Copyright © 2002, American Society for Microbiology. All Rights Reserved.
Regions of the Herpes Simplex Virus Scaffolding Protein That
Are Important for Intermolecular Self-Interaction
Valerie G. Preston* and Iris M. McDougall
MRC Virology Unit, Institute of Virology, Glasgow G11 5JR, United Kingdom
Received 9 July 2001/Accepted 1 October 2001
The herpes simplex virus type 1 (HSV-1) scaffolding protein encoded by gene UL26.5 promotes the formation of the icosahedral capsid shell through its association with the major capsid protein VP5 and through intermolecular interactions with itself. Inside the capsid shell, the UL26.5 product together with the matura-tional protease, a minor protein, form a spherical structure which is broken down and released from the capsid during packaging of the viral genome. Selected residues from four internal regions of the HSV-1 scaffolding protein that have significant conservation of amino acids within the scaffolding proteins of alphaherpesviruses were mutated, and the properties of the proteins were examined. Only the HSV-1 scaffolding protein with mutations in the conserved N-terminal domain showed reduced interaction with the varicella-zoster virus homologue in a cell-based immunofluorescence assay, providing the first evidence that this domain in the HSV-1 protein is likely to be involved in intermolecular self-interaction. Scaffolding protein with mutations in this domain or in either of two other domains failed to assemble into scaffold-like particles but retained the ability to self-interact, although the aggregates were significant smaller than most of the aggregates formed by the wild-type protein. These results suggest that there are multiple domains involved in the intermolecular self-association of the HSV-1 scaffolding protein that can act independently of one another. This conclusion was supported by the observation that none of the mutant proteins with lesions in an individual domain, including a protein with mutations in a central region previously implicated in self-interaction (A. Pelletier, F. Doˆ, J. J. Brisebois, L. Lagace´, and M. G. Cordingley, J. Virol. 71:5197–5208, 1997), interfered with capsid assembly in a baculovirus expression system. A protein mutated in the central region and another conserved domain, both of which had been predicted to form coiled coils, was impaired for capsid formation but still retained the capacity to interact with VP5.
The genome of herpes simplex virus type 1 (HSV-1) is en-closed inside a large T⫽16 icosahedral capsid, ⬃1,250 Å in diameter and⬃160 Å thick within the virus particle. The as-sembly pathway of the mature capsid has striking parallels with those of double-stranded DNA bacteriophages, such as T4 and P22 (2, 27). One common feature is that these viruses all have internal scaffolding proteins that are not present in the mature virion but are required for the formation of the precursor capsid prior to encapsidation of the genome. Two genes specify the scaffolding proteins of HSV-1. UL26.5 encodes the major component of the scaffold or protein core present in the cavity of the procapsid, the precursor capsid into which the DNA is packaged. UL26 specifies the minor constituent of the scaffold. The precursor protein of UL26 consists of two distinct func-tional domains. The N-terminal region, comprising 247 amino acids, is a serine protease, and the C-terminal region, of which the last 329 amino acids are identical to the full-length UL26.5 product, interacts with the major scaffolding protein (7, 15, 16, 34). In the absence of the scaffolding proteins, incomplete capsids are produced (5, 33, 35). The activity of the protease is not required for the assembly of the capsid shell, and large numbers of intact capsids are generated in the presence of the UL26.5 product only or the scaffolding portion of the UL26 protease (8, 25, 33, 35). Although the UL26 product can
par-tially replace the UL26.5 protein, its prime function is likely to be the release of the scaffold from the capsid through the action of the maturational protease (17, 30). During capsid assembly, the UL26 and UL26.5 products associate and inter-act with the major capsid protein VP5, which forms the 12 pentamers at the vertices of the capsid and the 150 hexamers on the triangular faces (4, 11, 13, 20, 34, 37). The capsid shell proteins VP19C and VP23 bind together to form a trimer, referred to as the triplex, consisting of two copies of VP23 and one copy of VP19C, which connects adjacent capsomers on the capsid (20, 31, 32). The maturational protease is required for the conformational transition of procapsids to mature angu-larized capsids in vivo (8, 21, 25, 29). The protease cleaves itself at an internal site, separating the proteolytic domain, VP24, from the scaffolding region, VP21 (6, 14). It also breaks the connections between the scaffold and the inner capsid shell wall by cleaving the carboxy-terminal ends of scaffolding pro-teins, detaching the region containing the VP5 binding site from the scaffolding core (38). This proteolytic step is essential for the release of the scaffold from the capsid and packaging of viral DNA (8, 25).
