0022-538X/82/030940-07$02.00/0
Molecular
Cloning
and Physical Mapping of Restriction
Endonuclease Fragments of Autographa californica Nuclear
Polyhedrosis Virus
DNA
MARK A.COCHRAN, ERIC B. CARSTENS, BRYAN T. EATON, AND PETER FAULKNER* Departmentof Microbiology andImmunology, Queen's University, Kingston, Ontario K7L 3N6, Canada
Received 4September1981/Accepted 2 November 1981
A
restriction fragment library containing Autographa
californica nuclear
poly-hedrosis virus
(AcNPV) DNA was constructed by using the pBR322 plasmid as a
vector. The
library, which is representative of more than
95% of the viral genome,
consists
of
2
of
the 7 BamHI
fragments,
12of
the 24
HindIII
fragments, and 23 of
the 24 EcoRI fragments. The cloned
fragments
werecharacterized
and used to
generate
physical maps
of
the
genome
by
hybridizing
nick-translated
recombinant
plasmid
to
Southern
blots of AcNPV DNA
digested with SmaI, BamHI,
XhoI,
PstI,
HindIII, and EcoRI
restriction
endonucleases. This information
wasused
todefine
our
strain
of AcNPV (HR3) with respect
toother strains for which
physical
maps have
been
previously
published.
The
hybridization
data also indicate that
reiteration of
DNA
sequences occurs at the HindIII-L and
-Q regions
of
the
genome.
The nuclear
polyhedrosis
virusof
thealfalfa
looper, Autographa californica (AcNPV), is a
prototype for molecular studies of insect
baculo-viruses. The replicative cycle of the virus in
permissive
insect cell lines is
complex but can be
divided
into
two
phases
(4, 6).
First,
singly
enveloped
nucleocapsids,
termed
nonoccluded
viruses (NOV), are budded from the cell
mem-brane
during
the
early
stages
of
infection.
Sec-ond,
later in the
infectious
cycle,
production
of
NOV is curtailed and occlusion
bodies (OB),
which
consist
of
enveloped bundles
of
nucleo-capsids
lying
within
aparacrystalline protein
matrix, form in the cell nuclei. The genome
of
AcNPV
consists
of
a
double-stranded,
super-helical,
circular DNA molecule of
approximate-ly
128
kilobase
pairs
which has been
physically
mapped for
several
restriction
enzyme
sites
(9,
12, 17). Wild isolates of AcNPV
derived frominsects can be
separated by plaque purification
into variant strains
having minor
alterations in DNArestriction
patterns (8, 11, 15).
As
a
prelude
togenerating
atranscription map
of the
AcNPVgenome,
wecloned
almost theentire
genome of AcNPV
asBamHI,
EcoRI,
andHindIII
fragments
in
theplasmid pBR322.
Therecombinant
plasmids
werecharacterized
andauthenticated
by restriction enzyme
digestion
and
by
hybridization
torestricted AcNPV
DNA.These data also
served
toidentify
ourwild-type
strain (HR3)
asbeing
quite
similar
to the E2strain of Smith and Summers
(12)
and the
Li
strain of Miller and
Dawes(9)
by
confirming
their
physical
maps for
most of theSmaI,
BamHI,
XhoI, and EcoRI
restriction
fragments.
Furthermore,
complete physical maps
for therestriction enzymes HindIII and
PstI arepre-sented.
MATERIALS
AND
METHODS
Purificationofvirusandviral DNA. A plaque-puri-fied MP (many OB/cell) strain of AcNPV, designated here asHR3, which grew at both 28 and 33°C (1) was propagated on monolayers of Spodopterafrugiperda cells at28°C(16). Cells were infected at a multiplicity of 5 to 10 PFU per cell, and extracellular NOV and OB were isolatedas described by Dobos andCochran (3). Viral DNA was isolated from gradient-purified NOV as described by Tjia et al. (15). Briefly, the NOV suspension(2 to 5 mgof protein/ml in 0.1 M Tris-0.01 MEDTA, pH 7.5),wasincubatedat37°C for t h in 1% (wt/wt) sodium dodecyl sulfate (SDS), 1% (wt/wt) sodiumlaurylsarcosinate, and 500 SLgof proteinase K per ml (Merck &Co., Inc., Rahway, N.J.). Subse-quently, pronase B was added to 500 ,Lg/ml, and incubation was continued for 1 h. The solution was extracted three times withbuffer-saturatedphenol and once with chloroform-isoamyl alcohol (24:1). The aqueousphasewasdialyzed extensively against0.1x
SSC (lx SSC is 0.015 Msodium citrate and 0.15 M sodiumchloride, pH 7.5). Toisolate DNA fromOB, gradient-purified OB (5 to 10 mg of protein/ml) were suspendedin 0.1 MNa2CO3, 0.17MNaCl, and0.01 M EDTA, pH 10.9, and incubatedat37°C for15minto liberate the occluded virions.DNAwaspurified from this solutionasdescribed above.
