCES 255
sizebetween 16 and 22 kb (27). Consequently,
from the summation (approximately 32 kb) of
the fragment sizes obtained with X1041, it
ap-pears that the 15.1-kb EcoRI fragment might represent an intramolecular recombinant, e.g.,
from looping out during amplification of the
DNAstock.
Hybridization of Southernblots of the
EcoRI-digested DNA with the 32P-labeled Kpn-Xba
probe (Fig. 3B) shows that Xl1 contains two
DNA fragments(8.7 and 2.2 kb)carrying
proto-amv sequences, and X533 and X1041 contain
onlyoneeach, 8.7 and 5.4 kb, respectively.The
same results were obtained with the HAX4
probe. Thus, the proto-amv-containing EcoRI
0
c)
0.35
1
AMV
0
-0 x 0.45
/ / LI
B
LI IS L2
t
2.2o
0
T5.4
0 I 19.6
A+----i
1-u--o 0
o 0
llJ LLI
1.4l
c C c
l t 5.4
5- -r
I I
E E
m 0
en e
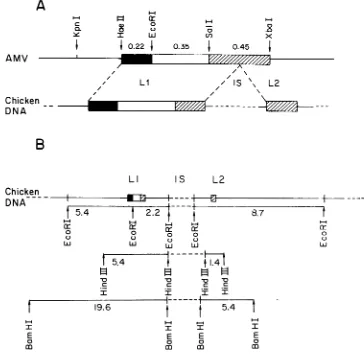
FIG. 4. Partial arrangementof the proto-amv sequencesin C/O chicken DNA.(A)Theproto-amv sequences
areinterrupted byatleastoneinterveningsequence(IS)in C/O chicken DNA.(B)The relativepositionofthe
EcoRI, Hindill, and BamHI fragments carrying proto-amv sequences was established from hybridization patterns obtainedwith theprobesdescribedin thelegendtoFig.1. Itis not known whether the2.2-and8.7-kb EcoRlfragments arecontiguous.Thesameuncertainty appliesto the twoHindIIIand BamHIfragments;i.e., theremaybeone ormorerestrictionenzymesites within the intronshown.Also,as discussedin thetext,there maybeasecondintron between the5.4- and 2.2-kb EcoRIfragments.
Chicken DNA
A
/is
\\.
L2
/
Chicken DNA
5.4
0
C.)
LUJ
8.7
I
cr
0LUJ
I
E
I
E
m
m
im
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.504.67.431.248.602.2]256 PERBAL AND BALUDA
DNAfragments from thesechicken-A
recombi-nant clones havethe same sizes as those
detect-ed in hybridization studies between the
Kpn-Xba probe and normal chicken DNA digested
withEcoRI.
DISCUSSION
Previous studies performed with different
AMV internal restriction fragments as
hybrid-ization probes indicated that theamv sequence presentin the AMVgenome washomologousto
a setof normal chicken DNAsequences
(proto-amv)from which itmighthaveoriginated (27).A
complex organization of the proto-amv
se-quences inchicken DNA was suggested by the
existence of several Hindlll and EcoRI
frag-ments complementary to the amv sequence (5, 27).
In this study, DNA fragments representing
different regions of the amv sequence having
little or no homology with MAV helper
se-quences were used as hybridization probes to
maptheproto-amv sequencesinchickenDNA.
Three EcoRI fragments (8.7, 5.4, and 2.2 kb),
twoHindIIIfragments (5.4 and1.4 kb)andtwo
BamHI fragments (19.6 and 5.4 kb)in chicken
DNA are homologous to amv. Since the amv
sequence contains only one EcoRI site but no
HindIII orBamHI site (26), these results
indi-cate that proto-amv information might not be
contained in asingle setof DNAsequences.
Hybridization with the AMV EcoRI 3.3-kb
fragment and with the AMV 1.3-kb fragment
locatedbetween the 3'-proximal EcoRI and
Hin-dIII sites had shown (27) that the 5.4-kb EcoRI
fragment was adjacent toeither the 8.7- or the
2.2-kb fragment. The hybridization patterns
ob-tained with the SalI-XbaI and EcoRI-SalI
probes now place the 2.2-kb EcoRI fragment
adjacenttotheEcoRI5.4-kbfragmentand
indi-cate that approximately one half of the amv
sequences located between the SalI and XbaI
sites originate fromtheEcoRI 2.2-kb fragment.
Therefore, it appears that the 3' region of the
proto-amv chicken sequences are interrupted by
atleastone interveningsequence.