In the absence of capsid shell proteins, the HSV-1 major scaffolding protein readily forms spherical particles approxi-mately 60 nm in diameter when extracted from purified capsids and renatured (18). In insect cells infected with a recombinant baculovirus expressing the major scaffolding protein, scaffold-like particles are also detected, especially when the C-terminal cleaved form (VP22a) is synthesized or produced by proteo-lytic cleavage. In addition, large aggregates of protein with a
* Corresponding author. Mailing address: MRC Virology Unit, In-stitute of Virology, Church Street, Glasgow G11 5JR, United King-dom. Phone: 44 141 330 4636. Fax: 44 141 337 2236. E-mail: v.preston @vir.gla.ac.uk.
673
on November 8, 2019 by guest
http://jvi.asm.org/
less defined structure are observed (24). Recently, a purified hybrid human cytomegalovirus (HCMV)/HSV-1 scaffolding protein was shown to form oligomers, ranging from dimers to complexes containing 20 to 30 or more copies of the hybrid protein (19).
The regions of the HSV-1 and HCMV scaffolding proteins required for intermolecular self-association have been investi-gated, and different results have been obtained for each pro-tein (4, 23, 36). In the HCMV scaffolding propro-tein, an N-ter-minal domain located between residues 34 and 52 and containing amino acid residues highly conserved among alpha-, beta-, and gammaherpesviruses was found to be required for intermolecular interaction of the protein in the yeast-two hy-brid system (36). A mutant protein with a single point mutation in this region failed to interact with the wild-type (wt) virus scaffolding protein, suggesting that this region is essential for self-interaction. In contrast, a study with the HSV-1 scaffolding protein using yeast two-hybrid and far-Western blot analyses implicated several regions in self-association (23). These in-cluded a predicted␣-helical domain lying in the center of the protein between residues 164 and 219, a region within residues 54 to 164 on the N-terminal side of the central domain, and another on the C-terminal side. Disruption of the central do-main did not abolish interaction of the mutant protein with the wt scaffolding protein in the yeast-two hybrid system, suggest-ing that there are multiple domains involved in intermolecular self-interaction. The region corresponding to the HCMV scaf-folding protein N-terminal domain was not required for self-association in the yeast-two hybrid analysis. A weak interaction with the wt protein, however, was detected using a mutant containing amino acid residues 1 to 120 of the HSV-1
scaffold-ing protein as a probe in far-Western blot analyses, whereas a strong interaction was obtained using a mutant protein span-ning the central core region only (amino acid residues 164 to 219).
We have investigated regions of the HSV-1 scaffolding pro-tein implicated in intermolecular self-interaction by using the intact protein not linked to any other protein or domain, unlike the approaches used in previous studies. In particular, we have explored whether the region in the HSV-1 protein, homolo-gous to the N-terminal region in HCMV, is important in this association. We have also determined whether disruption of any of these domains has any effect on capsid assembly and scaffold formation, an aspect that has not been studied previ-ously.
MATERIALS AND METHODS
Cells and viruses.Vero cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with penicillin, streptomycin, and 10% fetal calf serum. Sf21 AE cells were grown as described previously (13). The isolation and char-acterization of baculoviruses AcVZV33.5, AcUL26, AcUL26.5, AcUL35, and AcUL18/19/38, which express the varicella-zoster virus (VZV) scaffolding pro-tein, UL26 protease, the UL26.5 product, the UL35 product, and the essential outer capsid shell proteins, respectively, have been reported elsewhere (24, 26, 33).