PurificationofplasmidDNA. The bacterium HB101 (rk- mk -; obtained from B. N. White, Department of Biology, Queen's University),containing the plasmid pBR322orrecombinantplasmids,wasgrownin either 5to10ml(miniprep)or 1liter of Luria broth contain-940
on November 10, 2019 by guest
http://jvi.asm.org/
BACULOVIRUS ing20pg ofthe antibioticstetracyclineandampicillin
per ml, alone or together. Cell suspensions were grown at37°Cin ashakerbath,andtheplasmidswere amplified during logarithmic growthbytheaddition of chloramphenicol (200,ug/ml) (2). The cellswere har-vested, and aclearedlysatewas prepared as described byGuerryet al. (7)with somemodifications. Briefly, the pellets from miniprep and 1-liter cultures were suspended in 0.5 and 20 ml, respectively, of cold STEN buffer(25% sucrose, 50 mMTris, pH 8.0, 20 mMEDTA,and 100 mMNaCl).After addition offresh lysozyme (SigmaChemicalCo.,St.Louis, Mo.)to1.5 mg/ml,thesuspensionwasbroughtto1%(wt/vol)SDS and 1 M NaCl. Lysates from 1-liter cultures were incubated on ice overnight and then centrifuged at 17,000 x gfor 45 min at4°C.Miniprepswerelefton icefor45minandcentrifugedin aBeckmanmicrofuge (15,000 x g) at 4°C. The supernatant from these centrifugationswasdesignatedthe clearedlysate.
To purify candidate recombinant plasmids from minipreps, 0.5-ml volumes of cleared lysates were extracted sequentially with water-saturated phenol and with chloroform in Eppendorftubes. The DNA was then precipitated with 2 volumes of ethanol at -70°C for 30 min, and theprecipitate (5to 10 ,ug of plasmid DNA) was dissolved in 50 ,ul of TE buffer(10 mMTris-1 mMEDTA, pH 7.5).
Superhelical plasmid DNA was purified from the cleared lysates of 1-liter cultures, using the acid-phenol extraction method of Zasloff et al. (19) as described by Gordonetal. (5).
Construction ofrecombinant plasmids.Inthree sepa-rate experiments, AcNPV and pBR322 DNAs were each digested to completion with either BamHI, HindIII,orEcoRI.Eachsamplewas thenchloroform extracted, ethanolprecipitated,dried,resuspendedin TE buffer, and heated to 65°C for 5 min. A 50-,u ligation reaction mixture contained
0.5-p.g
ofdigested pBR322,1.0 to 3.5p.g
ofdigestedviral DNA, and 0.1 Uof T4 DNAligase(Bethesda ResearchLaboratories, Rockville,Md.) in50mMTris-hydrochloride, pH 7.6, 10 mMMgCl2,
0.1 mMEDTA, 1 mMdithiothreitol, and 1 mM ATP (J. Hassel, personalcommunication). Theligationreactionoccurredduringan18-h incuba-tion at 12°C for BamHI andHindIII ligations and at 0°CforEcoRI ligations.Competent HB101 cells were prepared as follows (B. White, personal communication). Cells were grown in 100 ml of Luria broth to anabsorbancy at 260 nmof 0.6, chilledonice,andcentrifuged at 2,500x g for 5 min at4°C. Thepellet wassuspendedin25 ml of cold 10 mM piperazine-N,N'-bis(2-ethanesulfonic acid)(PIPES), pH 6.8-10 mMRbCl, pelleted as be-fore,andsuspendedin 10ml of cold 10 mM PIPES, pH 6.8-10 mM RbCI-75 mM CaC12. After incubationat 0°Cfor30min, thecellswere pelleted again andgently suspended in 5 ml of ice-cold 10 mM PIPES, pH 6.8-10mM
RbCl-75
mMCaCl2-15%
glycerol. The cells were divided into equalportions, frozen in a dry ice-isopropanol bath, and stored at -70°C. Competent cells (200P,u)
were transformed by addition of the ligationmixture (100p.l)
containing 100 mMNaCl.The mixturewasmaintained on ice for 20min,incubated at 25°C for 10 min, and, aftersupplementationwith 1 ml ofgrowthmedium, incubatedfor 40minat 37°C.Cells werethenplated on medium containing the appropri-ateantibiotics. Under theseconditions,theefficiencyoftransformationwas typically about 2 x 106
trans-formants/,ug
ofuncutpBR322.
Ampicillin-resistant bacteria were selected and screened for susceptibility to tetracycline in the BamHIandHindllI ligation experiments. These
Apr
Tcs clones and the
Apr
Tcr clones from theEcoRI
ligation experimentswerescreenedby colony
hybrid-ization(14),using nick-translatedAcNPV DNAastheprobe.
Restrctn endonuceases, gel electrophoresis, blot-ting, hybridization, and nick translation. The SmaI, BamHI, XhoI, PstI, HindIII, and EcoRI restriction endonucleases werepurchased from eitherBethesda ResearchLaboratoriesorBoehringer Mannheim
(Dor-val, Quebec). The conditions forrestriction ofDNA were asdescribed in theprotocols suppliedwith the restrictionenzyme.
