0022-538X/88/082696-05$02.00/0
Copyright © 1988, AmericanSocietyforMicrobiology
Activity of
Avian Retroviral Protease
Expressed
in Escherichia
coli
MOSHE KOTLER,t RICHARD A. KATZ, ANDANNA MARIE SKALKA*
Institute for CancerResearch, Fox Chase Cancer Center, 7701 BurholmeAvenue, Philadelphia, Pennsylvania 19111
Received5January 1988/Accepted22April1988
The3' end of the aviansarcomaleukosis virus (ASLV) gag geneencodes a 124-amino-acid protease (PR)
responsibleforprocessingthegag andpolpolyproteinprecursorsinto the mature virion structuralproteinsand the reverse transcriptase. Herewereportthesynthesisof the mature ASLV PR and anucleocapsid(NC)-PR gag precursorfragmentinEscherichia coli.E. coli extractscontainingmature PRcorrectlycleaved asynthetic decapeptide homologoustoaknown ASLVcleavagesite.Also,the NC-PRprecursorfragment appearedtobe correctly processed to produceNC and PR in the bacterial cells. Thiscleavagewasblocked byamutationin the putative active site of PR. These resultsstronglysupportthehypothesisthat PR is involved incleavingitself from the gag precursor.
The retroviralgag,pol, and env genes encode precursor
polypeptides, which are cleaved to produce the mature
forms oftheproteins found in virions (6).The
processing
of gag and pol precursors is directed by a virus-encodedprotease(PR) (7, 8, 18, 26-28) thatbelongs tothe
family
ofaspartic proteases (20, 22, 23). (We have used the recently
proposed nomenclature for retroviral proteins [14]: PR
[aviansarcomaleukosis virus {ASLV}p15]andnucleocapsid protein [NC] [ASLV ppl2].) The PR-directed processing presumably occursjust before or after budding (6). PR is
essential forinfectivity, although viruses thatcontain muta-tionsin PR areabletoassemble intomorphologically imma-ture particles (4, 8, 9, 16, 29, 30).
Unlike the mammalian retroviruses in which PR is
en-coded in the pol gene and thus contained in the gag-pol
precursor (4, 13, 15), the PR ofASLV is encoded in gag.
Therefore, the PRdomain is contained inboth the C
termi-nusof thePr769'9precursor andthePr18Vag-p precursor
(6). Forboth avianandmammalianretroviruses, mature PR
isreleasedfromprecursorproteins by proteolysisduringthe
morphogenesis ofviralparticles. Since 20 to 50 times more
of thegag thanthegag-pol precursor isproduced,the bulk of mature ASLV PRis formedas aconsequenceofcleavage
betweenPRandtheadjacentNC(ppl2), locatedupstream in
thegag precursor (21). In contrast, release of PR in
mam-malian viruses, such asMoloneymurine leukemia virus and
humanimmunodeficiencyvirus,requires cleavage at both N
andC termini ofthe PR in thegag-polprecursor (13, 15). It is not known exactly how PR is released from these precursors. Atleasttwomechanisms are possible. (i)
Cellu-lar proteases may be
responsible
for the initiation of the process, resulting in the release of a few mature PRmole-cules. The releasedPRmightthen complete the processing,
producing more PR and the other mature viral proteins as well.
(ii)
PR might cleave itself from the precursor via either aninter-orintramolecularmechanism. Similar mechanisms ofactivationareknown for a number of asparticproteases,
in which an inactive zymogen is converted into an active enzyme by autocatalysis. Oneexample is the autocleavage ofpepsinogen to pepsin (22).
Inthisreport, we describe the expression in Escherichia
coli of the predicted ASLV PR-coding region. A protein
*Correspondingauthor.
tPermanent address: Department of Molecular Genetics, the HebrewUniversity, Hadassah Medical School, Jerusalem, Israel.
structurally andimmunologically indistinguishablefrom PR was produced, and a proteolytic activity analogous to the viral PR was detected in E. coli extracts. These results confirm the location of the ASLV PR-coding region. In
addition, we show that anNC-PR gag fragment
undergoes
processingin E. coli, resultingin theformation of amature PRthat appears tobe identical tovirion PR. Mutation ofa highly conservedaspartic acid residue that is thought to be partof thePR active site eliminates proteolytic activity. This
demonstratesthatthe ASLV PR is responsible for
releasing
itself from the NC-PR gag precursorfragment
in E. coli.MATERIALS ANDMETHODS
Bacterialcells. E.coliMC1061(3)wasused as a hostfor all expression vectors. The cells contained the plasmid pRK248cIts, which directs expression of a temperature-sensitive bacteriophage lambda repressor protein (5).
