Copyright01976 American SocietyforMicrobiology Printed inU.S.A.
Restriction
Enzyme Digests
of
Rapidly Renaturing Fragments
of Vaccinia Virus DNA
FRANK M. DEFILIPPESLaboratory of Biology of Viruses,NationalInstitute ofAllergyand InfectiousDiseases,
Bethesda,
Maryland
20014Received for publication 17 July 1975
Vaccinia virus DNAfragments that have been denatured by alkali and then neutralized contain a fraction that rapidly reforms duplex structures. The
fraction isenriched byfractionating onhydroxyapatite columns andserves as a
substrate
fordigestionby
tworestrictionendonucleases isolated fromHemophi-lusparainfluenzae, Hpa I and HPaII.Thepatternsobtainedby gel
electrophore-sis of thedigested fragments show the presence of three major bands after Hpa I
digestion and four majorbandsafterHpa IIdigestion. TheDNAthat is isolated
from some of these bands quickly reforms duplex regions after alkaline
denaturation. The size of the DNA segments in the major bands has been
estimated tobein the rangeof0.44 x 106to 3.2 x 106daltons. Thefragments which rapidly reform duplex chains after denaturation are sensitive to single-strand-specific nucleases. These results are consistent with a model of vaccinia
virusDNA which has acovalent linkconnecting complementary chains.
The linear, double-stranded DNA molecule isolated from vaccinia virus weighs 122 x 106
daltons(8).Thecomplementarystrands of this
DNA donotseparateunder alkaline
denaturing
conditions, indicating that covalent bonds link thetwostrands (3). If this DNA is
sheared,
then the strandsof mostoftheduplex
fragments willseparate after denaturation. The
fragments
which contain the covalent link connecting the
complementary
strands(covalently-linked
com-plementary
[CLCI
fragments), however,
will rapidly reformduplex molecules("snap-back")
when alkaline solutions are neutralized. The
CLC
fragments may be separated from single-stranded DNAby
hydroxyapatite chromatogra-phy. Restriction endonucleasedigestion
ofthese CLC fragmentsproduces specific
segments ofthe vaccinia DNA molecule. These segments
should be near the ends ofthe DNA since the covalent linksconnecting the strands have been locatedwithin 50nucleotidesofeach endofthe intact molecule (8).
One
model which has been proposed (8) for the structureofvaccinia DNA has thecomplementary chains joinedby
asmallloopofsingle-stranded DNAateach end ofthe molecule.
Therestrictionendonucleasesfrom
Hemophi-lus
parainfluenzae
have been used todigest
thesnap-back
fragments. Digestsof the entirevac-cinia DNA molecule
by
theseenzymesproducealarge numberofsegments that are difficult to resolveby gel electrophoresis.Thisprecludesan
227
analysis of the entire vaccinia genome by
methods which have been usedto study smaller
DNA molecules isolated from simian virus 40
and otherviruses (4, 10, 14, 18). However, the presence of CLC fragments, which may be
generated by shearing theintactDNA, presents an opportunity to analyze a fraction of the
whole moleculeby restriction endonuclease
di-gestion.
MATERIALS AND METHODS
Virus. HeLa S3-1 cells, grown as suspension
cul-tures in Eagle minimal essential medium with 5%
horse serum, werekindly supplied by B. Moss and N.
Cooper.Vaccinia virus (strain WR) was grown in HeLa cells inmedium with 5% dialyzed horse serum
contain-ing2 or 4
MCi
of['H]thymidine
per ml or 0.05gCiof["4C thymidine perml(New England Nuclear Corp.). The virus was grown in phosphate-free Eagle medium forlabeling with either 1 or 5
sCi
of [2P]orthophos-phate per ml (New England Nuclear). The procedure for virusinfectionhas been described previously (16). The virus was purified by the Joklik method (12), which wasmodifiedtoexclude sonic oscillation steps and the initial sedimentation through 36% sucrose. The crude cytoplasmic extract, obtained by Dounce homogenization of the cells followed by three low-speed centrifugations to remove nuclei, was incubated at 37 C for 5 h with 2x crystallized trypsin (Sigma ChemicalCo.) ataconcentration of1mg/ml. A band containing virus particles was isolated by
sedimenta-tion ofthedigested extract into a 25 to 40% sucrose gradient (12). After dilution of the band with an equal volume of 10 mM Tris-chloride, pH 9.0, the virus
on November 10, 2019 by guest
http://jvi.asm.org/
particles were pelleted by sedimentation at 21,000 rpm for 45min. The pellets were suspended in 10 mM Tris-chloride and repelleted through a 36% sucrose solution by sedimentation at21,000 rpm for60min. The pellet was suspended and stored in 10 mM Tris-chloride, pH 9.0.
DNA. Vaccinia DNA was prepared by digesting
virus particles with 100 ug of Proteinase K (EM Laboratories, Inc.) per mlat 37C for12h inabuffer containing 10 mM NaCi, 10mM sodium EDTA, 10
mM Tris-chloride, pH 8.3, and 1% sodium dodecyl sulfate(SDS). Solid sucrose was added to this digest togive a final concentration of 27% sucrose, andthe mixture wasextracted twice with phenol. The DNA was thendialyzed againstbuffer H (50 mMNaCl, 0.4 mMtrisodium EDTA and 10mM Tris-chloride, pH 7.8). 8H-labeled DNA had a specific activity of at
least 1.5 x 101 counts/min per
Ag
and "4C-labeledDNAhad an activity of 1.9 x 104counts/minper
Ag.
32P-labeled DNAhad specific activities of 1.1 x 105 and 2.1 x 105counts/min perjig.
Radioactivity was counted in aBeckman modelLS-250 liquid scintilla-tion counter inscintillation fluid containing toluene-Triton X-100-water (6:3:1) with 6.4% Spectrafluor(Amersham/Searle). T4 and T7 bacteriophages were grown according to standard procedures (19) and the DNAwasisolatedby the method described by Bautz and Dunn for T4 DNA (2).
DNA solutions containing 1 M NaCl in buffer H werebubbled with nitrogenfor5 min at 0C, and the DNA, at a concentration of20
jg/ml,
wasshearedby 10 passages through a 9-cm, 20-gauge needle. The DNA was then dialyzed against buffer H for 12 h. DNA was denatured in solutions containing 0.2 N NaOH at 37C for 10 min. The solution was neutral-izedby addition of an equivalent amount ofHCland diluted into 5volumes of80mM sodium phosphate (pH 6.8)at 0Corintothesamebuffer containing50%formamide.
