Copyright© 1975 AmericanSocietyforMicrobiology Printed in U.S.A.
Host-Phage Interaction
in
Agrobacterium tumefaciens
IV.
Phage-Directed
Protein
Synthesis'
PATRICIA A. WALLS2 AND CHRISTINE F. POOTJES*
Department ofMicrobiology, ThePennsylvania StateUniversity, UniversityPark, Pennsylvania 16802
Received forpublication 10 September 1974
Gel electrophoretic and autoradiographic techniques were used to detect the
temporal sequence of protein synthesis after infection of the sensitive strain
Agrobacterium tumefaciens with phage LV-1. Three classes of protein were
detected: early proteins, class I, which include a protein capable of shutting off
hostproteinsynthesis;class II, proteins which are detected after 30min;and late
proteins, class III, which include the phage-directed endolysin and five
addi-tional proteins that appear 45 min after infection
Agrobacterium tumefaciens is a
gram-nega-tive rod which causes crown gall, a neoplastic
disease affecting avariety ofplants.
Lysogeny has been shown to be relatively
common in strains of A. tumefaciens (12).
Phage LV-1,atemperatephageisolated fromA. tumefaciens V-1,issimilar in size and
morphol-ogy to coliphage X. DeLey et al. (4) havefound
that phage LV-1 is virtually indistinguishable from three other agrophage, including omega,
the first temperate agrophage to be described
(2). Phage LV-1 has a polyhedral head and a
longflexibletail (12), which places it intoclass
B of Bradley's phage classification (3). The linear, double-strandedLV-1 DNA hasa
molec-ularweightof 31 x 106(5).Agenomeofthissize should be able toencode 30to40proteins. The phage is composed of four major structural
proteins and possibly as many as seven minor
ones.It isestimated that the structuralproteins
constitute less than 25% of the total coding
capacity of thephage DNA.
One
phage-coded
protein,endolysin,
a lateenzyme responsible for lysis of the host cells
afterphage infection, has been described
previ-ously (10).
Synthesis
ofendolysin
can bede-tected at 45 min after infection, halfway
through the latent period.
In the present study, gel electrophoretic and
autoradiographic techniques were used to de-tectthe temporal sequence ofprotein synthesis
after infection of the sensitive strain A.
tumefaciens B6 withphageLV-1.
MATERIALS AND METHODS
Bacterialandphage strains. A. tumefaciens B6, a phage-sensitive strain, originated from R. Klein,
'This is paper no. 4658 in the journal series of the Pennsylvania Agricultural Experiment Station.
'Presentaddress: Blair Memorial Hospital, Huntingdon,
Pa. 16652.
University of Vermont. Phage LV-1 was obtained by mitomycin C induction of the lysogenic strain A. tumefaciens V-1, which wasoriginally isolated by R. Hamilton, the Pennsylvania State University. Phage wereconcentrated and purified by CsCl density gradi-entcentrifugation prior toelectrophoresis.
Media and growth conditions. Labeling experi-mentsutilized modifiedStonier medium consisting of (perliter): 0.1 g ofCaSO4,0.2gofMgSO4 .7H2O,0.2g ofNaCl, 2.7 g ofNH4NOS, 8.4 mgofFe(NO3) .9H2O,
0.1 mgofMnCl2, 0.5 mg ofZnCl2, 10 g of potassium citrate, 2 g of sodium glutamate, 0.34 g of
NaH2PO4.H2O, and 0.9 g of K2HPO4 (8). Glucose,
sterilized separately, was added to a final
concentra-tion of0.2%. Cultures were incubated at 30 C in a gyratory waterbath.
Aglucose-yeast extract broth was used for all other experiments(7). These cultures were grown at 30 C on areciprocalshaker.
Radioactive labeling of proteins. A. tumefaciens B6 was grown to mid-log phase (3 x 108/ml) in Stonier medium. Before and after infection with phage LV-1 (multiplicity of infection of 15), portions of the culture were pulse-labeled for 3 min with "4C-labeled mixed amino acids (New England Nu-clear) at adosage of2uCi/ml.Incorporationoflabel was stopped by adding chilled Casamino Acids (Difco) to a final concentration of 3% and immedi-ately placing themixture onice.The cells were then harvestedby centrifugingfor 10minat 4300 x g.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. (i) Preparation of samples for disc gels. The cells from 2.5-ml portions of culture
were harvested and lysed with 50 gliters of sample buffer. The sample buffer, a modification of that described by Laemmli (6), consisted of 0.0625 M sodiumphosphate buffer, pH 7.2, containing 2% SDS,
10% glycerol, and 5% f,-mercaptoethanol. The sam-ples were heated for 1 min at 100 C to facilitate dissociationofprotein subunits.
