Copyright© 1989,American SocietyforMicrobiology
Growth
and Survival of Reovirus in Intestinal Tissue: Role of the
L2
and Si Genes
DINAH K. BODKIN'* AND BERNARD N. FIELDS1'2
Department of Microbiology and Molecular Genetics and theShipleyInstitute of Medicine, Harvard Medical
School,'
and Divisionof Infectious Diseases, Department of Medicine, Brigham and Women'sHospital,2Boston, Massachusetts 02115 Received 9 May 1988/Accepted18 November1988
Reovirusserotype1Langcanbe recovered inhightiter from the intestinesof neonatal miceuptoday8after peroral inoculation. Bycontrast, reovirusserotype 3Dearingcannot be recovered from intestinal tissuepast day 4 after peroral inoculation. Thisdifference between the two reoviruseswasmapped by usingreassortants generatedfromnonmutagenized laboratorystocks. When the L2 andS1 genesof reovirusserotype3Dearing
were present in reassortants, the reassortants behaved like serotype 3 Dearing in exhibiting a decreased
capacityto be recoveredfrom intestinal tissue. Likewise, viruses which contained the L2 and S2genesfrom serotype1Lang exhibitedanenhancedcapacitytogrowandsurvive,which is characteristic ofserotype1Lang. Thus, the capacityof reovirustosurvive in intestinal tissuewasdeterminedbythe L2 andS1genes.
Theupperalimentarytractis the major portal ofentryfor
anumber of viruses. Little isknowneither about the factors
in the gastrointestinal tract that influence the capacity of virusestoutilize this pathwayorabout the viralcomponents that regulate the growth of viruses in the gastrointestinal tract.Inourstudieswehave usedthe mammalianreoviruses
as a model to define the genetic and biochemical factors influencing earlyeventsintheviral lifecycle occurringin the gastrointestinaltractaswellaseventsinvolving later virus-host interactions. The reoviruses have segmented double-stranded RNAgenomeswhich, by readily allowing reassor-tants tobe generated after mixed infection by two parents, provideausefulsystem foridentifying viralgenes involved inpathogenesis (reviewed in reference 22). The initial reas-sortants used in our studies were generated by crosses
between mutagenized temperature-sensitive stocks of reovi-rus serotype 3 Dearing and wild-type stocks ofserotype 1 Lang (13, 15, 19). These initial reassortants were used to show that a number of biologic properties, including type-specific neutralization and cell and tissue tropism, were
determinedby the Si gene, which encodes the viral
hemag-glutinin (24-26).
In contrast to the qualitative differences between the parental viruses that mapped to the viral hemagglutinin, otherviralproperties differedin aquantitative fashion. For
example, allreovirus strains examined have the capacityto inhibit protein synthesis or to grow in intestinal tissue
following peroral inoculation. However, differences exist in theextentof inhibition of protein synthesisortheextent of viral growth in the gastrointestinal tract among different
isolates of the same or different serotypes. In studies in which these quantitativedifferenceswereexamined, theuse
ofreassortants made from mutagenized temperature-sensi-tiveparental stocks oftengaveresults soambiguousthatno
cleargenetic link could be established. These findings ledus to isolate a second series of reassortants, generated by crossesbetweennonmutagenized stocks ofserotype3 Dear-ingandeitherserotype 1Langorserotype2 Jones (2, 4, 18).
*Corresponding author.
Thesereassortants were used inanumber of studies which confirmed and extended the previous Si mapping (see, for example, references 10 and23)orwhich allowedproperties tobe mappedtogenomesegmentsother than Si (2, 4, 5, 18). The study of Rubin and Fields (17) is one in which a determinant of virulence other than Si had been identifiedby using reassortants derived from the initial mutagenized stocks. In thisstudy, growthinintestinal tissuewasmapped to theM2gene. However, inconsistencies in the datawere
described in the initialreportwhichwere thoughttobe due to aberrant M2 gene products present in the temperature-sensitive stocks. In a recent study involving the more recentlyisolated reassortants thatwere made from nonmu-tagenized stocks, KeroackandFields(11) demonstratedthat viral shedding from the intestines of neonatal mice and transmission betweenlittermates both were determined by
the L2gene.Thisfindingraised thequestionof whether the L2gene mightalsoplaya role ingrowthand/or survival of
reovirusinintestinal tissue. We thusreexamined the genetic basis forgrowthand survival of reovirus in intestinal tissue by using the reassortants generated from nonmutagenized stocks. In the present study we found that the L2 and Si
genomesegmentsplayedarole indeterminingdifferences in
thegrowthand/or survival of thetwo viruses on days 4, 6, and 8postinoculation. Althoughboth the L2 and Si genome segments were very clearly implicated, the Si segment contributed more significantly to survival than the L2
seg-mentdid.