Hydroxyapatite chromatography. Hydroxyapa-tite columns were prepared in disposable plastic
syringes that had a porous polyethylene support. In the mostbasic of several procedures, hydroxyapatite (Bio-Gel HTP, Bio-RadLaboratories)wassuspended
in80mM sodiumphosphate, pH 6.8, and poured into the column. The neutralized, alkali-treated DNA solutionwasapplied tothe column thatwaswashed with2column volumes of80mM sodiumphosphate. Single-stranded DNA was eluted with 8 column volumes of 0.18 M sodium phosphate, pH 6.8, and duplex DNA was eluted with4column volumesof0.4
M sodium phosphate, pH6.8.Trisodium EDTAwas
added toduplex DNA which had been eluted from the column to giveafinal concentrationof5mM, and the DNA was dialyzed against bufferH for 12 h. This DNAwas alkali denaturedasecondtimeand neutral-izedasbefore, except that the dilution buffer was 80
mMsodiumphosphate, 50%formamide, pH6.8.The
hydroxyapatite wassuspended in this bufferaswell,
and single-stranded DNA was eluted with 0.18 M
sodiumphosphatecontaining50%formamide.Duplex DNA was eluted as before using a buffer without formamide anddialyzed against bufferH.TheseDNA
fragments served assubstrate inthe restriction
en-zyme digestion, and they are referred to as CLC fragments.
Restriction enzymes. The two restriction endonu-cleases from H.parainfluenzae, Hpa I and Hpa II, were isolated by a modification of a previously pub-lished procedure for the isolation of Hpa I (6). The modifications included breaking the bacterial cells openinapress(Manton GaulinLaboratory Homoge-nizer) and precipitating the proteins in the crude extract with (NH4)2SO4 at 64% of saturation. The final step, after concentration onthe phosphocellulose column, was to separate the two enzymes by gel filtration on a Sephadex G-100column. The enzymes were stored and used aspreviously described (6). The digestion reaction was stopped by the addition of trisodium EDTA and SDS to final concentrations of 30 mM and 0.5% and enough sucrose was added to give a 15% solution.
Gelelectrophoresis.Agarose (1.6%) gels (11 by 0.5 cm; Seakem) were formed in plexiglass tubes. The gels were made up in buffer E (36 mM Tris, 30 mM NaH2PO4, 1 mM trisodium EDTA, pH 7.9). Electro-phoresis was in buffer E with 0.02% SDS and the samples were runfor10to 11 hat 22 V at 24 C. After electrophoresis of3H-or "4C-labeled DNAsegments,
thegelswere cuttransversely into 1-mm sections and
the DNAwas hydrolyzed by heating in 0.5 ml of 1 N HCl at 80 C for 5 h. The samples were counted as previously described (5). Gelscontaining 32P-labeled DNA segments were cut longitudinally, dried on Whatman 3MM paper, and placed in contact with
RP/R2X-ray film (Eastman Kodak Co.) according to standard procedures for obtaining radioautographs (7). In some cases gels containing 32P-labeled DNA segments were cut transversely as above. Each agarose slice was immersed in 5 ml of buffer H, and the radioactivity was determined by Cerenkov counting. The slices that contained the DNA segments were selected, the DNA was eluted by electrophoresis into dialysis sacs, and the samples were dialyzed against
buffer H. The DNA from the individual fragments
was then alkali denatured, neutralized, and passed throughsmall columns (0.2 ml) ofhydroxyapatiteto
separate single-stranded and double-stranded DNA.
Generallyless than5min wasinvolvedinneutralizing andapplying the entire DNA solution to the column.
Some gels were not cut but were immersed in a
solution of ethidium bromide (1 ,ug/ml) for 1 h and then examinedbyUVlight which showed the
fluores-centethidium-stained DNAbands (18).
The distance which the restriction enzyme
frag-ments migrated in the 1.6% agarose gels was
mea-sured and compared to the migration distance of
segments ofT7 DNAwhich had been produced by
digestion with Hpa I. M. Simon and F. W. Studier kindly providedestimates ofthemolecularweightsof
T7 DNA segments after Hpa I digestion (personal
communication), and these were used to determine the relative molecular sizes of the vaccinia DNA
segments. The logofthereciprocalofthe molecular
weightof avaccinia DNAfragmentwasassumedtobe related tothe distancemigratedinthe usual fashion (11).
Nuclease digestion. S. endonuclease from
on November 10, 2019 by guest
http://jvi.asm.org/
gillusoryzae waspurified bythe method ofVogt (22)
and his definition ofa unit ofenzyme activity was
used. TheDNAwasdigestedat37or55 C for30orfor 60minineither50or270mMNaCi, 1.5 mMZnSO4,
5%glycerol, and30mM sodiumacetate,pH 4.65,with 25Asgofsheared denatured calf thymus DNAperml,
and0.2or2U ofenzyme.Thereactionwasstopped by
additionofSDSto0.5%and EDTAto5mM,and the
sample waseither alkali denatured, neutralized, and
passed through hydroxyapatite or precipitated with
7% perchloric acidat0C.Precipitated sampleswere
collected on GF/C filters (Whatman) which were
driedand counted for radioactivity.
A crude preparation ofasingle-strand specific
en-donuclease associated withvaccinia virions was
pro-vided by P. Geshelin and K. Berns. The enzyme
digestionwasat37or55 C for 1 h in buffercontaining
2.5 mM trisodium EDTA, 0.1 M Tris-chloride, pH
7.8,and250Agofsterilegelatinperml.Usually0.1to 1.1 MgofDNAwasdigestedwith10to20Alofenzyme
preparation. The reaction was stopped by the
addi-tionofSDSto0.5% and the condition of the DNAwas
testedbythetechniques used afterS,digestion.
RESULTS
Hydroxyapatite fractionation of digested DNA. Shear breakage of vaccinia DNA, using the conditions described, produced fragments thatsedimented inaneutralsucrosegradientas abroad band whosepeak moved slightly faster
than simian virus 40 form III DNA (data not shown). This indicated that mostofthepieces
were lessthan 7% of the original length. These
fragments were denatured, neutralized, and
passed through a hydroxyapatite column to separate single-stranded from double-stranded DNA. Hydroxyapatite chromatography was at 24C rather than 60C (13) to reduce random
bond breakage, and some hydroxyapatite
frac-tionation was done inthe presence of
formam-ide. Goodman et al. (9) have shown that the fractionation of nucleic acids by hydroxyapatite chromatography at room temperature using buffers containing formamide is similarto the fractionation achieved at elevated tempera-tures.