RestrictionfragmentsofAcNPV DNA and
plasmid
DNA wereseparatedby electrophoresisin0.8% agar-ose gels in 100 mMTris-hydrochloride, pH 9.0, 7.7 mM H3BO3, 2.5 mMEDTA, and 0.5 ,ugofethidium bromideperml at 1.5 V/cm for 16 h onhorizontalslab gels.DNAfragmentsusedasstandards for size deter-minations were the HindIII fragments of lambda DNA (Bethesda Research Laboratories), the HaeIlI frag-mentsof40OX174
(Bethesda ResearchLaboratories),
andtheHpaIIfragments
ofpBR322.After electrophoresis, the restricted DNA was transferredontonitrocellulose filters, using the South-erntechnique (13)aspreviously described (15).Viral sequences were detected by hybridization of 32p-labeledAcNPVDNA orrecombinant plasmid DNA to the Southern blots. The conditions of hybridization were as described byWahl et al. (18). Occasionally hybridizations were performed under more stringent conditions at 650C in 4x SSC, 0.1% SDS, 50 ,ug of sonicated anddenatured salmon sperm DNA per ml, plusDenhardt solution (Ficoll, polyvinylpyrrolidone, andbovine serum albumin; 2% each). Under eitherof these conditions, the amount of DNA used in thegel was 0.025p.g per well. DNAwas labeled with 32P by nick translation, using the method of Rigby et al. (10) asdescribedbyTjia et al. (15).
RESULTS
Restriction endonuclease
fragment
patterns of HR3 AcNPV DNA. The fragment patterns ofSmaI,
BamHI, XhoI, PstI,HindIll,
andEcoRI
are shown in Fig. 1. There were no
detectable
differences
between the restriction patterns of AcNPV DNA purified from OB or NOV(data
not
shown).
The DNA used in these studieswas from both sources. Individual fragments were assigned analphabeticaldesignation
onthebasis of size: the largest fragment for each enzyme was referred to as A, thesecond largest
was referred to as B, etc. The sizes of the AcNPV DNA fragments were estimated by comparing their mobilities in agarosegels
with standard fragments of known numbers of bases (Table1).
The sum of the sizes of the fragments
generated
by
the enzymes listed in Table 1 indicated that the AcNPV genome contained about 128 kilo-basepairs. TheXhoI-N and EcoRI-Xfragments
VOL. 41,1982 941
on November 10, 2019 by guest
http://jvi.asm.org/
Srr o'
APM_
B i C. f
BclrrHIP
A
E-G-.
; I
f..s 1. E FCI
L<
L
I
Psi ino
.4 Wm
A-B :,
I'). r. E
Ml
F GF
~i
H-K L
N
D)
K
PC
V w
P..A
r--r
.N
L.
PA
r.1
i
*-c tC!
FIG. 1. Agarose gel electrophoresis of AcNPV DNAafter digestion with SmaI, BamHI, XhoI,PstI, HindlIl, andEcoRI restrictionendonucleases. Condi-tions ofelectrophoresiswerethose described in
Mate-rials and Methods.
are in
addition
tothose
reported by Smith and
Summers
(12), and the HindIII-V, -W, and
-Xfragments
arein addition
tothose
reported by
Miller
and
Dawes(9).
Differences between
ourHR3strain,
the E2variant of
Smith and Summers (12), and the Listrain
of Miller and Dawes
(9)wereevident
uponHindIlI digestion (Fig.
2).Digestion with
SmaI,BamHI,
XhoI,
EcoRI,and
PstIshowed
identical
patternsamongthe three
strains,
exceptthatthe
PstI-J fragments of E2 and Li
were 200base
pairs (bp)
smaller than thatof
HR3. The soledifference between HR3 and E2
wasthat the
HindIII-L' fragment
of
E2 wasabout
200bp
smaller than
that of HR3 (Fig. 2). Differences
between
HR3
and Li included:
anextraHindIII
site in the HindIII-A
fragment generating the
HindIII-B' and -H' fragments of
Li;and
anLiHindIII-L'
that
was200
bp smaller than the HR3
HindIII-L
fragment.
Construction
ofrecombinant
plasmid
library.To
obtain
acollection of plasmids which
con-tained
sequencesfrom the viral
genome,total
restriction digests of AcNPV DNA
wereligated
to
pBR322 and used
totransform Escherichia
coli strain
HB101. DNA from three
setsof
digestions
wasused:
(i) AcNPV and
pBR322
[image:3.491.51.242.55.285.2]DNAs
cleaved
withBamHI; (ii) AcNPV and
pBR322 DNAs cleaved with HindIII; and
(iii)
AcNPV and pBR322 DNAs cleaved
withEcoRI.