MATERIALS ANDMETHODS
Cells and viruses. Mouse L cells were propagated in
suspensioninJoklik minimalessential mediumasdescribed previously (14). L-cell monolayers were used for virus
plaque formation and for subsequent propagation of virus. Viralpreparations inoculated into animalswere used atthe secondpassage level. Reovirus serotype 1 Lang and
sero-type3Dearingwerestandardlaboratory stocks provided by
KarenByers.
Reassortants used in this studywere prepared from
non-mutagenized laboratory stocks of reovirus serotype 1 Lang
1188
on November 10, 2019 by guest
http://jvi.asm.org/
4- EBB8I
H4115
3EB14nmal
=
EBi45
6- \>EB14lEB144
o 2 4 6 8 0 1 2 4 6 8
Day
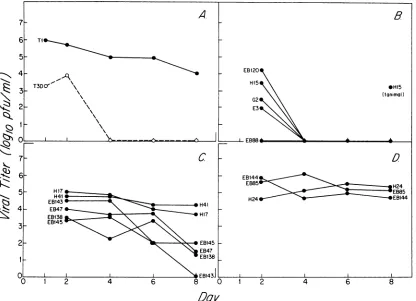
FIG. 1. (A) Recovery ofreovirus serotype 1 Lang and serotype 3Dearingin intestinal tissuefollowingperoral inoculation(105PFU) into neonatalmice. The virus was administered into the stomachs of neonatal mice following intragastric intubation. The intestines were harvested at days 1, 2, 4, 6, and 8, and the fluid contents werewashedout as describedpreviously(11). The intestinal tissueremainingafter the wash wasassayedfor the presence of virus. Since the lowest dilution thatcould be assayed was a 10-2 dilution,thelimit of detectability in this assay was 5 x 102 PFU. Each time point represents the mean of three or four samples. The standard deviation was less than 0.6log10atall time points. (B) Recovery of reassortants exhibiting an L2-S1 gene pair from serotype 3 Dearing in intestinal tissue following peroral inoculation(105PFU) of neonatal mice. The standarddeviationwas less than 1.5log10for day 2samplesand was 0 (given a limit of detection of 5 x 102PFU) for all samples on days 4, 6,and8. (C) Recovery ofreassortants exhibitinga heterologousL2-S1gene pair in intestinaltissue following peroral inoculation(105PFU) of neonatal mice. The standard deviation was less than 1.5 log10 at all time points. (D) Recovery of reassortantsexhibiting an L2-S1 gene pair from serotype 1 Lang in intestinal tissuefollowingperoralinoculation(105PFU) of neonatal mice. The standarddeviation was less than 0.5 log10 at all time points.
and serotype 3 Dearing. The reassortantsused in this study wereisolated and electropherotypedeither by D. Drayna (4) or by E. Brown and M. Nibert (2). Reassortants were
providedby M. Nibert.
Mice. Pregnant NIH Swiss mice were obtained from the
National Cancer Institute. Alladultmice werefed standard
laboratory chowandwater ad libitum. Neonatalmice were inoculated 2days afterbirth by intragastric intubation (17) and placed into cages with one mother per eight suckling mice. At specified timesafterinoculation, pairsof neonatal
mice weresacrificed by cervical dislocation. The abdomen
ofeachmouse wasopened, and theintestine wasdissected
from duodenum to anus. The intestinal contents were flushed as described previously (11). The intestinal tissue wasplaced in1mlofgelatin-salineand frozenat -70°Cprior
totitration.