When 0.1 ggorless ofshearedvaccinia DNA was denatured, neutralized, and diluted into
coldphosphate buffer containing 50% formam-ide and then loaded on hydroxyapatite, 12 to 15% of the DNA was recovered as duplex
material. If the sheared DNA had a uniform
molecular weight of 6 x 106 and was derived
from an intact molecule with a covalent link
connectingcomplementary strands ateach end ofthemolecule, then 10% of the DNA should be recovered asduplexmaterial after
hydroxyapa-tite fractionation. A typical result is shown in Fig. 1B. Less than5minwasneededto
neutral-6
4
2
0
6
4
In
r)
2
oL
6
4
2
0
in A
-N4
I
I
IAle
C
11
I,
L-B
--j I
-I I I
Br-I
I I
3 5 7 9 FRACTIONNUMBER
B~
B,
1,
_j I
Bn
B!
L~
RCTO UBE
9
0 0
ex
10 o
5
O
0
6 X
_
4 wl
,.
0
4 lw
12
Jo
FIG. 1. Chromatography of [3H]thymidine
vac-cinia DNA on hydroxyapatite columnsat 24C with phosphate buffer, pH 6.8, containing50%formamide.
All samples in these figures were elated by step increasesin the molar concentrationof buffer, which
was applied as indicated by the arrows. (A) Native
DNA was shearedby 10passages througha22-gauge
needleand thenapplied and elated from the column.
(B) DNA was sheared and then alkali denatured,
neutralized, diluted into phosphate buffer,and
imme-diately appliedtothehydroxyapatite. (C-F) Sheared, double-stranded vaccinia DNA was digested witha
nuclease, alkali denatured, neutralized, diluted into phosphate buffer, and appliedtohydroxyapatite. (C) DNA was digested with S1 nuclease; (D) DNA was
digested with a combination ofrestriction
endonu-cleases, Hpa IandHpa II, isolatedfrom H. parain-fluenzae; (E)DNAwasdigested with HpaI; (F)DNA was digested with Hpa II. T4 DNA, labeled with
[14C]thymidine, was sheared and applied to each
hydroxyapatitecolumntoserve as adouble-stranded
DNA marker.
ize the solution and adsorb the DNA to the columnsothat renaturationwasminimal.If the
shearedDNAwasdigested withasingle restric-tion enzyme and denatured and neutralized,
then less duplex material was recovered from the hydroxyapatite. This indicated that the sheared fragments contained the proper
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.491.252.441.57.367.2]quence for restriction enzyme digestion. After Hpa I digestion 4 to 7% of the DNA was recovered from the hydroxyapatite (Fig. 1E),
and after Hpa IIdigestion 3 to 5%ofthe DNA was recovered as double-stranded segments
(Fig. iF). Digestion of the DNA with both
restriction enzymes reduced the amount of
duplex DNA recoveredto 1 to 3% (Fig. iD). If
the DNAwasdigestedfor 60 min at 55 C with2
U of the
single-strand-specific
S1nuclease,
in buffer with either 50 or 270 mM NaCl, and denatured and neutralized, then less than1%ofthe fragmentswereretained by hydroxyapatite (Fig. 1C). In separate tests with native
"IC-labeled T7 DNA, 1 U of this S1 preparation nicked more than 90% of the molecules after
incubation for 60 min at 37 C in a reaction
mixture containing 270 mMNaCland 25
gg
ofdenatured calf thymusDNA per ml (see
refer-ence 22). Thus it is assumed that the enzyme
nicks double-stranded portions of the CLC
fragments in addition to digesting single-stranded regions.
Isolation of CLC fragments. As stated above, about5%of the input DNA isrecovered
asCLCsegments after restriction enzyme
diges-tion. However these enzymes cleave vaccinia
DNA at many sites and produce so many segments that it is difficult to tell when the
reaction has gone to completion. Also, to pre-pare microgram quantities of CLC segments
routinely, relativelylarge amountsofrestriction enzyme mustbe used.To conservethesupplyof enzyme an alternative procedure was adopted. Sheared fragments were denatured, neutral-ized, and passed through hydroxyapatite. Du-plex fragmentswererecovered from the column, and the cycle was
repeated
usinga secondcol-umn of
hydroxyapatite
with bufferscontaining
50%formamideasdescribed above. The double-stranded fragments obtained from the second columnwereusedas asubstratefor arestriction
enzyme. About 3 to5% ofthe original sheared DNA was recovered with the elution of the
double-stranded fragmentsfromthe second
col-umn. The size distribution ofthese fragments afterelectrophoresis through a1.6% agarose gel isshowninFig. 2. The peak of thisdistribution
contains fragments which move more slowly
than the largest fragments obtained after
re-striction enzyme digestion. Figure 3 contains
electrophoretic profiles of restriction enzyme
digests of CLC fragments obtained as duplex DNA from the second hydroxyapatite column.
Thetop frameshowsthe pattern after digestion of
[3H
]thymidine
DNA fragmentsbyHpa I.Themiddle frame is thepattern after digestion with
Hpa II and the bottom frame is the result
FRACTIONNUMBER
FIG. 2. Electrophoresis of sheared, duplex vaccinia DNA which is recovered from hydroxyapatite.
Sheared 3H-labeled DNA wasalkali-denatured, neu-tralized, passed through hydroxyapatite, and re-coveredasduplexDNA. Theprocedurewasrepeated, using bufferscontaining50% formamideasdescribed, and the duplex fraction from the hydroxyapatite,
called CLC fragments,wasdialyzedandanalyzed by electrophoresis through 1.6%agarosegels.In allgels
migration was toward the left (anode). Thegel was
sliced, digestedwith
HCl,
and countedasdescribed.obtained after digestion with HpaIandHpa
II.