After
transformation of the
E.coli
strain HB101,
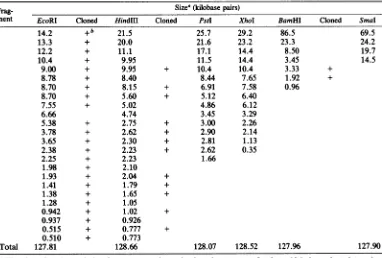
TABLE 1. Sizes of AcNPV DNArestriction fragments and enumeration of clonesFrag- Size0 (kilobase pairs)
ment EcoRI Cloned HindIII Cloned Pstl XhoI BamHI Cloned
SmaI
A 14.2 +b 21.5 25.7 29.2 86.5 69.5
B 13.3 + 20.0 21.6 23.2 23.3 24.2
C 12.2 + 11.1 17.1 14.4 8.50 19.7
D 10.4 + 9.95 11.5 14.4 3.45 14.5
E 9.00 + 9.95 + 10.4 10.4 3.33 +
F 8.78 + 8.40 8.44 7.65 1.92 +
G 8.70 + 8.15 + 6.91 7.58 0.96
H 8.70 + 5.60 + 5.12 6.40
I 7.55 + 5.02 4.86 6.12
J 6.66 4.74 3.45 3.29
K 5.38 + 2.75 + 3.00 2.26
L 3.78 + 2.62 + 2.90 2.14
M 3.65 + 2.30 + 2.81 1.13
N 2.38 + 2.23 + 2.62 0.35
0 2.25 + 2.23 1.66
P 1.98 + 2.10
Q 1.93 + 2.04 +
R 1.41 + 1.79 +
S 1.38 + 1.65 +
T 1.28 + 1.05
U 0.942 + 1.02 +
V 0.937 + 0.926
W 0.515 + 0.777 +
X 0.510 + 0.773
Total 127.81 128.66 128.07 128.52 127.96 127.90
aThesize of each restrictionfragmentwas
determined
astheaverage ofatleast 10independent
determina-tions,usingHindIll fragments of lambdaDNA, HaeIII fragmentsof (X174 DNA, andHpaII fragments of pBR322asstandards. Sizes offragments greater than 15 kilobaseswerecalculated from the sizes of restriction fragmentsgeneratedafterEcoRIdigestion.
b+, Cloned into
pBR322.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.491.63.445.370.628.2]HR3 En LiT
AB
a
FIG. 2. Agarose gel electrophoresis of HR3, E2 (11), andLI (9) AcNPV DNAdigestedwithHindIII restriction endonuclease. Fragments which differed in electrophoreticmobilitywith respecttoHR3are(i) L' of E2 andLi and(ii)B' and H'ofLi.
bacterial
clones
werescreened for the
appropri-ate antibiotic
phenotype, i.e., Aps Tcr
pheno-type
for
BamHI
ligations
and
Apr Tcs
phenotype
for
HindIII
ligations. Clones
from the EcoRI
ligation experiments
were
first selected for the
Apr
Tcr
phenotype
and then screened
along
withthe clones from the other
experiments
by colony
hybridization, using
nick-translated
32P-labeled
AcNPV DNA as
the probe. Clones
which gave a
positive hybridization signal
werethen
ampli-fied,
and recombinant
plasmid
DNA was
puri-fied. The DNA from these
preparations
was
digested
with
the
appropriate enzyme in order to
cut out
the inserted viral DNA and
analyzed by
gel electrophoresis
along with
similarly
digested
AcNPV DNA.
Analysis of
the
recombinant
plas-mids
showed that all
of
the 24 EcoRI
fragments
except
J,
12
of
the 24
HindIII
fragments,
and 2
of
the 7 BamHI
fragments
were
cloned
(Table 1).
Figure
3
shows examples of
recombinant
plas-mids
containing
AcNPV DNA
EcoRI
fragments.
Some
of
the recombinant plasmids
contained
more
than one
inserted viral
fragment,
e.g., the
plasmid
pAcEcoCX18 contained the EcoRI
frag-ments
C and X.
Confirmation of AcNPV DNA
sequences
inrecombinant
plasmids. AcNPV DNA sequences
in
recombinant
plasmids were identified and
characterized
by
thefollowing techniques:
colo-ny
hybridization; electrophoretic mobility of
cloned
fragments
with
respect to appropriately
digested
AcNPV DNA (as in Fig. 3); digestion
with another restriction enzyme; and
hybridiza-tion of
32P-labeled
recombinant
plasmid DNA to
Southern
blots of AcNPV digested with several
enzymes (as in
Fig. 4). The electrophoretic
MIU. 3.
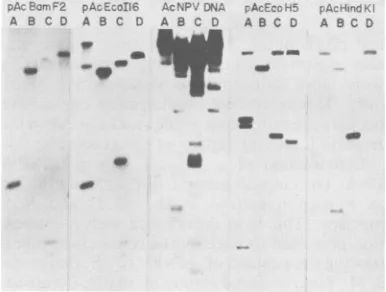
examples
o0recomoinant
plasmius
con-taining AcNPV DNA EcoRIfragments. Plasmid DNA prepared byacid-phenol extractionwasdigestedwith EcoRI and subjected to electrophoresis on a 0.8% agarose gel. Lane 6 contains fragments A to K of EcoRIdigested AcNPV DNA. All other lanes contain digests of plasmids containing AcNPV DNA frag-ments asfollows: (1)pAcEcoA14, EcoRI-A; (2) pAcE-coB20, EcoRI-B; (3)pAcEcoCX18,EcoRI-C; (4) pA-cEcoD19, EcoRI-D; (5) pAcEcoE7, EcoRI-E; (7) pAcEcoF22, EcoRI-F; (8) pAcEcoGX2, EcoRI-G; (9) pAcEcoH5,EcoRI-H; (10) pAcEcoI16,EcoRI-I;(11) pAcEcoKRW12, EcoRI-K. The bandcommon toall thedigested plasmidsis linearpBR322.Recombinant plasmid nomenclature is as described in Table 2.