(ii) Disc gels. Acrylamide gels (10%) were pre-paredandelectrophoresis performed accordingtothe
methodsof WeberandOsborn (11).Samples (25to 40
,uliters)wereappliedtoeachgel.
Afterelectrophoresis, gelsforscintillationcounting
372
on November 10, 2019 by guest
http://jvi.asm.org/
PROTEIN SYNTHESIS IN A. TUMEFACIENS
were frozen and sliced into 2-mm fractions. Each fraction wasplaced into a vial and solubilized in a 3% solution of Protosol (New England Nuclear). After incubation at 37C for 48 h, the samples were counted in aliquid scintillation counter.
(iii)Slab gels forautoradiography.
Polyacrylam-ide gels andbuffers were prepared using the methods of Studier (9). Samples (10 to 20
gliters)
were placed into each well of the slab gel. Electrophoresis was carried out at a constant voltage of 35 V for 12 to 14 h.Stainingand destaining werethe same as for the disk
gels. The slab gels were dried under vacuum and placed on Kodak Blue brand medical X-ray film for 10 to 14days before being developed.
RESULTS
Synthesis of phage LV-1 structural
pro-teins in infected cells. The four major
struc-tural phage proteins, Al, A2, A3, and A4,
described by De Ley et al. (4) are readily
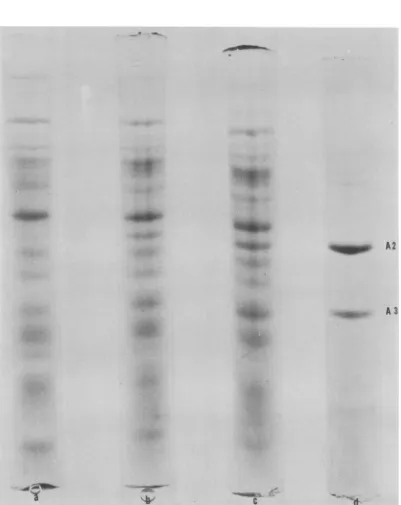
detected by SDS-polyacrylamide gel electro-phoresis ofpurified phage LV-1 (Fig. 1). Only
two of these proteins, A2 and A3, can be detected in infected cell extracts against the heavily strained background of host protein
(Fig. 2).Thesetwoproteins arefirstobservedat 45 min after infection, halfway through the latent period.
Synthesis of majorphage-directed proteins
in infected cells. Pulse labeling of cells for 3 min with
"4C-labeled
mixed amino acids (2iACi/ml)
atintervalsafter infection revealed thepattern ofphageproteinsynthesisininfectedA.
tumefaciens B6. Attemptsweremadetoshutoff
hostproteinsynthesistoinsure incorporation of
the labeled amino acids into phage protein rather than into host protein. UV irradiation,
evenin low
doses, rendered
A. tunLefaciens B6 virtually incapable of supporting phage-directed proteinsynthesis.
Another indirectattempt to suppress host
synthesis
was to treatthe cells with nalidixic acid (10
ug/ml) shortly
before phage infection, but nalidixic acid only partially suppressed host cellsynthesis.
Fortunately,
one of the early functions ofphage LV-1istoreduce the proteinsynthesisof
the hoststrain B6. Thisinhibitionisevidentas early as 1 min after infection and is quite pronouncedby30min(Fig. 3), asdemonstrated
by gel electrophoresis
ofthelabeledsamples. By
60 min postinfection, 10 to 12 protein peaks
appear, and the major structural proteins are
evident.
Synthesis
ofminorphage-directed proteins
ininfected cells.Autoradiographywasusedto detect phage-directed proteins which are
syn-thesized in smaller quantities than could be
readily detected by scintillation counting of fractionated disk gels.
"4C-labeled
extracts ofuninfected and phage-infected A.
tumefaciens
B6were run on 10 and 15%polyacrylamideslab
gels accordingtotheprocedures ofStudier (9).
An autoradiogram of a 10%gel isshown in Fig. 4. Uptake oflabel was lowest at 15 min after
infection, and it was slightly higher in later
samples. The intensity of the exposure of the
autoradiogram (Fig. 4) indicates that protein
synthesis did not return to the preinfection level.
Since hostproteinsynthesis was not entirely stopped, it was sometimes difficult to
distin-guish phage proteins from residual host
pro-teins. Thirteen new proteins appear to be
defi-nitely of phage origin. An additional seven
proteins, P1,P3, P5, P8, P10,P11, andP15 (Fig.
5a), could be either phage or host directed. Figure 5a provides an analysis of the time
sequence ofappearance ofthe phage proteins.
Synthesisofeightofthe 13phage proteins had begun by 15minafter infection. Phage proteins
P4, P7, P9, P13, P17, and P18 exhibited
inter-valsofmaximal synthesis followedby decreased synthesis, which suggests the existence of
con-trol mechanisms.