These types ofpatterns were consistently
pro-duced after
digestion
ofthree differentprepara-tionsof [3HHthymidine vaccinia DNA.TheHpa
I pattern always showed several large peaks
near the top of the gel (fractions 62-75), the largestofwhichalways movedmoreslowly than thematerialproviding the slowest moving peak
(fraction 65) found for the Hpa II digests. Two other peaks in the Hpa I pattern (fraction 46
and 36) show segments that migrate at faster
rates. However, in other gel patterns, the peak thatcorresponds totheonefoundatfraction36 is much less pronounced. The Hpa II digests always showed four major peaks which were
well separated in 1.6% agarose gels. The back-ground levelin some ofthese gelswashigh, and the definition of individual peaks was not as
good as that which would be found if the
original DNA substrate had beena
homogene-ous collection ofmolecules. Shearing the DNA
breakssomeofthepotentiallyuniform(specific size) segments which would be
produced
by
restriction enzyme digestion into fragments of random length.Afterdigestiononeend of these fragments isformedbyenzymatic cleavage andthe other end already existsdue to shear frag-mentation. Additional variation inthepatterns was probably due to radiation damage of the
DNA samples which had specific activities of
I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.491.265.458.60.244.2]0
x
0.
9 8 6 4 2
10 20 30 40 50 60 70 80 90 100
FRACTIONNUMBER
FIG. 3. Electrophoresis ofDNAsegmentsproduced by restriction enzyme digests of 3H-labeled CLC
fragments. (A) The fragments were digested with enzyme Hpa Ias described and then analyzed by
electrophoresis through 1.6%oagarosegels. (B)
Diges-tion withenzymeHpaII.(C) Digestionwithenzymes
Hpa I and HpaII.ThegelsweretreatedasinFig.2.
105counts/minperjgormore.Thetwo denatu-ration steps whichwereused toselect theCLC fragments allowed separation ofpartsofnicked fragments of DNA. Since single-stranded tails have little effect on the affinity of duplex
regions for hydroxyapatite (20), CLC segments with tails of differentlengths will be retained by the column and they will migrate through agarose gels atdifferentrates. The susceptibil-ity of 3H-labeled DNAtoradiation damage has
been cited(19). Radioautographs of electropho-retic separations of digests of "P-labeled DNA fragments provided better profiles. Figure 4 shows thatthe patterns obtained afterdigestion of 32P-labeled DNA fragments by Hpa I and
Hpa II agree with the major features of the profiles shown in Fig. 3, but that it is easier to
visualize the major bands. The Hpa I pattern has the three majorbands at thetop of the gel and one more easily distinguishable band,
la-beled E in Fig. 4A, as well as several minor bands. Bands A, B, C, and E in Fig. 4A correspondto thepeaks atfractions 74, 66, 62, and 46 in Fig. 3A. Bands D and F are always much less intense than bandE in the
radioauto-grams and I regard them as minor bands. The
Hpa IIprofile shows that the peak representing thelargest DNAsegmentsinFig. 3B, atfraction
65, can be resolved into 4 bands, designated
a-ay. This profile is very reproducible andthe
four bands are always present. It is not known
whetheranyofthese bands ariseasdegradation
products from any of the other bands. The patternremainsconsistent when thesame
sam-ple of 32P-labeled fragments is digested by Hpa II upto a week after the original digestion and
electrophoresis. Another one ofthe four peaks
shown inFig. 3B, atfraction 43, corresponds to twobandsdesignated cl andc2inFig. 4B. This
is slightly more variable than the presence of
the fourbandsnearerthetopofthe gel. Bands b and dcorrespondtothe peaks foundatfractions 50 and 30 inFig. 3B. Thesetypesofpatternsare
alsorepeated when CLC fragmentsareprepared
fromunlabeled DNA and the digestedsegments
areseparated by electrophoresis. Ethidium
bro-mide stained gels are shown in Fig. 5. They
confirm the patterns shown for previous gels. Relatively large amounts of vaccinia DNA (50 to100
Ag)
havetobepreparedtopurifyenough CLC fragments for digestion when the ei-thidium staining procedure is used. The back-ground staining due to the presence ofbrokenfragments of DNA limits the sensitivity of this procedure. The right hand gel in Fig. 5 was
prepared from 32P-labeled fragments which
were stored for 10 days at 0C. The pattern, although recognizable, has deteriorated because ofradiationdamage.
Fractionation of DNA from gel slices. It
wasgenerallymostusefultoprepare32P-labeled
CLCfragments. After digestion and electropho-resis the gel can be sliced and the radioactive
profile is determined by counting the slices in
dilute buffer. DNA was recovered from the
individualslices by electrophoresis and discrete
segments could be tested for the CLC property
by alkali denaturation, neutralization, and hy-C
17,
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.491.40.230.62.480.2]Ir
a
2a--4
9.
a-
3..
...
4_ b
.s--- I
F
A
B
FIG. 4. Electrophoresis of32P-labeledDNAsegmentsproduced by HpaIdigestion of CLC fragments.The DNAwasdigestedandanalyzed byelectrophoresis. Thegelwasthenslicedlongitudinallyintotwoslices,anda
radioautographwasprepared bytheprocedure ofFairbanksetal. (7). Electrophoretic migration is towardsthe
bottom (anode). The bands of the radioautogram have been designated by large letters. Gel segments
corresponding tothese bandsinsimilargelswereexcisedand the '2P-labeled DNA segmentswereelated by electrophoresisasdescribed.(B)Electrophoresis of "2P-labeled DNAsegmentsproduced by Hpa IIdigestionof
CLCfragments. Theprocedureisthatdescribed in (A). The fourcloselyspaced DNA bandsnearthe top ofthe
gelhavebeendesignateda,-a4,witha,beingtheband nearest the cathode.
droxyapatite fractionation. When this was
done, however, no one gel bandwas associated
with segments which gave 100% ofthe counts
eluting from hydroxyapatite as duplex DNA.
Thiswasexpectedhowever becauseofradiation
damage and the presence of random sheared fragments. In addition, the peaks obtained by slicing the gel were probably not pure, so that
individual segments were contaminated by
other segments. Despite the difficulties in-volved itwas found thatbands Aand B in the
Hpa I digest shown in Fig. 4 contained a
substantial fraction ofDNAwhichappearedto besnap-back DNA. Bandsaand d intheHpa II
digestof Fig. 4 alsoappeared tocontain snap-backDNA. The relative amountof DNAwhich
A
B
C
* D
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.491.58.449.65.494.2]FIG. 5. Electrophoresis of unlabeled DNA
seg-mentsafter digestion of CLC fragmentswith restric-tion enzymes. Thegels were stained with ethidium
bromideasdescribed. ThegelontheleftshowsHpaI
digestion products. The middle gel shows Hpa II
digestion products.ThegelontherightshowsHpaII
digestion products of "2P-labeled CLC fragments which had been stored at 0C for 10days prior to
digestion. Electrophoretic migration is toward the
bottom (anode).
is retained by hydroxyapatite after removing single-strand chains is given in Table 1. An
attempt was made to split the a band of the
Hpa II digest byusingalonger time for
electro-phoresis and dividing the increased number of discs cut from across this band into two frac-tions. Both fractions showed some segments eluting as duplex segments. After digestion of 'C-labeled CLCfragments the four bands in a
could beresolvedinethidium-stainedgels when viewed underUVlight. However, previous
expe-rience with other DNA samples has shown that this exposure to UV light (mineralight short
wave, model R-51) degrades the DNA and it is
also knowntocausecross-linking (15). For these
reasons no attempt was made to use this tech-nique toisolate theindividualbands, which are
shown in Fig. 5.