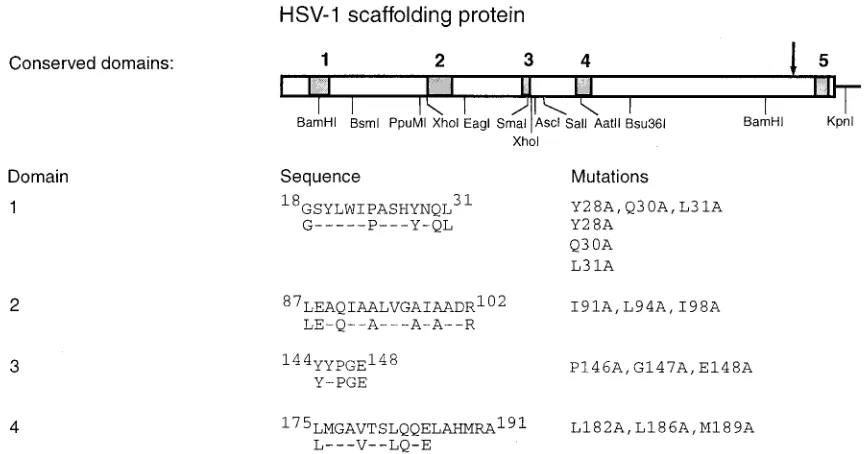
Plasmids.With the exception of the triple mutation in domain 3, missense mutations were introduced into conserved domain 1, 2, 3, or 4 of the UL26.5 gene within the plasmid pGX262 DNA by ligation of a short restriction nuclease fragment containing the mutation to the appropriate restriction endo-nuclease fragments (Fig. 1) (24). Two overlapping complementary synthetic oligonucleotides were annealed together to produce the mutant fragment. Mu-tant forms of the UL26.5 gene with defects in domain 1 were generated by ligating theBamHI-KpnI fragment, containing all the vector sequences from pGX262, to a BsmI-KpnI fragment encoding the carboxy-terminal portion of the UL26.5 open reading frame and aBamHI-BsmI synthetic double-stranded
oli-FIG. 1. Mutations constructed for analysis of HSV-1 scaffolding protein. At the top, the locations of domains within the HSV-1 scaffolding protein that are conserved in seven alphaherpesviruses and the restriction endonuclease sites used in the construction of missense mutations are shown. The alphaherpesviruses used in the alignment were HSV-1, HSV-2, VZV, equine herpesvirus type 1, equine herpesvirus type 4, pseudorabies virus, and bovine herpesvirus type 1 proteins. Below, the amino acid sequence and location of domains 1 to 4 is shown, with residues that are conserved in all seven viruses displayed on the line below (26). The amino acid changes in the domains used in this study are listed to the right.
on November 8, 2019 by guest
http://jvi.asm.org/
[image:2.587.75.508.73.300.2]gonucleotide containing mutations in domain 1. The mutant UL26.5 gene was identified by the presence of a novelNheI site that did not affect the wt virus UL26.5 amino acid sequence. The UL26.5 gene containing mutations in domain 2 was constructed by cleaving pGX262 withEagI andPpuMI and ligating the larger fragment to anEagI-PpuMI double-stranded oligonucleotide containing the missense mutations and a silent mutation, creating anNarI site. The muta-tion, E148A, was introduced into domain 3 by ligating aSmaI-SalI double-stranded oligonucleotide to the pGX262PpuMI-KpnI fragment containing all the vector sequences, thePpuMI-SmaI UL26.5 gene fragment, and theSalI-KpnI UL26.5 gene fragment. The presence of the mutation was confirmed by sequence analysis. A PCR product containing a triple mutation, P146A, G147A, and E148A, in domain 3 was made using a forward primer with anEagI site and a reverse primer containing the mutations and anAscI site. The PCR product was cloned into the vector pCR2.1 TOPO (Invitrogen), and the cloned DNA was sequenced. A gene containing a triple mutation in domain 3 was constructed by ligating theEagI-AscI fragment containing the domain 3 mutations from pCR2.1 TOPO into the largerEagI-AscI fragment of pGX262. A triple mutation, L182A, L186A, and M189A, was engineered into domain 4 by ligating anAatII-Bsu36I oligonucleotide containing the mutations to the largerPpuMI-Bsu36I fragment from pGX262 and thePpuMI-AatI fragment from pGX262, spanning domain 3 and part of domain 4. A mutant gene with lesions in both domains 2 and 4 was constructed by ligating the smallerXhoI fragment from the plasmid containing the domain 2 mutant gene to the largerXhoI fragment from the plasmid con-taining the domain 4 mutations. The gene with defects in domains 1, 2, and 4 was obtained by ligating the largerBsmI-KpnI fragment from the plasmid carrying the domain 1 mutant gene to the smallerBsmI-KpnI fragment from the plasmid containing the gene with mutations in domains 2 and 4. Mutant genes were transferred asBglII fragments into theBglII site of the mammalian expression vector CMV10Bgl (22) and the baculovirus transfer vector pAcCL29.1B (24).
Antibodies.Mouse monoclonal antibodies used were 406 (Serotec) and 5010 (28), which are specific for sequences within UL26.5, and a control antibody LP1, which is specific for the HSV-1 UL48 product, VP16. Rabbit antiserum raised against purified polyhistidine-tagged VP5 and a rabbit antibody raised against an oligopeptide corresponding to residues 471 to 486 of the VZV gene 33 product were also used (26). In indirect immunofluorescence experiments, the primary mouse and rabbit antibodies bound to antigen were detected using fluorescein isothiocyanate-conjugated goat anti-mouse (FITC-GAM) immunoglobulin G (IgG) (Sigma) and Cy5-conjugated goat anti-rabbit (Cy5-GAR) IgG (Amersham Pharmacia Biotech). The secondary antibodies used in immunoelectron micros-copy were GAM IgG coupled to 10-nm-diameter colloidal gold particles and goat anti-rabbit (GAR) IgG coupled to 30-nm-diameter colloidal gold (British BioCell International).