Viraltiterdetermination in mouse intestines. Mouse intes-tineswerefrozenand thawed three times anddisrupted by sonication for 30 to 45 s with the microprobe of a Heat
System-UltrasonicsW225R sonicator. Serialdilutions were
made in gelatin-saline, and 100 ,ul of the
10-1
to 1O-5 dilutions was used to inoculate L-cell monolayers. Theplaque assay procedure has been described
previously
(3).Titers are expressed as
log1o
PFU per milliliter of suspen-sion.Wilcoxon ranksumdistribution test.Titers fromdays 2, 4,
6, and 8 were used to rank the viruses (parental and
reassortant) accordingtotiter: 16wasthehighestand 1 was the lowest. Ranksumsforeach gene from each parentwere
referredtoatableofcritical values (12). RESULTS
Recovery of serotype 1 Langand serotype3Dearing from intestinal tissue of neonatal mice. To reevaluate the
genetic
determinant(s) of recovery of reoviruses from intestinal
tissue, we initially used conditions that mimicked those of the study of Keroack and Fields (11) in which it was
demonstrated that the L2 gene determined transmission among littermates. Mice were
given
aperoral
inoculum of105
PFU of either serotype 1 Lang or serotype 3Dearing,
andtheir intestinal tissues were harvestedat
days
1,2, 4,
6,
and 8 after infection. The titers of virus recovered from
intestinal tissue are shown in
Fig.
1A.Serotype
3Dearing
could be recovered from intestinal tissueat
day 2,
butby
day
4 and on subsequent days, virus was not detected. In
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.106.522.69.370.2]TABLE 1. Reovirusreassortants usedtomap theability of reovirus to grow in intestinal tissue
Origin ofgenome segmentencoding':
Clone Outercapsid Core Rank
L2c Si S4 M2 Li L3 Mi S2 M3 S3
EB85 1 1 1 1 1 1 1 3 3 1 16
H24 1 1 3 1 1 1 1 1 1 1 15
Ti 1 1 1 1 1 1 1 1 1 1 12.5
EB144 1 1 1 3 1 1 1 1 3 3 12.5
H17 3 1 1 3 3 3 1 3 3 3 12.5
H41 3 1 1 1 1 3 1 3 3 3 12.5
EB143 1 3 1 1 3 1 1 1 1 1 10
EB47 3 1 1 1 1 1 1 1 1 1 9
EB138 1 3 1 3 3 1 1 3 1 1 8
EB145 3 1 3 3 3 3 3 3 1 3 7
G2 3 3 1 1 1 1 1 1 1 1 3.5
E3 3 3 3 1 3 3 3 3 3 3 3.5
EB88 3 3 3 1 3 3 3 3 3 3 3.5
EB120 3 3 1 1 3 1 1 3 1 1 3.5
H15 3 3 1 3 1 3 1 3 3 3 3.5
T3 3 3 3 3 3 3 3 3 3 3 3.5
1/3 6/10 8/8 11/5 10/6 8/8 9/7 12/4 6/10 8/8 8/8
aGenomesegmentsfrom serotype3Dearingorfrom serotype 1Langare
designated3 or 1.Thereassortants arelisted fromhightolowaccordingto
their viral titers inintestinal tissueonday4(Fig. 1).
bThe rank numberofeach reassortantisgivenin theright-handcolumn.
Tied values received the mean of the rank numbers at the positions
con-cerned. Thesignificance levelsattainedbythe rank sumforagivengene are
shown inFig.2.
c L2,a geneencoding a core protein, is here listedas an outer capsid
component simplytoemphasize its association with SI.