mobility of a cloned fragment along with
thecolony
hybridization
datawasusually enough
toidentify it as
aspecific viral fragment. However,
restriction digestion and hybridization data
weregenerally obtained for unambiguous
identifica-tion and were critical in the construcidentifica-tion of
thephysical map. Table 2 summarizes the results of
hybridizations of 32P-labeled recombinant
plas-mids
to
Southern blots of AcNPV
DNAdigests.
Some of the
clones, in addition to hybridizing to
tAcF.rrm -2 pA---oIIE
A B CD A B C D
_ __
_
AcNPvDNA
A B C D
m
AcE~?5
AB C D
- ~O
fAcBCII
A B C D
FIG. 4. Hybridization of
32P-labeled
recombinant plasmidstoSouthern blots of AcNPV DNA digested withBamHI(A), EcoRI (B),HindIII (C), andPstI(D) restriction endonucleases. The32p
probe is indicated atthe topof each panel. Conditions of electrophoresis, blottransfer, and hybridization were as described in Materials and Methods. The results to this figure are summarized in Table 2.VOL.41,
on November 10, 2019 by guest
http://jvi.asm.org/
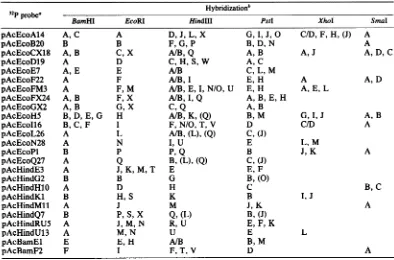
[image:4.491.114.176.71.235.2] [image:4.491.258.444.72.175.2] [image:4.491.254.447.446.592.2]TABLE 2. Summary of the results ofhybridizations of 32P-labeled recombinant plasmids to AcNPV DNA digests on nitrocellulose filters
Hybridization'
BamHI
EcoRIHindIII
PstI
XhoISmaI
pAcEcoA14 A,C A D, J, L, X G,I, J, O C/D, F, H, (J) A
pAcEcoB20 B B F,G, P B, D, N A
pAcEcoCX18 A, B C, X A/B,Q A, B A,J A, D,C
pAcEcoD19 A D C, H, S, W A,C
pAcEcoE7 A, E E A/B C, L, M
pAcEcoF22 A F A/B, I E, H A A, D
pAcEcoFM3 A F, M A/B, E, I,N/O, U E, H A,E, L
pAcEcoFX24 A, B F, X A/B,I, Q A, B, E, H
pAcEcoGX2 A, B G, X C, Q A, B
pAcEcoH5 B, D, E,G H A/B, K, (Q) B, M G,I,J A, B
pAcEcoI16 B,C, F I F,N/O, T, V D C/D A
pAcEcoL26 A L A/B,(L), (Q) C,(J)
pAcEcoN28 A N I, U E L, M
pAcEcoPl B P P,Q B J, K A
pAcEcoQ27 A Q B,(L), (Q) C, (J)
pAcHindE3 A J, K, M, T E E, F
pAcHindG2 B B G B,(0)
pAcHindH1O A D H C B,C
pAcHindKl B H,S K B I,J
pAcHindMll A J M J, K A
pAcHindQ7 B P, S, X Q, (L) B, (J)
pAcHindRU5 A J,M, N R, U E, F, K
pAcHindU13 A M, N U E L
pAcBamEl E E, H A/B B, M
pAcBamF2 F I F, T, V D A
aCloneswerenamedaccordingtothefragmentstheywerefoundtocontain. Forexample,aclone containing
the EcoRI-A fragment was named pAcEcoA14: p for plasmid, Ac for AcNPV DNA, Eco for the EcoRI restriction enzyme, A for theEcoRI-Afragment, and14for the isolate number.
bSome clones in additiontohybridizingtotheexpectedregionsof the genomehybridizedtootherregions. Hybridizationtoadditionalregionsis indicatedby parentheses. A slant line indicates that itwas notpossibleto
distinguish betweentwofragments because ofcomigrationin agarosegels.
the
expected regions
of the
genome,hybridized
to
other
regions. Most frequently,
additionalhybridization
was tothe HindIII-L
and-Q
re-gions with
plasmids
pAcEcoL26,
pAcEcoQ27,
and
pAcHindQ7. Binding
tothese
regions
wasalso observed when hybridization conditions
were more
stringent (see
Materials and
Meth-ods).