Constructionof thebacterialexpressionclones.The bacte-rial expression clones were constructed by using the
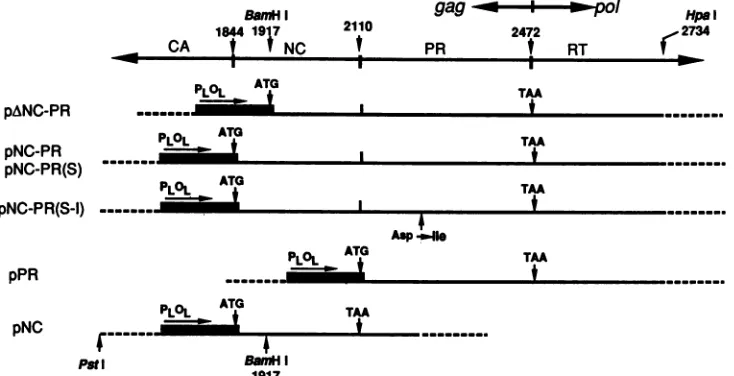
pEV-vfrexpression vectors which supply a lambda phage promot-er-operator region (PLOL), a ribosome-binding site, and a translational initiation codon (5). The pANC-PR clone was constructed by replacement of a pEV-vrf2 BamHI-PvuII
fragment with a viral 817-base-pair BamHI-HpaI fragment
(Fig. 1) derived from an infectious clone of the avian sarcoma virus PrC strain, pATV-8 (11). The viral DNA fragment contains the C-terminal two-thirds of the
NC-coding region, all of the PR-coding region including the translational terminationcodon for Pr76 gag, and the 5' end of thepol gene. The construction of pNC (formerly called
pRKP12) was described previously (10). The pNC-PR plas-mid wasconstructed by substitution ofaca. 1-kilobase-pair
pNC-derived PstI-BamHIfragmentinto pANC-PR(Fig. 1). Thisfragmentcontained thePLOLregion andthe ribosome-binding site ATG region properly aligned for expression of thenative Nterminus of NC (10). The pPR expression clone was constructed from pANC-PR by using oligonucleotide-directed, site-specificmutagenesis (10, 19) to delete the NC region and to precisely align the ATG codon in the vector with the first codon of the mature PR, a strategy used
previouslyto construct pNC (10). Theoligonucleotide used for deletion mutagenesis was 5'-AGGAGGAATTAAT ATGTTAGCGATGACAATGG-3'. A mutation was intro-ducedinto theputativeactive site region of PR (20, 23) by using site-directed oligonucleotide mutagenesis (19). The construct coding for the mutant PR [pNC-PR(S-I)] was
2696
on November 10, 2019 by guest
http://jvi.asm.org/
BamH I
1844 1917
^A a ..
pANC-PR pNC-PR pNC-PR(S) pNC-PR(S-I)
gag
I
p1
2110 2472
--a_. _
Hpi I
ZO2734
_CA
I
NC v PR v RT I _P OL ATG
i ~~~~TAA
PLLAT
TM
a...M. , ...I * ____
PLOL
-i.
ATG TMi _
Asp-m.419
PL0L
,
ATGi
pPR
pNC r--- -- * ________.