contrast,we recovered serotype 1 Langthrough day8after inoculation. These sampleswere also assayedforthe pres-enceof virusinintestinalwashings,asdescribedby Keroack
and Fields(11; datanotshown).Wefound that titers of virus in intestinal washes and in intestinal tissue were roughly equivalent. The 4-to
5-log1o
PFUdifferencein the recovery of serotype 1 Lang and serotype 3 Dearing from intestinal tissueindicated that this phenotypewas suitable forgenetic mapping.Recovery of reassortants from intestinaltissue.Toidentify
the genesresponsiblefor the differencebetween serotype 1 Lang and 3Dearing,wetested reassortantsfortheircapacity
to grow and survive in intestinal tissue. Because of the reportof Rubinand Fields (17) thatthe M2 gene plays a role in determining the differences in growth of reoviruses in
intestinal tissue and the report ofKeroack and Fields (11) that the L2 genome segmentis responsiblefordifferencesin
transmission ofvirus between neonatal mice, we initially assessed the independent contributions ofthe M2 and L2 genome segmentstogrowth and survival inintestinal tissue. E3(isolated byD.Drayna) and EB88(isolated by E. Brown) were independent isolates containing the M2 gene from serotype 1 Lang and all other genes, including L2, from serotype 3 Dearing (Table 1). Both of these isolates were similar to serotype 3 Dearing in the extent to which they were recovered from intestinal tissue (Fig. 1B), indicating that theM2genefrom serotype 1 Lang did not confer on the reassortants the capacity to grow and survive in intestinal
tissue. The opposite reassortant, EB144, which has an M2 gene segment from serotype 3 Dearing and an L2 gene segmentfromserotype 1Lang (Table 1), grewlike serotype 1 Lang (Fig. 1D), again indicating thatunder these
condi-tions, the M2 gene did not determine thecapacity of reovirus to grow in intestinal tissue. Thus, the use of these newer reassortantsindicated that the M2 gene of serotype 1 Lang did not confer on reassortants containing other serotype 3 genesthe capacity to grow in intestinal tissue.
Since Keroack and Fields(11)demonstrated, by using the reassortants generated from nonmutagenized stocks, that the L2 genome segment determined the transmission of reovirus, we further evaluated the role of the L2 gene in intestinal growth. To do this it was necessary to choose reassortants with an L2 genefrom oneparentandas many genes as possible from the other parent. EB47 contains an L2 gene segmentof serotype 3Dearing againstabackground of nine serotype 1 Lang genes (Table 1). EB47 was recov-ered in significantly higher titer from intestinal tissue than serotype 3 Dearing was, but it did not grow as well as serotype1 Lang,indicating that L2played somerole in the growth and survival of the virus but didnot convertthe virus from a serotype 1 Lang level ofgrowth tothat of serotype 3 Dearing(Fig. 1B). G2, however, wasidenticaltoserotype 3 Dearing in itsinabilitytoberecovered from intestinal tissue (Fig. 1B). Since G2 contains both the L2 and Si of serotype 3 Dearing against a serotype 1 background (Table 1), the
comparison of G2 with EB47 indicated that although L2 played a role in viral survival in intestinaltissue, bothL2and Siwerenecessarytoconferon a reassortanttheinabilityto survive in intestinal tissue that is characteristic of serotype 3
Dearing.
To confirm that the L2 and S1 genome segments deter-mined viralgrowth and survival in intestinal tissue and to assess whether other genes played a role, we analyzed a largercollection ofreassortants(Table 1;Fig. 1BtoD).The recoveryof thesereassortantsfrom intestinal tissue is shown inFig. 1B toD. In each case, the serotype 3Dearing L2-S1
genepair conferred onall reassortantsaninabilitytosurvive
inintestinal tissue(Fig. 1B). Reassortants withtheserotype 1Lang L2-S1 gene pair grewaswellasserotype 1Lang(Fig.
1D), whereas reassortants with a heterologous L2-Sl gene
pairingexhibited intermediate levels ofrecovery (Fig. 1C),
indicating that both the L2 and Si genes of serotype 1Lang
werenecessarytoachieveaserotype1Langlevel ofgrowth
in intestinaltissue.