Denaturation of DNA segments produced
by a restriction enzyme. Another approach, used initially, wasto digest the CLCfragments
with a restriction enzyme and then alkali dena-ture the entire reaction mix and apply it
di-rectly to agel. Those unique segments produced
by therestriction enzyme which do not contain the covalent link connecting complementary
strands remain single-stranded after
denatura-tion. Any random size duplex fragments
with-out the covalent link also remain single
stranded after denaturation. In addition the unique segments which contain the covalent linkmayby nicked, andpart of thechain which isbrokenatthe nick willbecome single stranded. Figure 6B shows that denaturation produces single-stranded DNA, which migrates more
quickly than duplex DNA (Fig. 6A). Some of
this isdue to the migration ofdegraded single strands, although intact single-stranded DNA moves fasterthan double-stranded DNA ofthe
same length in these gels. Those restriction enzyme segments which retain the covalent linkage should move as double-stranded
mole-culesaftertheyenterthegel and migrateinthe pH7.9buffer. The lowerpanel shows the result
of denaturation, neutralization, and passage
throughhydroxyapatite. The duplexDNA from
the
hydroxyapatite
gives the electrophoreticprofile shown in Fig. 6C. Most ofthe smaller
(single-strand)
DNA is removed by thisproce-dure, leaving only the DNA initially seennear
the top of the gel, with a shoulder on the low-molecular-weight side. Definition of indi-vidual peaks inpanel C isreduced by the high background. This indicates the presence of some degraded CLC segments, which are only partial
duplexes
that have been retainedby
the hydroxyapatite column. If the CLC segments are in the fractions withpeaks at 80 and 74inpanel
A, thenthey
wouldprovide
a source ofpartially
degraded
segmentswhichareresponsi-ble for the background in panel C. This is not
surprising since these experiments were
per-formed with [3H
]thymidine-labeled
DNAfrag-ments which are particularly susceptible to radiationdamage. Figure 7 shows the same type
experiment except that [3H
Ithymidine-labeled
fragments were digested with Hpa II. The
middle panel shows that denaturation leaves one high-molecular-weight segment which movesataboutthe same rateasthemajor peak
in panel A. The bottom panel demonstrates
again that hydroxyapatite treatment removes
thesingle strands, leavingthe major peak found
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.491.91.198.67.400.2]TABLE 1. Fraction of recovered CLC segments retained by hydroxyapatitea
Total counts 0.4M phosphate counts/0.08 M + 0.18 Mphosphate counts Sample
Expt 1 Expt 2 Expt 3 Expt1 Expt 2 Expt3
Hpa I
A 1,399 883 1.02 0.71
B 1,366 621 0.62 0.51
C 1,160 471 0.18 0.14
E 509 231 0.16 0.09
Hpa II
a,-a4 2,481 2,326 0.92 0.74
b 1,148 863 0.13 0.14
c-cC2 900 527 0.09 0.15
d 999 850 1.55 0.65
a,-a2 933 0.71
a3-a4 666 0.77
a32P-labeled CLC segments, produced by digestion with restriction enzymes Hpa I and Hpa II, were
separatedby electrophoresis throughagarosegels. Gelsweresliced into 1-mmdisks,and theradioactivitywas measuredby the Cerenkov method. Selected gel fractions, corresponding to those bands which are labeledin
Fig. 4A andB, were placed in electrophoresis buffer and the DNA segmentswererecovered byelectrophoresis intodialysis sacs. The DNA was denaturedin0.3NNaOHat 37C for10minand then neutralized withHCl anddiluted with bufferto afinal concentration of80mM sodiumphosphate, pH6.8,with 50% formamide. The
sampleswereloadedon0.25ml ofhydroxyapatite columns and eluted with 0.18 M phosphate-50% formamide followed by0.4Mphosphate-50% formamidebuffer, pH 6.8, at 24C.Radioactivitywascounted in the triton fluorgiveninthetext.Thespecific activityofthe DNA inexperiment1 was 1.1 x 101counts/minperggand in experiments2and3 was 2.1 x 101counts/minper fig. Adigest of this latter DNAby Hpa II was separatedby electrophoresisand the mainabandwascut intopiecesthoughttocorrespondapproximatelytoa,plusa,and
a,plus a4.
nearthe top of thegel and a smaller peak on the
low-molecular-weight end. The data presented in Fig. 6 and 7 suggest that the larger DNA segments containspecieswhich haveacovalent link connecting complementary chains. The HpaIIdigest also showsa peak
(about
fraction29, Fig. 7 panel C) with DNA which runs
slightly faster than thesegments producing the peak at fraction 34 in panel A. When native
C-labeled T7 DNA was denatured and
neu-tralized,
it was eluted from hydroxyapatite inthe 0.18 M phosphate buffer. Since single
chains of T7 DNA are longer than thesheared vaccinia DNA strands, it is clear that the
hy-droxyapatiteis not retaininglong single chains
ofvaccinia DNA.
Size of the DNA segments. The size of the enzyme segments derived from the CLC frag-ments has beenestimated by comparison with
an Hpa I digest of T7 DNA which was run
simultaneously, usingidentical conditions.The
molecular weights of the T7 fragments have been determined by M. Simon and F. W. Studier (personal communication) who kindly provided their data. This indicated that bands A, B, C, D, E, and F of the Hpa I digest shown
in Fig. 4A had DNA segments with molecular weights which wereabout 3.2 x
106,
2.2 x 106,1.9 x 106, 1.35 x 106, 0.9 x 106 and 0.56 x 106.
The Hpa II digestof Fig. 4B has bands a,b, c, and dwithDNAwith molecularweights of 2.1 x 106. Although these sizes are onlyestimates, it seemscertain
that
the relative sizes of themajor segments arereliable
sincethe patterns arere-producible despite the high background levels.