PLOL ATG
PstI
TM
i
TM
i
I
1917
FIG. 1. Maps of PR, NC-PR, and NC E. coliplasmidexpression clones. All clones are derivatives of pEV-vrf vectors (5). A partial map of theviral DNA clone pATV-8 (11), from which PR and NC sequences were derived is shown at the top; the flankingcapsid (CA) andreverse transcriptase (RT) regions are indicated. Nucleotide positions indicated correspond to the avian sarcoma virus sequence described by Schwartzetal. (21). Symbols: ---, pBR322-derived vectorsequences; _,bacteriophage lambdapromoter-operator region (PLOL)(not drawntoscale). The initiation codon provided by the expression vector is indicated (ATG). The position of thetermination codon forthe PR-coding regionisindicated (TAA). The site of the Asp-*Ile mutationintroduced into the putative active site of PR isshown. Numbers indicate nucleotide positions.
provided by V. M. Vogt. It was derived by site-directed
mutagenesisattheGAC codon that encodes aspartic acid 37, which converted this codon to an ATA (isoleucine). In this construct, a portionof NC-PR sequences are derived from the Schmidt-Ruppin strain of avian sarcoma virus. A wild-typeversion of this clone was also constructed [pNC-PR(S)]. Analysis of NC and PRexpressed in E. coli. The ASLV NC and PRexpressed in E. coli were detected by
immunoblot-ting with rabbit sera directed against NC (10) and PR.
Anti-PR antibody wasprepared by immunization of rabbits with purified viral PR provided by J. Leis. Expression in cellscontainingthevarious vectors was induced by shifting
cultures from 30 to 42°C for 2 to 4 h, and virus-related proteins were detected by immunoblotting as described
elsewhere (1, 10, 24).
Preparation of bacterial extracts. Induced and uninduced E.coli cultures (1.75 ml)werepelletedandsuspendedin 400
,Iu of 1%toluene-10 mMTrishydrochloride (pH8.0)-S5mM
mercaptoethanol-0.1
mMEDTA-0.1%Triton X-100.Bacte-rial cellsuspensionswerevigorously agitated by vortexing (3
min)andwereincubated for20 min at37°C. Unbrokencells andcell debriswerepelleted by centrifugation. Supernatants
were freeze-dried and dissolved in 50
RI
of water, from which 2RI
was assayed.Synthesis and cleavage ofdecapeptidesubstrates. The
syn-thesis and
purification
ofthedecapeptides
Thr-Phe-Gln-Ala-Tyr-Pro-Leu-Arg-Glu-Ala (wild
type) and Thr-Phe-Gln-Ala-Ala-Pro-Leu-Arg-Glu-Ala(variant)
are described elsewhere (M. Kotler, R. A. Katz, W.Danho,
J.Leis,
and A. M. Skalka, Proc. Natl. Acad. Sci. USA, in press). ASLV PR cleaves thewild-typepeptide
betweenTyrand Pro(Kotler
et al., inpress).Proteolytic
reactionswerecarriedoutin 10,ul
of0.1 Msodium citrate buffer
(pH 5.5) containing
1.1 mMofdecapeptide substrate and 2
pul
of bacterial extract. Thereactions were incubated at 37°C for 10 h. The
cleavage
productswereanalyzed bya
thin-layer
electrophoresis
assay(Kotler et al., in press).
Samples
of4pul
werespotted
oncellulose plates (Art 5577, 20
by
20 cm; E. M.Reagents,
Federal Republic of Germany). Plates were subjected to 45 mA for 1 h in pyridine-acetic acid-acetone-distilled water (volume ratio, 1:2:8:40). They were then air dried and
sprayed first with 1% triethylamine (Pierce Chemical Co.,
Rockford,
Ill.)
inacetoneand thenwithfluorescamine (Hoff-mann-La Roche Inc., Nutley, N.J.)at0.1 mgof acetone per ml. Photographswere taken under UV light.RESULTS
Expression of PR in bacteria. ASLV PR is an abundant
virionprotein formed byproteolytic processing;itis
derived
from the Cterminus ofthe Pr769'9 precursor. Forexpressionof the mature PR inE. coli,a translationalinitiation codon
provided by theexpression vector wasintroduced immedi-ately
preceding
the PRreading
frame(Fig.
1, PR). Noalterationwasnecessaryatthe3'end, since the termination
[image:2.612.132.499.65.253.2]codon ofPRcanalsofunctioninbacteria(Fig. 1). Thesame vectorwas also used forexpression ofa viral NC-PR gag precursorfragment derived fromSR-A aviansarcomavirus
[pNC-PR(S)]
or PR-C avian sarcomavirus(pNC-PR)
(Fig.