Relative contributions of L2 and Si genes to intestinal growth. Reovirus genetic studies have classically yielded
data in which thereassortantsfallintotwoclasses clustered about theparentalphenotypes. Inthosestudies,in which the
association oftwo discontinuous factors are examined, we have usedchi-square analysistoconfirmthat theassociation ofagenewith aphenotype is statistically significant. Inthe presentstudy, the levelsofgrowth andsurvival inintestinal
tissue thatwereexhibitedbythereassortantsdid not fall into two classes clustered about the parental phenotypes,
but,
rather, exhibited a continuum. This distribution would best be described as discontinuous
(Ti
or T3 gene) versuscon-tinuous(rangeof growth) and is moreappropriately analyzed
by the Wilcoxon rank sumdistribution test. The viral titers inintestinal tissue for each reassortantfromdays 2, 4, 6,and 8 were used torank the reassortants from highest to lowest
(Table 1). Theonly two genes whose rank sums consistently fell outside alevel ofP > 0.02orless were L2 and
Si
(Fig. 2). It can beseen that, as predictedfrom thecurves inFig. 1A, the most statistically significant data were generated from theday 4, 6, and 8 titers. It was clear that by days 6 and8,Siwas the mostsignificant determinantof the recoveryof reovirus from intestinal tissue following peroral inoculation
(Fig. 2). Althoughgenes (M3, S3, and
Mi)
other than L2 andon November 10, 2019 by guest
http://jvi.asm.org/
Day8
Day6
Day4
Dqy2
PValue
FIG. 2. Relative contributions of the 10 reovirus segments as
estimated bytheWilcoxonrank sumdistributiontest.Theaverage
titers from days 2,4, 6,and8wereusedtorankeachreassortant. Theranksumsforeachgeneweredetermined for days2, 4, 6, and 8. When thesumof ranks fell outside the limitsatagivenPvalue, the next highest P value was examined, until the P value was
identified within whichthesumof ranks fell. The confidence limits
foreachgene aregivenalong thexaxis. Onlygeneswhich attained
Pvaluesof0.02orlessare shown.
Si appearedtoshowstatistical significanceondays2and6,
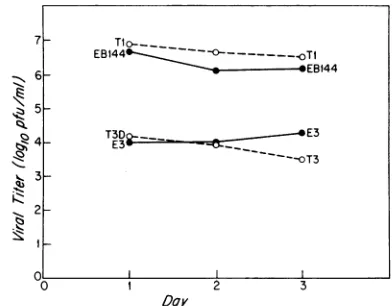
they didnotappeartoconsistently contributeto the pheno-type,and therefore their role hasnotbeenexplored further. Growth of reassortants given a high-dose inoculum. The
studyof Rubinand Fields(17) implying that M2 playsarole
in thegrowth of reovirus in intestinal tissuediffered from the present study not only in that the reassortants used were
generated from mutagenized stocks butalso in the dose of virus that was used and the times at which the intestinal tissueswere harvested. Therefore, it was possible that the
discrepancyinthegeneticassignmentin thetwostudieswas
due to the differences in the dose of virus or time of
harvesting between the studies. Therefore, werepeated the evaluationofgrowth ofreassortantsgenerated from nonmu-tagenized stocks by using doses that mimicked the previous conditions as much aspossible. Mice were given aperoral
inoculum of i07PFU, and their intestines wereharvestedat days 1, 2, and 3, as done by Rubin and Fields (17). Only reassortants whichsegregated the M2gene(Table 1, EB144 and E3) were examined. Again, as in the experiments performedwithalowdoseofvirus, itwas seenthat M2did notplayarole in determining the capacityof reoviruses to
grow in intestinal tissue (Fig. 3). Thus, when the newer
collection ofreassortants was used to study the growth of reovirusserotype1Langand serotype3 Dearinginintestinal tissue,the L2andS1 genome segments,ratherthan the M2 segment, wereresponsible for determining the capacity of thereassortants togrowtomaximal titers.