The isolation of a X Charon 4A recombinant
(Xlii)
DNA carrying the twoEcoRI
8.7- and2.2-kb fragments in the same insert indicates
thatthesetwo fragments are relatively close to
each other and certainly are not located on
different chromosomes. Comparison of the
in-serts of
Alii
and X533 reveals that theEcoRI
8.7-and 2.2-kb fragments are either contiguous or separated by 1.8 kb of chicken DNA at the most.
Sincewedidnot find achicken-A recombinant
carrying both the EcoRI 5.4- and 2.2-kb
frag-mentsamong the 18 clones obtained so far, the possibility exists that another intervening
se-quence might be present between these two
proto-amv fragments.
These results, along with those obtained
pre-viously (5, 27), permitapreliminary arrangement
of theproto-amv sequencesin H&NC/O
chick-en DNA (Fig. 4). Two different domains of
proto-amvsequences(Li andL2)areseparated
in chicken DNA by an intervening sequence
which contains at least one BamHI site, one
EcoRI site, and one HindIII site. The relative
positions of the EcoRI, BamHI, and HindIlI
sites with respect to each other within the
in-tron, and the size of this intron, are being investigated.
ACKNOWLEDGMENTS
We thank L. Souza for helpfuldiscussions.
This research was supported by Public Health Service research grantCA-10197from the National Cancer Institute. B.P. was partly supported by the Centre National de la Recherche Scientifique and was the recipient ofa travel fellowshipfrom the North AtlanticTreaty Organization.
LITERATURE C1TED
1. Backman, K., M. Ptashne, andW. Gilbert. 1976. Con-struction ofplasmids carrying the cI gene of bacterio-phage X. Proc.Natl. Acad.Sci. U.S.A. 73:4174-4178. 2. Bahl, C.P., R. Wu, K. Ikakura,N.Katagusi, and S. A.
Narang. 1976. Chemical andenzymatic synthesisof lac-toseoperator of Escherichia coli and itsbindingtolactose repressor. Proc.Natl.Acad. Sci. U.S.A. 73:91-941. 3. Baluda, M.A., and W. N.Drohan. 1972. Distribution of
deoxyribonucleicacidcomplementarytothe ribonucleic acid of avianmyeloblastosisvirus in tissues of normal and tumor-bearingchickens. J.Virol. 10:1002-1009. 4. Benton, W.D.,and R. W.Davis. 1977. ScreeningofAgt
recombinantclonesby hybridizationtosingle plaquesin situ.Science196:180-182.
5. Bergmann, D.G., L. M.Souza,and M. A. Baluda. 1981. Vertebrate DNAs contain nucleotide sequences related to the putative transforminggene ofavian myeloblastosis virus. J. Virol. 40:450-455.
6. Bimboim, H. C., and J. Doly. 1979. A rapid alkaline extractionprocedure forscreening recombinantplasmic DNA. Nucleic Acids Res. 7:1513-1523.
7. Blattner, F. R., B. G. Williams, A. E. Blechl, K. D. Thompson,H.E.Faber,L. A.Furlong, D. J.Grunwald, D.0. Kiefer, D. D. Moore, J. W. Schuman, E. L. Sheldon, and0. Smithies. 1977.Charonphages:saferderivatives of bacteriophagelambdafor DNAcloning. Science 196:161-169.
8. Boyer, H. W., and D. Rouland Dussoix. 1969. A comple-mentationanalysis of the restriction and modification of DNAinEscherichia coli. J. Mol. Biol. 41:459-472. 9. Clewel,D.B.1972. Natureof ColElplasmid replicationin
Escherichia coli in the presence of chloramphenicol. J. Bacteriol. 110:667-676.
10. Curtiss, R., M. Inoue, B. Pereira, J. C. Hsu, L. Alexander, and L.Rock. 1977.Construction and use of safer bacterial hoststrains for recombinant DNA research, p. 99-111. In W. A. Scott and L. Rock (ed.), Molecular cloning of recombinant DNA. Academic Press, Inc., New York. 11. Goff,S. P., E. Gilboa,0. N. Witte, and D.Baltimore.
1980. Structure of the Abelson murine leukemia virus genome and the homologous cellular gene: studies with theviral DNA. Cell 22:777-785.
12. Heffron,F., M. So, and B. J. McCarthy. 1978. In vitro mutagenesis of a circular DNA molecule by using synthet-icrestrictionsites. Proc.Natl.Acad. Sci. U.S.A. 75:6012-6016.
13. Kedes, L. M. 1979. Histone genes and histone
messen-J.VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
SEQUENCES
gers.Annu.Rev. Biochem.48:837-870.