1). A truncated version of thepNC-PR
clone which ismissingtheN-terminalone-thirdof NCwasalsoconstructed (pANC-PR).
E.colitransformed withpPRwasgrowntomid-log
phase
at30°C. Expressionfrom the PL promoterwastheninduced byshiftingtheculturesto
42°C.
Cellswereharvested 3to4 hpostinduction,
andproteins
wereanalyzed by
polyacryl-amidegel
electrophoresis
andimmunoblotting,
with antiseradirected
against
thepurified
ASLV PR and NC. Aprotein
that comigrated with viral PRwas
produced
in the induced but not in the uninduced cells(Fig. 2A,
lanes 1 and2).
Neither the viral PRnorthebacterially
produced
PRreacted with anti-NC serum(Fig.
2B, lanes 1 and 2), asexpected.
FromCoomassie bluestaining,
it canbe estimatedthat PR represents 5 to10%
of the totalprotein
in inducedbacteria (datanotshown).Synthesisandcleavage ofNC-PR inbacterial cells. Immu-noblot
analysis
oftheproteins
produced
after induction of.mn.mm- m I I
I
"mm ...m
on November 10, 2019 by guest
http://jvi.asm.org/
A
-ANC-PR
7 -PNC-PR
1 2 3 4 5 6 7 8
B
a: a: <0 cr
> EL z z
-L a Ca
Zn U I U I U I U I
43-25- .. ..:.:_...§-PR
1ANC-PR
25-12 ..=t-NC-
-
6-12 345 678
FIG. 2. Immunoblot analysis ofPRand NC productsexpressedinE. coli.E. colilysateswerefractionatedona15% polyacrylamide-sodiumdodecylsulfate gel,and virus-relatedproteinsweredetectedbyusingrabbit antiserum directedagainstASLV PR(A)orNC(B)and 1251I-labeledprotein Gasdescribed elsewhere (1).LysateswerepreparedfromE.colicontaining pPR (lanes1 and2),pANC-PR (lanes3 and 4),pNC-PR(lanes5and 6), andpNC (lanes7 and8).Lanes1, 3, 5, and7,lysatesfromuninduced(U)cultures;lanes2, 4, 6, 8, lysatesfrom cultures induced(I)for3 h at42°C. Molecularsize markersareindicatedinkilodaltons. Viral NCand PRmarkersareshown. The anti-PR serum (A) cross-reacts withanumberofE.coliproteins,as seeninlysates from uninduced cultures(lanes 1, 3, 5,and7).Theanti-NC serum (B)cross-reacts with an E.coli protein ofca. 35kilodaltons.
thepNC-PR vectoridentifiedaproteinof themasspredicted for the NC-PR precursor fragment (ca. 23 kilodaltons); it reacted withboth anti-PR and anti-NC (Fig. 2AandB,lanes 5and 6). Two smaller proteins which had migration coeffi-cientscorrespondingtothose ofproteins produced by pNC andpPR and those ofthe analogous purified viral proteins werealsoidentified. One of these two proteins reacted with anti-NC, while the other reacted with anti-PR, indicating that the NC-PR precursor underwent cleavage at (or near) the siterecognized duringvirusmaturation(Fig. 2A and B,
lanes5 and6). Analysis ofproteins produced after induction ofpz&NC-PR gave similar results (Fig. 2AandB, lanes 3 and 4). As expected, both the precursor fragment and the
NC-related productwere shorter than those produced by pNC-PR, while the PR product was unaffected. This result
con-firmed the identities of the small anti-NC and anti-PR
reactive bands.