DISCUSSION
In the present studywehavefound that thedifferencein
the capacity of reovirus serotype 1 Lang and reovirus serotype 3 Dearing to be recovered from intestinal tissue following peroral inoculation was determined primarily by the S1 gene segment and, to a lesser extent, by the L2 segment. Forthesestudies,reassortantswereused thatwere
generated from crosses of nonmutagenized parental virus
stocks. Previous studiesinwhich theanalysis ofgrowthof reovirus in intestinal tissue had been performed by using reassortants generated from mutagenized
temperature-sen-I
Z
Q
(.$
0 1 2 3
[image:4.612.63.301.66.212.2]Day
FIG. 3. Growth of reovirus Ti (Lang), T3 (Dearing), and two reassortants atdays 1, 2,and3 followinginoculationwithahigher
dose (107PFU)of virustoneonatalmice.Theelectropherotypes of thereassortantsaregiveninTable 1. Procedureswere asdescribed forFig.1.Thestandarddeviationwasless than 0.5logloatall time
points.
sitiveparental stocks suggested thatthe differencebetween reovirus serotype1Lang andreovirusserotype3 Dearingis due to the M2 gene (17). We now believe that this earlier
report is incorrect, most probably as a result of mutations present in the original reassortant stocks. Although the presenceof mutations inourinitial collection ofreassortants wasnoted (3, 17),itwas notthoughttoaffecttheassignment ofviralintestinalgrowthtothe M2gene.Theearlier collec-tion of reassortants has not been used for genetic studies since 1980, and thus ourrecent assignments have all been based on reassortants generated from nonmutagenized stocks.
We would liketo note twodifferences betweenourstudy and that of Rubin and Fields other than the reassortants used. Rubinand Fields used BALB/cmice,whereaswehave
used NIH Swiss mice. In addition, in the previous study reassortants were purified by CsCl gradient centrifugation priorto beinginoculated intomice, whereas in the present study we inoculated stocks consisting of cell lysates. It is unlikelythat thegenetic determinant(s)ofreovirusgrowthin intestinal tissue isspecifictoaparticularmousestrain,since other mapping experiments that were done with BALB/c mice(24)havebeenreproduciblewith NIHSwiss mice(10). Moreover, in recentbiochemical studies (D. K. Bodkin et al., manuscript in preparation), we did not detect any
difference in therecoveryof virus from the intestine follow-ing inoculation into mice either as purified virus or as cell lysates.
Thus, itwouldappearthat theprior mappingofintestinal growthtotheM2generepresentsaninstance ofanincorrect genetic assignment as a result of the use of mutagenized stocks forpreparing viral reassortants. Earlierexperiments with reassortants generated from mutagenized stocks mappedthe inactivation of reovirusinvitrobychymotrypsin to M2 (17). This result was subsequently confirmed in a
genetic analysis oftranscriptional activation (which corre-lates with loss of infectivity) by using the reassortants generatedfromnonmutagenizedstocks(5).Otherstudiesby Hrdyetal. (8) mappingdifferences in neurovirulencewithin a serotype to M2 involved reassortants generated from
nonmutagenizedstocks.Asnoted in theintroduction,earlier
studieswith reassortantsgeneratedfrommutagenizedstocks
!- L2 Si
M3
L2 Si
53
- L2 S1i
,-_ L2 Si
002 001 0.005 0.002 0.001 <0001
EB144 =o= .Tl
6 EB144
5-4_ T3D2__ t E3
4~ ~
E3*E----O3
~- ~ ~3
2
I)
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.338.534.67.220.2]involving themapping of propertiestotheS1 genehave been confirmed in a number ofsubsequent studies.
Thesefindings underscore potential problems arisingfrom thehigh frequency of mutation of RNA virusesandillustrate
the need to minimize the generation of mutations in viral stocks that might alter biologic properties ofthe viruses in question. The interpretation of data from experiments per-formed with reassortants generated from mutagenized stocks may bedifficultbecause thereassortantsmaycontain mutations not present in the original, nonmutagenized vi-ruses.Using bunyaviruses, Rozhonetal.(16) also found that temperature-sensitive stocks generated by chemical
muta-genesis contain non-temperature-sensitive mutations which
influence viral virulence. Further, given the high mutation rate ofRNA versus DNAviruses (7), theproblemof silent mutations which influence biologic properties may be
par-ticularly acuteforRNA viruses. The fact that the reported
cases of non-temperature-sensitive mutations which influ-ence viral virulence involve RNA as opposed to DNA viruses is consistent with this hypothesis.