14. Klein, R. D., E. Selsing, and R. D. Wells. 1980. Arapid microscale technique for isolation of recombinant plasmic DNA suitable for restriction enzymeanalysis. Plasmid 3:88-91.
15. Lawn, R. M., J. Adelman, A. E.Franke, C. M. Houck, M. Gross, R. Najarian, and D. V. Goeddel. 1981. Human fibroblast interferon gene lacks introns. Nucleic Acids Res.9:1045-1052.
16. Manlatis,T.,A.Jeffrey, and H. Van de Sande. 1975. Chain length determination of small doubleandsinglestranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry 14:3787-3793.
17. Marians, K.J.,R.Wu,J.Stawinski, T.Hozumi,and S.A. Narang. 1976. Cloned synthetic lac operator DNA is biologically active. Nature (London) 263:744-748. 18. Nagata, S., N. Mantel, andC. Weissmann. 1980. The
structureofoneoftheeightor moredistinctchromosomal
genesfor human interferon-y-.Nature(London) 27:401-408.
19. Oskarsson, M., W. L. McClements, D. G. Blair, J. V. Maizel, andG. F. VandeWoude. 1980. Propertiesofa
normalmousecell DNAsequence(sarc)homologousto the src sequence of Moloney sarcoma virus. Science 207:1222-1224.
20. Rigby, P. W.,M. Dieckmann, C. Rhodes, andP. Berg.
1977. Labeling deoxyribonucleic acid to high specific activity in vitro by nicktranslation with DNApolymerase I.J. Mol. Biol. 113:237-251.
21. Scheller, R. H., R. E. Dickerson, H. W. Boyer, A. D. Riggs,and K.Itakura. 1977. Chemicalsynthesisof restric-tionenzymerecognition sites useful forcloning. Science 196:177-180.
22. Shalloway,D., A. D. Zelenetz, and G.M.Cooper.1981. Molecular cloning and characterization of the chicken
genehomologoustothetransforminggeneof Rous sarco-mavirus. Cell24:531-541.
23. Southern, E. M. 1975. Detectionofspecific sequences amongDNAfragments separated by gelelectrophoresis.
J.Mol. Biol.98:503-517.
24. Souza,L.M.,and M.A.Baluda.1978.Qualitativestudies oftheendogenousprovirusin the chickengenome, p. 217-229.InJ. Stevens, G.J. Todaro,and C. F. Fox(ed.),
Persistent viruses. AcademicPress, Inc.,NewYork. 25. Souza,L. M.,andM.A.Baluda.1980.Identification of
theavianmyeloblastosisvirusgenome.I.Identification of
restrictionendonucleasefragmentsassociated withacute myeloblasticleukemia. J. Virol. 36:317-324.
26. Souza,L.M.,M.J.Brislin, R. L.Hillyard, and M. A. Baluda. 1980. Identification of the avianmyeloblastosis
virus genome. II. Restriction endonuclease analysis of DNAfrom Aproviralrecombinants andleukemic
myelo-blast clones. J. Virol.36:325-336.
27. Souza, L. M., J. N. Strommer, R. L. Hillyard, M. C.
Komaromy,andM. A. Baluda. 1980. Cellularsequences
arepresentin thepresumptiveavianmyeloblastosisvirus genome.Proc. Natl. Acad. Sci. U.S.A.77:5177-5181. 28. Sutcliffe, J.G.1978.Completenucleotidesequenceofthe
Escherichia coli plasmid pBR322. Cold SpringHarbor Symp. Quant.Biol.43:77-90.
29. Ullrtch, A., J. Shine, J. Chirgwin,R.Picket, E.Tischer,
W. J. Rutter, andH. M. Goodman. 1977. Rat insulin genes: construction ofplasmids containing the coding
sequences. Science196:1313-1319.
30. ViUla-Komaroff,L., A. Efstratiadis, S. Broone, P. Lome-dico,R.Tizard,S.P.Naber, W. L.Chick,andW.Gilbert. 1978. A bacterial clone synthesizing proinsulin. Proc. Natl. Acad. Sci. U.S.A. 75:3727-3731.
31. WahI, G.M., M.Stern,andG.R.Stark.1979.Efficient transfer oflarge DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridizationby using dextran sulfate. Proc. Natl. Acad. Sci. U.S.A. 76:3683-3687.
32. Yamada,Y., V. E. Avvedimento, M.Mudryj,H.Ohkubo, G.Vogeli, M. Irani, I.Pastan,andB. deCrombrugghe.
1980. The collagengene: evidence for itsevolutionary assembly by amplification ofaDNAsegmentcontaining
anexonof 54bp.Cell22:887-892.
257
on November 10, 2019 by guest
http://jvi.asm.org/