Thus fragment A ofthe Hpa I digest is larger
thanfragment A of the Hpa II digest. It is not
likely that the presence of a small
single-stranded end region would cause abnormal
migration of a segment which ispredominantly
duplex.
Digestion of CLC fragments
by
single-strand-specific nucleases.A
single-strand loop
in a CLC fragment should be digested by single-strand-specific nucleases. Very smallloops would be resistant to such nucleases but
they should expand with increasing tempera-tureandbemoresusceptibletodigestion. Since both the
S,
(1) and the vaccinia nuclease(17)
are active at 60C, they have been used byothers to determine the nature of the covalent linksconnectingcomplementarychains (8, 21).
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.491.59.456.81.262.2]0
x 10
_M 5
3
2
1
0
20 40 60 80 100
FRACTION NUMBER
FIG. 6. Electrophoresis of denatured 3H-labeled
DNA segmentsproduced by HpaIdigestion of CLC fragments. (A) Electrophoresis of native digestion products. (B) A 50-,ul digestion reaction was
termi-nated bythe additionofEDTA andSarkosyltofinal
concentrations of30mMand 0.5%, and then NaOH
was added to 0.2 N. After 10 min at 37C the
0
x
CL
y
[image:9.491.40.439.55.588.2]FRACTION NUMBER
FIG. 7. Electrophoresis of denatured 'H-labeled
DNAsegmentsproduced by HpaIIdigestion ofCLC
fragments. (A) Electrophoresis of native digestion products. (B)A50-,gldigestionwas treatedasinFig.
6B. (C)A 50-uldigestwastreatedasinFig. 6C.
denatured digest wasapplied directly toan agarose gelfor electrophoresis. (C)Theprocedureused in(B) wasfollowed but the alkalinedigestwas neutralized andpassed through hydroxyapatite. TheduplexDNA retained by thehydroxyapatite was dialyzedagainst buffer H and analyzed by electrophoresis. All gels
were cutandanalyzed as described in the legendto
Fig.2. 17,
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.491.47.237.68.599.2]Breaking the covalent link converts the duplex to a denaturable form which is eluted from
hydroxyapatite by 0.18 M sodium phosphate. Results ofthe hydroxyapatiteassay aregivenin
Table 2 as the relative amount of DNAwhich
remains double stranded after enzyme treat-ment and denaturation. As anticipated, en-zymaticdigestion eliminates duplex DNA and
is more effective at temperatures above 37C. Thisagreeswith theresultreportedby Geshelin andBerns (8), who used the vacciniaenzyme to
digest native DNA to produce molecules with
openends.Theiranalysis of their sedimentation
data indicated that there were no
single-strandedregions in nativevaccinia DNA except at theends of the molecule. TheSlendonuclease
fromA.oryzaealsodigestsCLCfragments when
[image:10.491.53.249.312.382.2]tested by the same criterion used for the vac-cinia nuclease. The digestion buffer contained
TABLE 2. Effect of nucleases on spontaneous
renaturationofCLC fragments
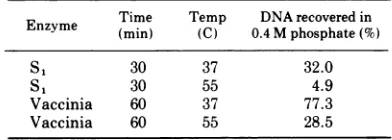
Enzyme Time Temp DNArecoveredin
Enzyme (min) (C) 0.4 M phosphate(%)
S 1 30 37 32.0
S1 30 55 4.9
Vaccinia 60 37 77.3
Vaccinia 60 55 28.5
a
0.27
,g
of[14C
]thymidine-labeled
CLC fragments(19,000 counts/min per ,ug) were incubated at the temperature and timeindicated inareaction volume of0.12ml in buffersdescribedinthe text, with and without enzyme. The
S1
reactionbuffer contained 270 mMNaCiand 0.2 U(22) of enzyme and the vaccinia nucleasecontained 10 Ill of a crude preparation. Each reactionwas terminatedby addition of SDS and thenchilling the tube. Afteradjusting the Si tubes to 5
mM EDTA all samples were denatured in 0.3 N NaOH at 37C for 10 min. The sampleswere neutral-ized with HCl and diluted with buffer to a final concentration of80mM sodiumphosphate, pH6.8, in 0.81ml.Thesampleswereloadedon0.25-ml
hydrox-yapatite columns and eluted by 0.18 M phosphate
followed by 0.4 M phosphate buffer, pH 6.8.
Radio-activefractionswerecountedinthe triton fluor. The
results aregiven as the ratio of the number ofcounts
elutedin0.4Mphosphateintheenzyme-treated
sam-ple to the number of counts in the 0.4 M
phos-phate fractioninthe controlsample without enzyme, expressedaspercentage.84% ofthe totalcounts inthe controlsample elutedinthe0.4Mphosphatefraction. Denatured[3HIthymidine-labeledHeLacell DNAwas 96% acid soluble after digestion with
S,.
The sameDNA was 70% acid soluble aftertreatment with the
vaccinia nuclease (see text for details). 97% of the radioactivity of these controls was eluted from
hy-droxyapatite columnsby0.18 Mphosphatebuffer be-foreenzyme treatment andall the radioactivitywas
eluted by this buffer aftertreatment.
270mM NaClsince Vogt (22) has reported that
the specificity of purified
S,
preparations forsingle-stranded DNA is greatly increased at
high salt concentrations. In 270 mM NaCl at 55C, theamountof enzymeusedtodigest
CLC
fragments inthe reaction given in Table2willdegrade over 95% of sheared, denatured, 3H-labeled HeLa cell DNA(2.5
,ug)
toacid-soluble materialin 30min.With thesameconditionsat 37C the S enzyme willdigest 76%ofthisDNA to acid-soluble material. The amount ofvac-cinianuclease usedintheexperiment forTable
2, in the Tris-EDTA buffer, causes 70% ofthe denatured HeLa cell DNA (1.1 ,g) to become
acid soluble in 60 min at 55 C. At 37 C 58% of
this DNA becomes acid soluble after60 min of digestion.
One would expect these enzymes to digest single-strand tails on degraded, hook-shaped CLC fragments. If the covalent link is at the
extremeendofthefragment, then it ispossible that digestion of the tail might leave a very
small hairpinstructure that hasasmall, intact
single-stranded loop. This loop would resist
digestion because it was extremely small,
per-haps consisting of one or twounpaired
nucleo-tides. The small duplex fragment which
re-mained would not be retained by hydroxyapa-tite. Wilson and Thomas (20) estimate that
duplex
chains withamolecularweight less than18,000 will be eluted from hydroxyapatite by 0.14M phosphate buffer. It mightappearthen that digestion had split the link connecting strands when the actual resultwasthe produc-tion ofaveryshortduplex CLC fragment.