The data presented here, combined with other studies
performedinthislaboratory, implythatit is
relatively
late inthe infectionthat the L2 and Si genesdetermine the differ-ences in the recovery of reovirus serotype 1 Lang and serotype 3 Dearing from intestinal tissue. Upon entry into theintestinal lumen, the intact virusoftheinputinoculum is converted to intermediate subviral particles identical to those generated invitro with chymotrypsin (D. K. Bodkin,
M. Nibert, and B. N. Fields, unpublished data). In these
particles the
u3
protein, (the S4 gene product) has been removed and theplc
protein, (the M2 gene product) hasbeen cleaved. However, this early cleavage eventdoes not determine the differential growthof serotypes 1 Langand 3
Dearing, since the genetic analysis presented in this study indicates that the differences in recovery of the virus after
day 4 between reovirus serotype 1 Lang and serotype 3
Dearingaregovernedbythe L2 and
Si
genesrather thanbythe S4 and M2 genes.
Howmightthe L2and
Si
genesbeoperatingtodetermine thedifferencein survival andpossibly growthof serotypes 1 Lang and 3 Dearing?Onepossibility is that it iscleavage ofthe viral crlprotein (the
Si
geneproduct) that accountsfor the difference between serotype 1 and serotype 3. Earlystudies implied that the serotype 3 Dearing al may be cleaved in vitro (9, 20). Nibert (unpublished data) has confirmed that the T3Dearing
a-l
proteinis cleaved in vitro by chymotrypsin, whereas Sturzenbeckeretal. (21) demon-strated that the serotype 1 Lang hemagglutinin is more resistant to proteolytic digestion. Given the role of theX2
protein (the L2 gene product) as an anchor to the viral attachment endoftheclprotein (1, 6), it is possible that the L2 and
Si
genetic compounds we have identified involveaccessibility to a cleavage site on the hemagglutinin to intestinal or cellular proteases. Evaluation of the process by which serotype 3 Dearing loses infectivity in the intestine mayfurther elucidate the interaction ofX2and crl inreovirus andprovide insightsinto factorsdetermining the capacity of reovirus to grow and survive in intestinal tissues.
ACKNOWLEDGMENTS
MaxNibertis acknowledged forproviding reassortants and for extremely useful discussion.
This work was supported by Public Health Service program project grant 2 P50 Ns11998-07 from the National Institute of Neurological and Communicative Disorders and Stroke and by
Public HealthService research grant 5 R37 A113178-12 from the National Institute of Allergy and Infectious Diseases. Dinah K. Bodkinwassupported by postdoctoralfellowshipsfrom the Amer-ican Cancer Society and the National Institute of Allergy and Infectious Diseases.
LITERATURECITED
1. Bassel-Duby, R., A. Jayasuriya, D. Chatterjee, N. Sonenberg,
J. C.Maizel, Jr.,andB.N. Fields. 1985.Sequenceof reovirus haemagglutinin predicts a coiled-coil structure. Nature (London)315:421-423.
2. Brown,E.G.,M.L.Nibert,andB.N. Fields.1983. The L2 gene of reovirusserotype 3controlsthecapacitytointerfere,
accu-mulate deletions, and establish persistent infection, p. 275-287. In R. W. Compans and D. H. L. Bishop (ed.), Double-stranded RNA viruses. Elsevier SciencePublishing,Inc.,New York.
3. Cross, R. K., and B. N. Fields. 1976. Use of an aberrant polypeptide in 3factorcrosses: furtherevidence for
indepen-dent reassortant as the mechanism of recombination between
temperature-sensitive mutants of reovirus type 3.
Virology
74:345-362.
4. Drayna, D., and B. N. Fields. 1982. Genetic studies on the mechanism of chemicalandphysicalinactivation ofreovirus.J. Gen.Virol. 63:149-159.
5. Drayna,D., and B. N. Fields. 1982.Activation and characteri-zation ofthe reovirustranscriptase: genetic analysis. J. Virol. 41:110-118.
6. Furlong,D. B., M. L.Nibert, and B. N.Fields. 1988. Sigma 1 protein ofmammalian reoviruses extends from the surfacesof viralparticles.J. Virol. 62:246-256.