The majorphage structural proteins, A2and A3, areindicated inFig. 4. Synthesis of A3 had begun by 15min,whereas A2wasfirstdetected
inthe30-minsample. Either P12orP13(Fig. 4)
probably corresponds to phage structural pro-tein Al. A4 could not be located because the Tris-glycine buffersystem doesnot allow
sepa-ration ofproteinshavingmolecularweights less
than 25,000 on a 10%gel (9).
When theproteinswere
separated by
electro-phoresis on a 15% slab gel, four new proteins
migrating in advance of P18 could be
distin-guished on the autoradiogram (Fig. 6). These
weredesignated P19a, P19b, P20, andP21.The
times oftheir appearance are
presented
inFig.
5b. Protein P19a is an
early protein,
which isbeing synthesized
by
15 minpostinfection.
The other three proteins did not appear untilhalf-waythrough the latent
period.
DISCUSSION
We have found that, after infection of A.
tumefaciens B6 with phage LV-1, one of the
early functions of the phage is to reduce host protein synthesis. This suppression is evident as
early as 1 min after infection, and it is very
pronounced by 30 min (Fig. 3). This was a
fortuitousdiscoverysince ourattempts to
artifi-cially suppress host synthesis either had little
effect or were so effective that phage protein
synthesiswasalsoblocked.However, since this
phage-directed suppression was not complete,
some of the proteins being synthesized after
infection could still represent residual host
VOL.15,1975 373
on November 10, 2019 by guest
http://jvi.asm.org/
A
I
_Ip
A2
tulip
A3
MP
A
4
synthesis. Such doubtful proteins are desig-nated by a question mark in Fig. 5.
Sixteen, and possibly asmany as 24, phage-directed proteins weredetected by autoradiog-raphy. This represents a substantial portion of
theestimated30 to 40proteinswhich the phage
DNA should be able to encode. The major
structural proteins described by De Ley et al. (4) are tentatively identified on the
autoradio-grams (Fig. 4 and 6).
A plot of the times of appearance of new
proteins suggests that control mechanisms are
important in directingsynthesis ofphageLV-1
proteins (Fig. 5). Proteins P17, P18, and P19a
areactivelysynthesized from 15 to45min after
infection, afterwhichtheir synthesis appears to
decline steadily. P14and P16 appear at 30 min
whereasP2, P6, P20, and P21 are delayed until
45 min. It isinteresting to notethatonstained
gels (Fig. 2), the two mostabundant structural proteins, A2 and A3, were not
distinguishable
until 45minafterphage
infection.However,by
autoradiographic
methods,
A3 can be detectedat 15 min and A2 at 30 min (Fig. 4and6).
Proteins P19a andP19b, seen inFig. 6, may
betwodistinct proteins, or
they
mayrepresentdifferent forms of the same protein. P19b is
vaguely discernible at the leadingedge ofP19a
in the 45-min sample. By 60
min,
P19b ispredominant, and P19aisveryindistinct. Only P19b is evident at 90 min. This suggests that P19b may be a cleavage product of P19a. As
P19a is being cleaved at 45 min, the new,
smaller protein,P19b,isabletomigrate
farther,
thus appearing at theleading edgeofP19a. Asmore P19a is cleaved and no more is
synthe-sized, it disappears and
P19b
predominates.
EitherP19borP20 isprobably equivalenttoA4,oneofthephage structural proteins.
Although the exact size of agrophage
en-dolysin
hasnot yet been determined (10), it isprobably similarto thelyticenzymescoded by the T phage and by coliphage X. Ifthis is the
case, the enzyme
probably
has a molecularweight of 15,000 to 20,000, and it would be expected to migrate in the lower third of the
15% slab gel. The in vitro assay revealed that
endolysin begins to be synthesized at 45 min
after phage infection (10), the same time that
P19b, P20, and P21 are firstdetected(Fig. 6).
Different classes of protein synthesis can be
detected in phage-infected cells. The early
proteins, class
I,
include one capable of shuttingFIG. 1. Structuralproteins of phage LV-1. Phage
particles, purified by banding in CsCl, were
dena-tured, and thecomponent proteins were separated by
electrophoresison 10%SDS-polyacrylamidegels.The
gels were stained with Coomassie Blue. MP, Minor
protein.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.503.69.260.72.680.2]PROTEINSYNTHESISINA. TUMEFACIENS 375
~ _
_ .W
_
_
,
~~~~~A2
4
A 3FIG. 2. Theappearanceof phageLV-l structuralproteins inextractsof infected A. tumefaciens B6. Proteins inthe crudecellextractswereseparated byelectrophoresison10%SDS-polyacrylamidegels and stainedwith Coomassie Blue. (a) Uninfected; (b)45minafterinfection; (c) 60min; (d) phageLV-1 structural proteins.