This
additional
hybridization
could have
been due
toreiteration of
sequencesbetween the
HindIII-L and
-Q regions
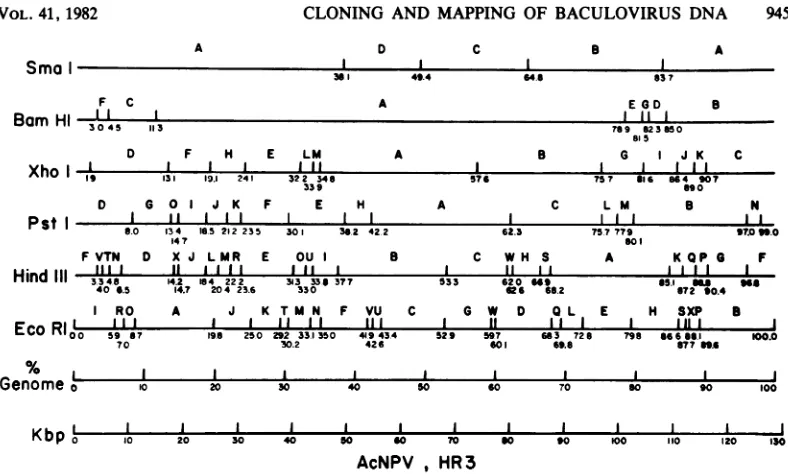
of the genome.Construction of a physical map of AcNPV-DNA.
The
circular
genomeis
described in
Fig.
5 as a maplinearized
at the EcoB and EcoIjunction.
This is in
accordance with
aconven-tion
proposed
tobaculovirus
researchersinves-tigating
the
genomeof AcNPV
(G.
E. Smith andJ.
M.Vlak,
personal
communication).
Comigrat-ing restriction fragments
aredesignated
as asingle letter in
alphabetical
order
starting
fromthe
zeropoint
(e.g., Hind-N
andHind-O).
Posi-tions
onthis
map aredefined
asthepercentageof the total
length
of the
genome and werecalculated from
the size of eachfragment
(Table
1)
together with the
setsof
datarequired
toorder
thefragments.
The
hybridizations detailed in Table
2usually
gave
enough information
todeduce the
order
and
approximate position of each fragment. The
physical
mapshown in
Fig.
5 wasderived from
these
hybridizations
togetherwith the following
supportive data: (i) the calculated sizes of the
restriction fragments (Table
1);(ii) reciprocal
double
digestions of viral
DNA;(iii) digestions
of cloned
DNAfragments
with other
enzymes;(iv)
digestions of isolated SmaI DNA fragments;
and (v)partial digestion with
HindIIl
of clonedand isolated
DNAfragments.
Thefollowing
ex-ample illustrates how
theorder of
theHindlIl
fragments
F, V, T, and N wasdeduced.
Therecombinant
plasmid pAcEcoI16
hybridized
to(i) BamHI-B, -C, and
-F;(ii) EcoRI-I; (iii)
HindIII-F,
-N, -T, and
-V; and
(iv) PstI-D (see
Fig.
4).
Digestion of pAcEcoI16 with HindIII
resulted in
intact HindIII-V and -T fragments.
The
clone
pAcBamF2hybridized
to(i)
BamHI-F;
(ii)EcoRI-I;
(iii) HindIII-F, -T, and
-V;and
(iv) PstI-D (Fig. 4). Digestion of pAcBamF2
with
HindIll
resultedin
anintact HindIII-V
fragment.
Theseresults, together with
thecalcu-lated
sizes
of therestriction
fragments wereon November 10, 2019 by guest
http://jvi.asm.org/
[image:5.491.55.442.93.350.2]CLONING AND MAPPING OF BACULOVIRUS DNA
A
Sma
ID
361 49.4
F C
Bam HI
,II
Xho
I Pst IHind
IIIC
64.
D F H E LM A 8 G I J K C
II I I IIf I III
19 131 19.1 241 32 2 34 8 576 757 816 664 907
339 890
D G 0 I J K F E H A C L M B N
I II I II I I I III II
6.0 134 165. 212235 301 36.2 42.2 62.3 75779 9706W.0
14 7 So
FVTN D X J LMR E OU I a C WH S A KQP G F
III I If I I I I II I I It II I I
3348 14.2 184 222 313 336 377 533 620 "9 65.1 a" 666
406.5 14.7 204 23.6 330 666 66.2 672 80.4
I RO A J K TMN F VU C G W D Q L E H SXP B
*~~~~~~~~~..~l ..a Al.. a. so. .. I a .
Eco RI0 -0C 59 87III
ol
I I II II IIII
19.6 250 292 33.1 350 49 43.4
30.2 426
I If I I
529 597 683 72 6
601 69.8
I I I I I I I I I j
10 20 30 40 s0 60 70 t0 100
I I I I I I I I
Kbpo o10 20 30 40 so 70
AcNPV , HR
3
I I I I I
60 90 100 110 120 1SO
FIG. 5. Physicalmapof AcNPV HR3DNA for theenzymesSmaI, BamHI, XhoI, PstI, HindIII,EcoRI.The
map was linearized at thejunction of the EcoRI-I and -Bfragments. Each cleavage site is indicatedwith a
horizontal line and is labeledasapercentage of thegenome.Comigratingrestrictionfragmentsaredesignatedas
asingleletter inalphabeticalorderstartingfromO.0o (e.g.,Hind-Nand-0).
enoughtoorder the HindIl fragments asF, V,
T, and N, and to mapthem withrespect to the otherfragments (Fig. 5).