How-ever, S,
digestion
of CLC fragments reduces theamountofduplexDNAretainedby hydrox-yapatite to abackground
level of about 5%.This implies that most ofthe CLC fragments
would have the unusual hooked-shaped struc-ture described above, which is unlikely. The most plausible explanation oftheresultsofthe
S,
digestion
is that this amount of enzyme issplitting all the single-stranded regions,
includ-ing those that contain the covalent link.
Eighty-nine percent of the CLC fragments
(not denatured) remain acid insoluble after
S,
treatment. If these fragments, inbuffer H, are
heldat 100Cfor 10min and then cooled to 0C anddigestedwith 0.2 U ofS,at 55 Cfor 30min, then 39% of the fragments remain acid
insolu-ble, although this amount of enzyme will de-grade over 95% of five times this amount of
single-stranded DNA toacid-soluble material.
DISCUSSION
Hydroxyapatite
chromatography
has beenused to isolate a rapidly renaturing fraction of
J.- VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
thesheared vaccinia DNAmolecule. Restriction
enzyme digests of this fraction yield a set of DNA segments which have been separated by
gel electrophoresis. The main features of the
electrophoretic profiles are reproducible and
characteristic of the enzymeusedfordigestion, indicating that the original substrate contained
aset offragments that had a specific nucleotide sequence. It is assumed that the ends of this DNA molecule which have a covalent link con-nectingcomplementary chains (8)arethesource ofthe specific fragments. Berns andSilverman
(3) noted the possibility that such cross-linked DNA was biologically inactive, and that infec-tious virus containeda small percentage of the
DNA which was free of links. IfCLCDNA has thesameduplex basesequence asthepresumed
free DNA plus a small number ofbases in the links ateach endofthemolecule, then
restric-tion enzyme digests of either DNA should produce nearly identical sets of segments. If
single-stranded loops link theendsthey mustbe less than 50 nucleotides long, since electron micrographs of intact vaccinia DNA do not
revealany single-stranded regions (K. I. Berns, personal communication).
The DNA segments which are produced
by
restriction enzyme cleavage are those which include and are adjacent to the covalently linked region. If the target sequence for theenzyme is located near the link, then the
segment containing the link could migrate
through the gel during electrophoresis and be lost, as is the case with any small hairpin
structure. Conversely, ifthetarget sequence is
located at a great distancefrom the link, then
the
duplex
chain would be broken at somerandom position
between
the link and thetarget sequenceduring
the shearing process. Since shearing reduces the molecular size below 8 x 106daltons these fragments wouldnotbe largeenough to migrate as a single band at some
slow, limitingratethrough theagarosegel (11). Also, theelectrophoretic profiles obtained with 1.6% agarose gels show the same pattern as
those obtained with 1% agarose gels (not
shown), where larger DNA segments are more
easily separated. These arguments, plus the
continuous natureoftheelectrophoretic distri-bution ofnondigested sheared fragments (Fig.
2), indicate that the largest DNA segments
produced by restriction enzyme digestion are
specific segments. Random sheared fragments
which have not been removed by the second
hydroxyapatite step do contaminate these gels
but theyarespread through the gel.
The most abundant restriction enzyme seg-ments should be those which contain the link,
and theabundanceofadjacent segments should
decrease with their distance from the link. I
expected two bands in each gel to contain the linked end segments and this seems to besofor the Hpa I digest. The HpaII digest, however, has two regions ofthe gel with a total of five
bands whichcontainsnap-backDNAsegments.
The fourbands
(a-a,)
ofFig. 4could be several forms or variations of one or more linkedseg-ments.Breakingthe covalentlinkmightcause a
conformational change that alters themigration rate. Radioautogramsofgelsafter electrophore-sis of alkali-denatured 32P-labeled DNA
seg-ments produced by Hpa II digestion show a
single band in a region corresponding to the a region. These gels have a considerably higher background level than do gels of
double-stranded DNA so it is harder to distinguish multiple peaks. The possibility remains that
some ofthe a segments are adjacent to linked
end segments and migrate at nearly the same rate. Ifthearegioncontainsbothend segments
then band d ofFig. 4B may be either aspecific degradation product ofthe a segmentsor may come from the interior region ofthe molecule. Band d isalways present as asharp peakafter
electrophoresis of Hpa IIdigests andcontains 20 to 25% of the counts in band a, so it does not seem tobegenerated from banda.One possible
structure however is a predominantly single-stranded molecule which is
generated by
abreaknearthe covalent linkinthe endsegment onthe strandcomplementarytothelong single strand. This
hooked-shaped
structure would have enough double-stranded length to allow retentionby hydroxyapatite but would migrate as asingle chain.Vaccinia doesnothave small repetitive DNA
sequenceswhich wouldrapidly reanneal due to
theirhigh concentration (3). Since thereare no
fixed internal cross-links (8), duplex DNA
formedby rapid reannealing would havetoarise
from single chains folding backon themselves. This would be the case if single chains
con-tained inverted repetitions. Ofcourse, the ends of the vaccinia DNA molecule could be de-scribed as a single chain with a turn-around. However there may be other, internal regions, where the duplex DNA is palindromic and consists ofsingle chains which can
self-associ-ate. Single strands ofeukaryotic DNA which
form duplex molecules with a small
single-stranded hairpinturnhavebeen described(21).
My hydroxyapatite procedure would retain
partial duplex structures which have large or
smallsingle-stranded loops(20). Iftheloopwas very large then this fragment would probably
nothaveenough duplex structure to contain the
on November 10, 2019 by guest
http://jvi.asm.org/
restriction enzyme target sequence. If these putative hairpins contain long duplex regions and a moderately large single-stranded loop, then the loop should be easily digested by the
single-strand-specific
nuclease from vaccinia.Theevidence presented here agrees with earlier results (8) and indicates that the single-stranded turn-around region is small since it is
not readily digested by the vaccinia nuclease. Finally, if the internal hairpins contain long duplex regions and a small turn-around, then there is very little physical difference between them and the ends of vaccinia DNA. These
internal structures would also define specific regions of thevaccinia genome.