7. Holland, J., K. Spindler, F. Horodyski, E. Brabau, S. Nichol,
and S. VandePol. 1982. Rapid evolution of RNA genomes. Science215:1577-1585.
8. Hrdy, D. B.,D. H. Rubin, and B. N. Fields. 1982. Molecular basis of reovirus neurovirulence: role of theM2 genein aviru-lence. Proc. Natl. Acad. Sci. USA79:1298-1302.
9. Joklik, W. K. 1972. Studies onthe effectofchymotrypsin on
reovirions. Virology49:700-715.
10. Kaye, K.M., D. R.Spriggs, R. Bassel-Duby,B.N.Fields,and K.L.Tyler. 1986. Geneticbasis for altered pathogenesisofan
immune-selectedantigenicvariant ofreovirusserotype3
(Dear-ing). J. Virol.59:90-97.
11. Keroack, M., and B. N. Fields. 1986. Viral shedding and transmission between hosts determined by reovirus L2 gene. Science232:1635-1638.
12. Lentner,C.(ed.).1982.Geigyscientifictables,vol.2. Introduc-tiontostatistics,p.156-162.Ciba-Geigy Corp.,WestCaldwell,
N.J.
13. Mustoe,T. A., R. F.Ramig, A. H. Sharpe, and B. N. Fields. 1978. Ageneticmapof reovirus. III.Assignmentofthe double-strandedRNA positivemutantgroups A, B,and Gtogenome segments. Virology85:545-556.
14. Ramig, R. F., R. S. Cross, and B. N. Fields. 1977. Genome RNAs and polypeptides of reovirus serotypes 1, 2, and 3. J. Virol. 22:726-733.
15. Ramig, R. F., T. A. Mustoe,A. H. Sharpe, and B. N. Fields. 1978.Ageneticmap of reovirus. II.Assignmentof the double-strandedRNAnegative mutantgroups C, D,andE togenome segments. Virology85:531-544.
16. Rozhon,E. J.,R. Gensemer,R.E.Shope, and D.H. L.Bishop. 1981. Attenuation ofvirulence ofabunyavirus involvinganL RNA defect and isolation of LAC/SSH/LAC and LAC/SSH/ SSHreassortants.Virology111:125-138.
17. Rubin, D.,andB.N. Fields. 1980. Molecularbasis of reovirus virulence:roleofthe M2 gene. J. Exp. Med. 152:853-868. 18. Sharpe, A. H., and B. N. Fields. 1982. Reovirus inhibition of
cellular RNA and protein synthesis: role of the S4 gene. Virology122:381-391.
19. Sharpe, A.H., R. F. Ramig,T. A. Mustoe, and B. N. Fields. 1978. A genetic map of reovirus. 1. Correlation of genome RNAsbetween serotypes1, 2,and 3.Virology84:63-74.
on November 10, 2019 by guest
http://jvi.asm.org/
20. Shatkin, A. J., and A. J. LaFiandra. 1972. Transcription by infectious subviral particles of reovirus. J. Virol. 10:698-706.
21. Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987.Intracellular digestion of reovirus particles requiresalow
pHand isanessentialstepin the viralinfectious cycle.J.Virol. 61:2351-2361.
22. Tyler, K.L., and B. N.Fields. 1985. Reovirus and its replication, p.823-862. In B. N. Fields(ed.), Virology. Raven Press, New York.
23. Tyler, K. L., D. A. McPhee, and B. N. Fields. 1986. Distinct
pathways of viral spread determined by the reovirus Si gene
segment. Science233:770-774.
24. Weiner,H. L., D. Drayna,D.R.Averill,Jr., and B. N.Fields. 1977. Molecular basis ofreovirus virulence: role of the Si gene. Proc. Natl.Acad. Sci. USA 74:5744-5748.
25. Weiner, H. L., and B.N.Fields.1977. Neutralizationof reovi-rus:thegeneresponsible for theneutralization antigen. J. Exp. Med. 146:1305-1311.
26. Weiner, H. L., R. F. Ramig, T. A. Mustoe, and B. N. Fields. 1978. Identification of thegenecoding for the hemagglutininof reovirus. Virology 86:581-584.