One explanation for the production of small,
discrete-sized products is that the NC-PR and ANC-PRprecursors werecleaved specifically by PR. To test this hypothesis, we used a mutated clone in which the codon for the highly conserved Asp residue(amino acid 37 in PR) located in the
putativeactive site of PR (20) was changed to an Ile codon
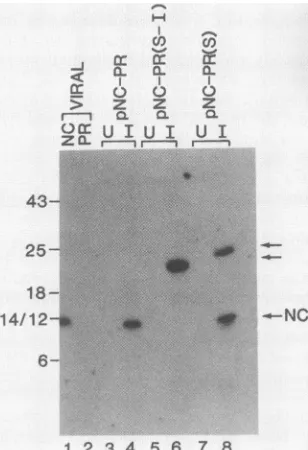
[pNC-PR(S-I), Fig. 1]. Expression of proteins from this
plasmid was assayed by immunoblotting with the anti-NC serum. The results showed that the NC cleavage product wasproduced from the wild-type construct, pNC-PR(S), but notfromthe mutant,pNC-PR(S-I) (Fig. 3, compare lanes 6
and 8). Thus, we conclude that bacterially produced PR
activity is responsible for specific cleavage of the NC-PR gag precursor fragment. The protein which contained the Asp toIle mutation migrates more rapidly than the wild-type NC-PR protein (Fig. 3, lane 6; see arrows), indicating that this change may have a significant effect on the protein
conformation.
Detection of PR activity in cell extracts. To confirm that an active PR was produced in E. coli, proteolytic activities
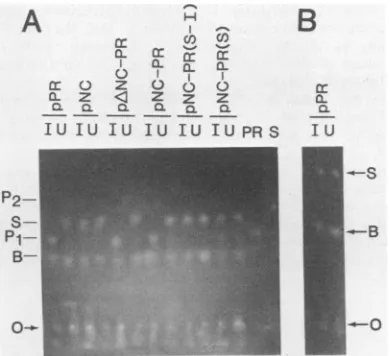
encodedbyallsix plasmids described in Fig. 1 were tested in cell extracts by using a synthetic peptide cleavage assay
(Kotler et al., in press). We have previously shown that
decapeptideswhich are composed of a known ASLV
prote-olytic processing site in pol (between the ot and pp32PoI
domains) are precisely cleaved by ASLV PR in vitro. A change inone of the amino acids which flanks thecleavage
site(e.g.,TyrtoAla) abolishes thiscleavage(Kotleretal., in press).
Extractsprepared from induced cells which express proc-essed PR [from plasmids pPR, pNC-PR, pANC-PR, and pNC-PR(S)] contained activities which cleaved the suscep-tible peptide. Extracts from cells which express pNC or
pNC-PR(S-I), which donotproduce processedPR, showed
00
0- C
a-> z z z
z
0-~
u
43- l Q
25
-18- .. .!...
14/12-f;,
NC
6-FIG. 3. Immunoblotanalysisof NC-PRproteins expressedin E. ccli.Analysis was carriedoutas described in thelegend toFig. 2, except that cultureswereinduced for 4 h.NC-relatedproteinswere
detectedbyincubation with rabbitanti-NC,followedby "'5I-abeled
protein G. Lysates were prepared from cells containing pNC-PR (lanes3 and4), pNC-PR(S-1) (lanes5and6),andpNC-PR(S) (lanes 7 and 8). Lanes 4, 6, and 8 contain lysates from cells that were
induced forexpression (I); lanes 3, 5 and 7 contain lysates from uninduced(U)cells. Lanes1and 2 contain authentic viralproteins.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.141.469.65.217.2] [image:3.612.357.511.405.630.2]no cleavage (Fig. 4A). As a control, a peptide containing a single amino acid change at the cleavage site (Tyrto Ala) was incubated with the same E. coli extracts. No cleavage was seen even after a 10-h incubation. Byusingthis sensitive peptide assay, low levels of PR activity could be detectedin some extracts prepared from uninduced cells which con-tained plasmids with the wild-type PRsequence,althoughno PR-related proteins could be detected by immunoblotting (Fig. 2 and 3). We presume that this is duetolowexpression (leakiness) of the respective plasmids under uninduced con-ditions. Since the peptides are quite stable incrudeextracts, the reaction mixtures can be incubated for very longperiods of time, thus further increasing the sensitivityofthe assay.