The model forvaccinia DNA (8) allows more
than one link at each end of the molecule, but I
can see no reason for having a single-strand hairpin and aninternal cross-link next to each other. A clover leaf arrangement at the end would have several
turn-arounds
butthere is no indication of extensive clover leafbranches inelectron micrographsofvaccinia DNA. The procedurepresented inthis paper shows how restriction enzymes may be usedto cleave limited regions of large viral DNAs. CLC seg-ments maybeobtained
by
digestionofvacciniaDNAwith different restriction enzymes. These
segments can be
redigested
with the H.parainfluenzae
enzymestogivenewelectropho-reticpatternsthatcan be compared with those presented here. This
technique
willidentify
newsegmentsthatareassociated withCLC
regions.
Although no genetic functions may be associ-ated with the CLC
fragments,
they
mayserveasreferencesequencesthat will
help
toestablishaphysical map of vaccinia DNA.
ACKNOWLEDGMENTS
This work was initiated after discussions with B. Moss who provided manyhelpfulsuggestionsduringthisstudyaswell asviral stocksand cells. I also thank NormanCooperforhis assistance and M.Simon and F. W. Studier for their dataon restrictionenzymedigestsofT7DNA. I also appreciate the gift ofvaccinia nuclease from P.Geshelin and K. Berns.
LITERATURE CITED
1. Ando, T. 1966. A nuclease specific for heat-denatured DNA isolated from a product ofAspergillus oryzae. Biochim.Biophys.Acta 114:158-168.
2. Bautz, E. K. F., and J. J. Dunn. 1971. DNA-cellulose
chromatography of proteins, p. 743-747. In G. L.
Cantoni and D. R. Davies(ed.), Proceduresinnucleic acidresearch, vol. 2. Harper and Row, New York.
3. Berns,K.I., and C. Silverman. 1970. Natural occurrence
ofcross-linkedvacciniavirusdeoxyribonucleic acid. J. Virol. 5:299-304.
4. Danna, K. J., G. H. Sack, Jr., and D. Nathans. 1973.
Studies of simian virus40DNA. VI. A cleavage map of theSV40 genome. J. Mol. Biol. 78:363-376.
5. DeFilippes, F. M. 1972. In vitro RNA synthesis from
unique pieces of simian virus 40 DNA produced by a restriction endonuclease. Biochim. Biophys. Acta 272:125-129.
6. DeFilippes, F. M. 1974. A newmethod for isolation of restriction enzyme from hemophilus parainfluenzae. Biochem.Biophys.Res.Commun. 58:586-596. 7. Fairbanks, G.,Jr.,C.Levinthal,and R. H. Reeder. 1965.
AnalysisofC'4-labeledproteinsby disc electrophoresis.
Biochem.Biophys.Res.Commun.20:393-399.
8. Geshelin, P.,and K. I.Berns. 1974.Characterization and
localization of the naturally occurring cross-links in vaccinia virusDNA. J. Mol. Biol. 88:785-796.
9. Goodman, N. C., S. C. Gulati, R. Redfield, and S.
Spiegelman. 1973.Room-temperaturechromatography
of nucleic acids on hydroxylapatite columns in the presence offormamide. Anal. Biochem. 52:286-299. 10. Griffin, B. E., M. Fried, and A. Cowie. 1974. Polyoma
DNA:aphysicalmap.Proc. Natl.Acad. Sci. U. S. A.
71:2077-2081.
11. Helling, R.B.,H.M.Goodman,andH.W.Boyer. 1974.
Analysis ofendonuclease R-EcoRfragmentsofDNA
from lambdoid bacteriophages and other viruses by
agarose-gel electrophoresis.J.Virol. 14:1235-1244.
12. Joklik,W. K. 1962. Thepreparation and characteristics
ofhighly purified radioactively labeled poxvirus. Bio-chim.Biophys.Acta61:290-301.
13. Kohne, D. E., and R. J.Britten. 1971. Hydroxyapatite
techniquesfor nucleic acidreassociation, p. 500-512. In
G. L. Cantoni and D. R. Davies(ed.), Procedures in nucleic acid research, vol. 2. Harper and Row, New York.
14.Lee,A.S.,and R. L.Sinsheimer. 1974. A cleavage map of
bacteriophage OX174 genome. Proc. Natl. Acad. Sci.
U. S. A. 71:2077-2081.
15. Marmur, J., and L. Grossman. 1961. Ultraviolet light
induced linkingofdeoxyribonucleic acid strands and its reversal by photoreactivating enzyme.Proc. Natl. Acad. Sci. U. S. A. 47:778-787.
16. Moss,B., and N. P. Salzman. 1968. Sequentialprotein
synthesis followingvaccinia virus infection. J. Virol.
2:1016-1027.
17. Pogo, B. G. T., and S. Dales. 1969. Two deoxyribonucle-aseactivities within purified vaccinia virus. Proc. Natl. Acad.Sci. U. S. A. 63:820-827.
18. Sharp, P. A., B.Sugden, and J. Sambrook. 1973. Detec-tion of two restriction endonuclease activities in H.
parainfluenzae using analytical agarose-ethidium
bro-mideelectrophoresis. Biochemistry 12:3055-3063. 19.Thomas, C. A., Jr., and J. Abelson. 1966. The isolation
and characterization of DNA from bacteriophage, p. 553-561. In G. L. Cantoni and D. R. Davies (ed.), Procedures in nucleic acid research. Harper andRow, New York.
20. Wilson, D. A., and C. A. Thomas, Jr. 1973.
Hydroxyapa-tite chromatography of short double-helical DNA.
Biochim.Biophys.Acta331:333-340.
21. Wilson, D. A., and C. A.Thomas,Jr. 1974.Palindromes inchromosomes. J. Mol. Biol.84:115-144.
22. Vogt, V. M. 1973. Purification and further properties of single-strand specific nuclease fromAspergillus oryzae. Eur. J. Biochem.33:192-200.
J.
![FIG.1.phosphateDNADNADNAfluenzae;phosphate(B)Allneedlehydroxyapatitediatelydouble-strandednuclease,digestedwasneutralized,wascleases,increasescinia[14C]thymidine, Chromatographyof[3H]thymidinevac- DNA on hydroxyapatite columns at 24 C with buffer, pH 6.8,](https://thumb-us.123doks.com/thumbv2/123dok_us/1565179.109118/3.491.252.441.57.367/phosphatednadnadnafluenzae-allneedlehydroxyapatitediatelydouble-strandednuclease-digestedwasneutralized-increasescinia-chromatographyof-thymidinevac-hydroxyapatite.webp)