Restriction fragments not previously report-ed, HindIII-W and -X and EcoRI-W and -X,
were mapped as follows. HindIII-W was
mapped between HindIII-C and -H because of thehybridizationpatternfor
pAcEcoD19
(Table 2) and because an intact HindIII-W fragment was found when SmaI-C was digested withHindIII. HindIII-X was mapped between HindIII-D and -J because of the hybridization datafor
pAcEcoA14,
together with the partial digestion data of pAcEcoA14 withHindIII,
and because HindIII-X wasgenerated when PstI-O was digested with HindIll. EcoRI-W wasmapped between EcoRI-G and -D because di-gestion of SmaI-C and XhoI-B generated EcoRI-W.EcoRI-Xwasmappedbetween EcoRI-S and -Pbecause of thehybridization characteristics of pAcHindQ7 and clones containing EcoRI-X and because digestion of
pAcHindQ7
with EcoRIgaveEcoRI-X.
Thephysicalmapderivedhere isquite similar
to the E2physical map of Smith and Summers (12) for the enzymesSmaI,BamHI, XhoI, and EcoRI exceptfor the positions of EcoRI-S and -P and XhoI-J and -K. The positions for these fragments in Fig. 5 were derived from the
hy-bridization characteristics for the clones pAcE-coH5,
pAcHindKl,
andpAcEcoPl (Table 2).
DISCUSSION
Sequences representative ofmorethan 95%of
the AcNPV genome were cloned as EcoRI,
Hindlll,
and BamHI restrictionfragmentsinthe plasmid vector pBR322. The cloned fragmentswereidentified by their
electrophoretic
mobility withrespect toappropriately restricted AcNPV DNA (Fig. 3) and by hybridization of nick-translated recombinant plasmid to Southern blots of AcNPV DNA restricted with BamHI,EcoRI, HindIII,
PstI,XhoI, and SmaI endonu-cleases(Fig. 4). The hybridization data obtained from these analyses (Table 2), together with restriction analysis of the individual SmaI frag-ments,also servedtoconfirm thephysicalmapsofour wild-type strain of AcNPV (HR3) with respect tothosepreviously published (8, 11) for the restriction enzymes SmaI, BamHI, XhoI,
EcoRI. These datawere also used toestablish thecompletephysicalmapsfor PstI and HindIII
endonuclease-generated DNA fragments. The physical map for HindIll endonuclease was ofspecial interest because differences
be-tween our HR3 strain, the E2 variant of Smith
and Summers (11), and Li strain of Miller and Dawes (9) wereevident afterHindlIl digestion
(Fig.
2). The onlydifference between
HR3 and E2isthat theHindIII-L'
fragment of E2 is about 200bp smaller than that ofHR3. However, this difference is smallenoughnot tobe reflectedina A
EGD
I I I B
TO9 823 50
61 5
/O L
Genome
D798 6 666.1
6776n6 00.0
VOL. 41,1982 945
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.491.53.447.57.294.2]the physical
maps.The
Li strain has the
same-sized L'
fragment
asthe E2
variant and has
an extraHindIII site in the HindIII-A
fragment
generating HindIII-B' and
-H'fragments
whencompared
with
HR3and E2. That is,
theHindIII-B' and -H' fragments of the
Li
strain
appear to
consist of the
sameregion
of the
genome as
the
HindIII-A
fragment
of
HR3 and
E2.
The
extraHindIII
site in the
Li
genomeis
probably
atthe
78 to79% region
onthe HR3
genome
(Fig. 5).
The data presented demonstrate that
frag-ments not
previously reported
aregenerated:
anXhoI-N
fragment of about 350
bp;
anEcoRI-X
fragment of
about500
bp; and
HindIII-V,
-W,and
-Xfragments of about 900, 800, 800 bp,
respectively.
Eachof these
fragments
wasfound
upon
digestion of the
E2and
Li
strains. Except
for XhoI-N
fragment,
the
locations of these W
fragments and EcoRI-W
areincluded in the
physical
map(Fig. 5).
The
additional
hybridizations (Table
2)
sug-gest
that
HindIII-Q
and -L
regions
contain
com-monsequences.
This
possibility
is
supported
by
the
following
observations.
Restriction
frag-ments
of AcNPV
DNAappear to occur at onemolar
frequency
onthe
basis of the ethidium
bromide
stain
(Fig. 1). However,
hybridization
of nick-translated AcNPV DNA
toSouthern
blots of AcNPV DNA
digests
confirmed the
general
patternof
equivalence
exceptthat
HindIII-L and PstI-J
fragments
hybridized
asthough they
weredouble
bands
(Fig. 4).
Both
fragments
correspond
tothe
samelocation
onthe
physical
map.This
binding
pattern
was notseen
with other
enzymedigests
because of the
size
ordisposition
of the
fragments
in that
region; i.e., SmaI-A,
BamHI-A,
and EcoRI-A
are too
large
and XhoI-F
and -H
fragments
migrate
tooclosely
tothe -G and -I
fragments
todetect
anyincrease
in
hybridization.