DISCUSSION
In this report, we describe expression of a retroviral protease in bacteria. This enzyme isactiveinbacterial cells as well as in cell extracts. Amutationintheputative active site of PR (Asp to Ile) in the pNC-PR construct resultedina failure to release NC and PR. Thisresultsuggests thatPRis responsible for releasing itself from the gag precursor. However, the experiments with the mutated PR cannot exclude limited participation ofcellular proteases. For ex-ample, it might be imagined that limited proteolysis by a cellular protease could release some maturePR molecules, and these might then be responsibleforadditionalprocessing
A
cc
cc z
-LZ <
a
IU IU IU
--l
CD) C
z z aD-z
IU IU IUPRS
B
c:
0-IU
P2-
S-
P1-
B-FIG. 4. DetectionofPR-specificactivity in bacterialextractsby
using a thin-layer electrophoresis assay. Synthetic decapeptides
containingTyr-Pro(wildtype)(A)orAla-Pro(mutant)(B)cleavage
siteswereincubatedwithbacterialextractspreparedfrom induced
(lanes I)anduninduced(lanesU)cultures. Asacontrol,theTyr-Pro
peptide was subjected to purified viral PR (lane PR) or was incu-batedwithnoPR(laneS).Theextractswerepreparedfromcultures containingthe indicated clones. After incubation ofpeptides with
extracts,themixtureswerefractionated by thin-layer
electrophore-sis(seeMaterialsandMethods). The mobilitiesofpeptidesubstrates
(S), products (P1andP2),andanunidentified spotassociatedwith thebacterialextract(B)areindicated. Theoriginis indicated(0).P1 isapentapeptidecleavageproductcorrespondingtotheN terminus
of the decapeptide substrate. P2 (dark spot) corresponds to a
pentapeptidederivedfrom theC terminus.Productswerelocatedby
stainingwithfluorescamine,followed byvisualizationwith UVlight
(see MaterialsandMethods). The P2spotappears dark becauseof theN-terminal Pro whichproduceschromophores that absorbUV light.Detailsofthemethodologyand characterization of the
prod-uctsidentified bythethin-layerelectrophoresis assayareprovided
elsewhere(Kotleretal., inpress).
which would release more active PR molecules. If the
processing is due entirely to PR
activity,
it would be of interesttodeterminewhether thefirstmaturemoleculesare released as aconsequence of inter- or intramolecular inter-actions. Introduction of mutationsintotheNC-PRcleavage
site mayprovide
someinsight
into thisquestion.
Our
finding
that the PR-NC gag precursorfragment
is cleavedintracellularly
is consistent with earlier reports ofproteolytic
processing
ofthe ASLV gag precursor inE.coli
(17) and human
immunodeficiency
virusgag-polprecursors in yeastcells(13). In contrast, the ASLVPr76fia
precursorsynthesized in vitro
by
using
a rabbitreticulocyte
system does notundergo
processing
unlessPRis added(28). Also,
Pr769' is not
processed
in mammalian cells infectedby
avian sarcoma virus
(25),
and these cells are unable to supportassembly
of infectiousparticles
(2,
12).
There areseveral
possible
explanations
for these differences.(i) Avian,
bacterial,
and yeastcells,
but not mammaliancells,
might
containaprotease which is abletoactivatePR
by
releasing
it. The active PR molecules
might
thencomplete
theproc-essing as mentioned above.
(ii)
Mammalian cells maycon-tain aninhibitor of ASLV PR which prevents
autocleavage
or
subsequent
enzymatic
activity.
(iii)
The releaseofPRmayrequire
dimerization ofthe precursor(20)
or an intermolec-ularreactionwhichoccursonly
whenahigh
concentration ofprecursor is present, as in
bacteria,
yeast cells, and thepermissive
avian cells.Thresholdconcentrationsmaynotbeachieved in vitro or in mammalian cells.
(iv)
ThePr769'9
may be folded in aspecific
structure which preventsauto-cleavage
until the time ofvirionbudding.
Subsequent
acti-vation of the proteasemight
occur in virionsproduced
in avian cells but not in mammalian cells. Precursorproteins
producedinbacteria
might
notfold in themannerrequired
to preventcleavage.