VOL.15, 1975
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.503.45.446.68.573.2]^ Ek
V
S
t
r::
z.[fIi..
t
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~-
r...
(
CW.
-o
cvQ
X
0n
CAI
-ti
t.
n x X~~~~C
E t)o
'4-x, Q Q.U
< e
CCA
et
^U *
*
e
m-C
'm
.E---~ 03n
ll i *>X
(i0l X)SGIDV ONIkNV-J1-kNd9
376
.4dia
... -.. ..-.W.ww. r
%.I.. - ..;
-.4
-
on November 10, 2019 by guest
http://jvi.asm.org/
.3 ?C
-
-w~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~r (Ac>
o o
0Q
*Jh, _,IC-0
0.0.0.0.0. 0.0.0.0.0.0. 0.0. 4 _.40. 0. 0.0. ~ ~.~4-~ C
3"
tit1i
F:s2
>2
-0
_~~~~~~~11 I At Z ~~-&.
II~~~~~~~~~~~~~~~~~~~~~~~~~~~~c
~~~~~IQ)~~~ ~ ~ ~~~~~~r
_ *~~~~~~~~~~~~~~~~~~~~~~~~~~~~bO
^.~~~~~~~~a
4Qn
e8
0.~~~~~~~~~~~~~~~~~~~~~~~0
COD C*
4.4~ ~ ~ * ^ o
° -N)Cj rl, 0IDP-)OD c\°C"N ° 0yl N>* E*
cr O.a.0. a. 0.CL a.0. (L 0. 0.0La. a. >S Q N
X1 I\ /1 1111
W~~~~~~~~~~~~~~~~~~~-4-U ; ! 0szz°~~~~~~~~~~~~~~~~~~~~~~L % <<aew
L
i
i
lo
g t = e:
rE >2~~~~~~~~~~~~4
377
on November 10, 2019 by guest
http://jvi.asm.org/
offthe host protein synthesis as early as 1 min
after infection. Fourteen proteins can be
de-tected by radioautography within the first 15
min after infection. Anadditional five proteins
appear in a second class, class II, which are
detected after 30 min. The late proteins, class
III, which include the endolysin, consist ofan
additional five proteins thatappear45minafter
infection and these continue to be synthesized
until the end of the latent period. One of the
class I proteins and one ofthe class II proteins
appear to be shut off after 48 min which
indicatesaregulatory mechanism for turning off
protein synthesisaswellasinitiation. Fromthe
concentrations of the bands, periods of
in-creased and decreasing amounts or protein
synthesis are observed with the suggestion of
the cleavage ofone ofthe proteins, P19.
Further experiments with temperature-sensi-tive mutants will be needed to determine the function of these proteins.
LITERATURE CITED
1. Beardsley, R. E. 1955. Phage production by crown-gall bacteria and the formationofplanttumors. Am.Nat. 89:175-176.
2. Beardsley, R. E. 1960. Lysogenicity in Agrobacterium
tumefaciens.J.Bacteriol.80:180-187.
3. Bradley,D.E. 1967.Ultrastructure ofbacteriophagesand bacteriocins. Bacteriol. Rev. 31:230-314.
4. DeLey, J., M. Gillis, C. F. Pootjes, K. Kersters, R. Tytgat,and M. VanBraekel.1972.Relationshipamong temperate Agrobacterium phage genomes and coat proteins.J.Gen. Virol. 16:199-214.
5. Korant, B. D., and C. F.Pootjes. 1970.Physiochemical properties ofAgrobacterium tumefaciens phage LV-1 and itsDNA.Virology40:48-54.
6. Laemmli, U. K. 1970. Cleavage of structural proteins duringtheassembly of the head of bacteriophage T4. Nature(London)227:680-685.
7. Pootjes, C., and B. Stemberger. 1974. Host-phage in-teraction inAgrobacterium tumefaciens.II.Hoststrain
responsetomitomycin C induction.Can. J. Microbiol. 20:367-370.
8. Stonier,T. 1956.Labelingcrowngall bacteriawith P32 for radioautography. J. Bacteriol. 72:259-268.
9. Studier, F. W.1973. Analyses ofbacteriophage T7early RNA's and proteins on slab gels. J. Mol. Biol.
79:237-248.
10. Walls, P. A., and C. F. Pootjes.1974.Host-bacteriophage interaction inAgrobacteriumtumefaciens. III.
Phage-codedendolysin.J. Virol. 13:937-938.
11. Weber, K., and M. Osborn. 1969. The reliability of molecular weight determinations bydodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412.
12. Zimmerer, R. P., R. H. Hamilton, and C. Pootjes. 1966. Isolation andmorphologyoftemperateAgrobacterium tumefaciens bacteriophage.J. Bacteriol. 92:746-750.