For the
same reasons,
it is difficult
to seethe
effect of
additional
hybridizations
tothe
HindIII-Q
re-gion
of the
genome.However,
in well-resolved
blots, HindIII-Q, EcoRI-P,
and
XhoI-J also
hy-bridize
asthough each
wasadouble band.
The
availability
of
alibrary
of
AcNPV DNAcloned
sequenceswill make the
analysis
of the
viral
genomeeasier
aslarge
amountsof these
sequences can now
be
generated.
Our
currentwork indicates that the cloned
fragments
will be
particularly useful
asspecific probes
for
hybrid-ization selection of
mRNAspecies
in
thedevel-opment
of
atranscription
map. Thecloned
frag-ments
will
also be useful in
determining
the
maplocation of AcNPV
mutantsby
marker
rescue.ACKNOWLEDGMENTS
Wethank M. D. Summers and L. K.Miller formakingtheir
strains ofAcNPVavailable forcomparison.Wearegratefulto
B. N. White, D. C. Riddell, J. Hassel,and R. Deeley for advice in developing the cloning procedures and Dorothy Agnewfor technical assistance.
This work was supported by a grant from the Medical Research Council of Canada.
LITERATURECITED
1. Brown, M., A. M. Crawford, and P. Faulkner. 1979. Geneticanalysis of a baculovirus, Autographacalifornica
nuclearpolyhedrosis virus. I. Isolation of temperature sensitive mutants and assortmentintocomplementation
groups.J.Virol.31:190-198.
2. Clewell, D. B. 1972. Nature of ColEl plasmidreplication
in the presence ofchloramphenicol. J. Bacteriol. 110:667-676.
3. Dobos, P., and M. A. Cochran. 1980. Protein synthesisin cellsinfected by Autographacalifornicanuclear
polyhe-drosis virus (Ac-NPV): the effect of cytosine arabinoside. Virology 103:446 464.
4. Faulkner, P. 1981. Baculovirus, p. 3-33. In E. W. David-son(ed.), Pathogenesis of invertebrate microbialdisease.
Allenhead,Osmun andCo.,Totowa, N.J.
5. Gordon,J. L., A. T. H. Burns, J. L. Christmann, and R. G.Deeley.1978.Cloningofadouble-stranded cDNAthat codes for aportion of chicken preproalbumin. J. Biol. Chem.253:8629-8639.
6. Granados, R. R. 1980. Infectivity and mode of action of baculoviruses. Biotech. Bioeng. 22:1377-1405.
7. Guerry, P., D. J. LeBlanc, and S. Falkow. 1973. General methodfor isolation of plasmiddeoxyribonucleic acid. J. Bacteriol. 116:1064-1066.
8. Lee, H. H., and L. K. Miller. 1978.Isolation ofgenotypic variants of Autographacalifornica nuclear polyhedrosis virus. J.Virol. 27:745-767.
9. Miller, L. K., and K. P. Dawes. 1979.Physical map of the DNA genome ofAutographa californica nuclear
polyhe-drosisvirus. J.Virol. 29:1044-1055.
10. Rigby, P. W. J., M. Diechmann, C. Rhodes, and P.Berg. 1977. Labeling DNA to high specific activity in vitroby
nick translation with DNA polymerase I. J. Mol. Biol. 133:237-251.
11. Smith, G. E., and M. D. Summers. 1978. Analysis of
baculovirusgenomeswith restriction endonucleases. Vi-rology 89:517-527.
12. Smith, G. E., and M. D. Summers. 1979.Restrictionmaps of fiveAutographacalifornicaMNPVvariants. J.Virol. 30:828-838.
13. Southern, E. M. 1975. Detection of specific sequences among DNAfragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517.
14. Thayer, R. E. 1979. Animproved method for detecting foreign DNA in plasmids of E. coli. Anal. Biochem. 98:60-63.
15. Tjia, S. T., E. B. Carstens, and W. Doerfier. 1979. Infection ofSpodopterafrugiperda cells with Autographa
californicanuclearpolyhedrosis virus. II. The viral DNA andthekinetics of its replication. Virology 99:399-409. 16. Vaughn, J. L., R. H. Goodwin, G. L.Tompklns,and P.
McCawley. 1977. The establishment of two insect cell linesfromthe insectSpodopterafrugiperda (Lepidoptera:
Noctuidae).InVitro 13:213-217.
17. Vlak, J. M. 1980. Mapping of Bam HI and Sma I DNA restriction sites on the genome of the nuclear polyhedrosis virus of the alfalfa looper, Autographa californica. J. Invertebr.Pathol. 36:409-414.
18. Wahl, G. M., M.Stern, and G. R. Stark. 1979. Efficient transfer oflarge DNA fragments from agarose gels to
diazobenzylmethyl-paper and rapid hybridization using dextransulfate. Proc. Natl. Acad. Sci. U.S.A. 76:3683-3687.
19. Zasloff, M., G. D. Ginder, and G.Felsenfeld. 1978. An
acid-phenol method for thepurification of superhelical
plasmidDNA. Nucleic Acids Res.5:1139-1151.
J. VIROL.