Some ofthesemodels may be testableby
using
methodsdescribed inthis paper.The bacterial
expression
systemsdescribed here will allow ustoproduce
large
amountsofnormal andalteredASLV PRproteins.
These should be usefulforstudying
thebiochemi-cal
properties
ofPRandforproviding
material forbiophys-ical
analyses.
The gagprecursorfragment (NC-PR)
will beauseful substrate to test the effect of various
cleavage
site mutations. Sincecleavage
occurs inbacteria,
it should bepossible
tostudy
the effect of such mutations withoutpurification
ofthe PR or substrates.ACKNOWLEDGMENTS
Wewishtothank W. Danhoforthe
synthesis
ofpeptides,
J. Leis for supplying purified viral PR, F. Erlitz forthesynthesis
ofthe oligonucleotides, R. Crowl forpEV-vrf
DNA,and D. Markham for adviceon extractpreparation.
Wegratefully acknowledge
V.Vogt,
K. Jones, J. Kulkosky, and P. Tsichlis for critical reviews ofthe manuscript. We also thank V.Vogt
and G. Schatz forproviding
mutantconstructs and for
helpful
andstimulating
discussions. This work wassupported
by
Public Health Service grants CA-06927and RR-05539 fromtheNational Institutes ofHealth, agrantfrom the Pew Charitable Trust, and an
appropriation
from the Commonwealth ofPennsylvania.
Partial support for R.A.K. wasprovidedby the W. W. Smith CharitableTrust. LITERATURE CITED
1. Alexander, F., J. Leis, D. A. Soltis, R. M. Crowl, W. Danho,
M.S. Poonian, Y.-C. E.Pan, andA. M. Skalka. 1987. Proteo-lyticprocessingof aviansarcomaandleukosisviruses
pol-endo
recombinant
proteins
revealsanotherpolgenedomain.J.Virol.
61:534-542.2. Altaner, C., and H. M. Temin. 1970.
Carcinogenesis
by RNAsarcoma viruses. XII. A
quantitative study
ofinfection ofraton November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.90.285.348.526.2]cellsin vitroby avian sarcomaviruses. Virology 40:118-134. 3. Casadaban, M. J., and S. N. Cohen. 1980. Analysis ofgene
control signals byDNAfusion andcloning inE. coli. J. Mol. Biol. 138:179-207.
4. Crawford, S.,andS. P. Goff.1985.Adeletionmutation in the 5' part of thepolgene ofMoloneymurine leukemia virus blocks proteolyticprocessing ofthe gag andpolpolyproteins.J. Virol. 53:899-907.
5. Crowl, R.,C. Seamans,P. Lomedico,andS. McAndrew. 1985. Versatile expression vectorfor high-level synthesis of cloned geneproductsin Escherichia coli. Gene 38:31-38.
6. Dickson, C., R. Eisenman, H. Fan, E. Hunter, and N. Teich. 1984. Proteinbiosynthesis and assembly, vol. 1, p. 513-648. In R.Weiss,N.Teich,H.Varmus, andJ.Coffin(ed.),RNA tumor viruses. Cold Spring HarborLaboratory, ColdSpring Harbor, N.Y.
7. Dittmar, K. J., and K. Moelling. 1978.Biochemicalproperties of p15-associatedprotease in anavianRNAtumorvirus.J.Virol. 28:106-118.
8. Eisenman, R.N., W. S. Mason, and M. Linial. 1980. Synthesis andprocessing of polymerase proteins ofwild-type andmutant avianretroviruses. J. Virol. 36:62-78.
9. Katoh, I., Y.Yoshinaka,A.Rein, M.Skibuga,T.Odaka, and S. Oroszlan. 1985. Murine leukemia virus maturation: protease region required for conversion from "immature" to "mature" coreform and forvirusinfectivity. Virology 145:280-292. 10. Katz, R. A., X.-D. Fu, A. M. Skalka, and J. Leis. 1986. Avian
retrovirus nucleocapsid protein, ppl2 producedin Escherichia coli has biochemical properties identical tounphosphorylated viral protein. Gene50:361-369.
11. Katz, R. A., C. A. Omer, J. H. Weiss, S. A. Mitsialis, A. J. Faras, and R. Guntaka. 1982. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned uninte-grated avian tumor virus DNA: structure of large terminal repeatsincircle junctions. J.Virol. 42:346-351.
12. Kotler, M. 1971. Interaction of avian sarcoma virus with rat embryo cells in culture.J.Gen.Virol. 12:199-206.
13. Kramer, R. A., M. D.Schaber, A. M. Skalka, K. Ganguly, F. Wong-Staal, andE. P. Reddy. 1986. HTLV-III gag protein is processedinyeastcellsby the viruspol-protease. Science 231: 1580-1584.
14. Leis, J., D. Baltimore, J. M. Bishop, J. Coffin, E. Fleissner, P. Goff, S. Oroszlan, H. Robinson, A.M. Skalka, H. M. Temin, and V.Vogt. 1988. Astandardizedand simplifiednomenclature forproteinscommon toall retroviruses.J.Virol.62:1808-1809. 15. Levin, J. G., S. C. Hu, A. Rein, L. I. Messer, and B.I.Gerwin. 1984. Murine leukemia virus mutant with a frameshift in the
reverse transcriptase coding region: implication for pol gene
structure. J. Virol. 51:470-478.
16. Lu,A.H.,M. M.Soong,and P. K. Y.Wong.1979.Maturation ofMoloneymurine leukemia virus. Virology93:269-274. 17. Mermer,B.,M.Malamy,andJ.M.Coffin. Roussarcomavirus
contains sequenceswhichpermit expressionof the gag gene in Escherichia coli. Mol. Cell. Biol. 3:1746-1758.
18. Moelling, K.,A.Scott,K. E.J. Dittmar,andM. Owada. 1980. Effect of p15-associated protease from an avian RNA tumor virus on avian virus-specific polyprotein precursors. J. Virol. 33:680-688.
19. Morinaga,Y.,T.Franceschini, S., Inouye,andM.Inouye.1984. Improvementofoligonucleotide-directedsitespecific mutagen-esis using double-stranded plasmid DNA. Bio/Technology 2: 636-639.
20. Pearl,L.H.,andW. R.Taylor.1987. A structural model for the retroviralprotease. Nature (London) 329:351-354.
21. Schwartz, D.E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequenceofRous sarcoma virus. Cell32:853-869.
22. Tang, J.,and R. N. S. Wong. 1987. Evolution in the structure and function ofaspartic proteases.J. Cell. Biochem.33:53-63. 23. Toh, H., M. Ono, K. Saigo, and T. Miyata. 1985. Retroviral protease-like sequence in the yeast transposon Ty-1. Nature (London)315:691.
24. Towbin, H.,T.Staehelin,andJ.Gordon. 1979. Electrophoretic transfer ofprotein from polyacrylamide gels to nitrocellulose sheets:procedureand someapplications.Proc.Natl. Acad. Sci. USA76:4350-4354.
25. Vogt, V. M., D. A.Bruckenstein, and A. P. Bell. 1982. Avian sarcoma virus gag precursorpolypeptide is notprocessed in mammalian cells.J. Virol. 44:725-730.
26. Vogt, V. M., R.Eisenman, and H. Diggelmann. 1975.Generation of avianmyeloblastosis virus structural proteins byproteolytic cleavage of a precursor polypeptide. J. Mol. Biol. 96:471-493. 27. Vogt, V. M., W. Wight, and R. Eisenman. 1979. In vitro
cleavageof avianretrovirus gag proteins by viralproteasep15. Virology 98:154-167.
28. Von der Helm, K. 1977. Cleavage of Rous sarcoma viral polypeptide precursorintointernal structural proteins in vitro involvesviralprotein p15.Proc. Natl.Acad. Sci. USA 74:911-915.
29. Voynow, S. L., and J. M. Coffin. 1985. Truncated gag-related proteins are producedby large deletion mutants of Rous sar-comavirus andform virus particles.J.Virol. 55:79-85. 30. Witte, 0.N., and D. Baltimore. 1978. Relationshipofretrovirus
polyprotein cleavagestovirion maturationstudied with temper-ature-sensitive murineleukemia virusmutants. J. Virol. 26:750-761.