Confocal immunofluorescence microscopy. Linbro wells containing sterile glass coverslips were seeded with 7⫻104Vero cells per well and transfected the
following day with plasmid DNA using lipfectamine and Plus reagent (Life Technologies). Plus reagent (4l) was added to a tube containing 21l of plasmid DNA (0.5 to 1g) diluted in opti-mem (Life Technologies), and the sample was incubated for 15 min at room temperature. Diluted Lipofectamine (1
l of Lipofectamine added to 24l of opti-mem) was mixed with the DNA solution treated with Plus reagent, and the sample was incubated at room tem-perature for 15 min. Prior to transfection, cells were washed twice with Dulbec-co’s medium lacking supplements and serum and overlaid with 200l of the same medium. After the transfection mixture was added to the cells, the sample was incubated at 37°C for 3 h. Complete Dulbecco’s medium supplemented with 10% (vol/vol) fetal calf serum (1 ml) was added to the sample, and incubation continued at 37°C. The next day, cells were fixed in 5% (vol/vol) formaldehyde in phosphate-buffered saline (PBS) containing 2% (wt/vol) sucrose, permeabil-ized with 0.5% (vol/vol) Nonidet P-40 in PBS containing 10% (wt/vol) sucrose, and incubated at room temperature with diluted primary antibody for 2 h. After extensive washing with PBS containing 1% (vol/vol) calf serum, cells were incu-bated with FITC-GAM IgG, cy5-GAM IgG, or both secondary antibodies for 1 h at room temperature. Cells were washed in PBS containing 1% calf serum, mounted, and examined under a Zeiss LSM 510 confocal microscope, using a 63⫻oil immersion objective lens (NA 1.4) and lasers with excitation lines at 488 and 633 nm.
Electron microscopy.Immunoelectron microscopic studies were carried out as described by Preston et al. (26). Thin sections of baculovirus-infected cells embedded in Epon 812 resin were prepared as outlined by Addison et al. (1).
Purification of capsids.Sf21 cells (3⫻108) were infected with AcUL26.5 or
a recombinant baculovirus expressing UL26.5 domain 2⫹4 mutant scaffolding protein, together with AcUL18/19/38 and AcUL35 at a multiplicity of infection of 5 PFU per cell for each virus. At 64 h postinfection, the cells were harvested and capsids were analyzed essentially as described previously (13) except that the
samples were centrifuged on a 10 to 40% (wt/wt) sucrose gradient at 40,000 rpm for 20 min in a TST41 Sorvall rotor. Successive fractions were collected using a BioComp fractionator, and the proteins were analyzed by sodium dodecyl sul-fate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were stained with ammoniacal silver essentially as described by Harlow and Lane (10).
Analysis of scaffold-like particles.Sf21 cells (108) were infected with a
multi-plicity of infection of 5 PFU per cell of recombinant baculovirus expressing wt or mutant HSV-1 scaffolding protein. At 72 h postinfection, the cells were har-vested, washed with PBS, frozen, and thawed five times in the presence of 300l of PBS. After the extract had been clarified by centrifugation for 5 min at 13,000 rpm, the sample was layered onto a 10 to 40% (wt/vol) sucrose gradient and the material was centrifuged for 1 h at 40,000 rpm in a TST41 Sorvall rotor. Fractions were collected using a BioComp fractionator, and the samples were analyzed by Western blotting or dialyzed against PBS to remove the sucrose. The material was screened for the presence of scaffold-like particles by treating the dialyzed samples with phosphotungstic acid and examining the material absorbed on to parlodion-covered grids under the electron microscope.
Detection of scaffolding proteins by Western blotting.Proteins were separated by SDS-PAGE, the separated proteins were blotted onto nitrocellulose paper, and Western blot analysis was carried out using the ECL system, a luminol-based detection method (Amersham Pharmacia Biotech).
RESULTS
Construction of mutant scaffolding proteins.Alignment of
the amino acid sequence of the HSV-1 scaffolding with those of other alphaherpesvirus homologues has revealed five re-gions showing significant conservation of amino acid residues (23, 26). Site-directed mutagenesis was carried out on the four internal conserved regions of the HSV-1 scaffolding protein, referred to as domains 1 to 4 (Fig. 1) on the grounds that these parts of the protein were likely to be functionally important. Furthermore, several of these domains had already been im-plicated in intermolecular self-interaction (23, 36). The fifth region, near the C terminus, which encompasses the VP5 bind-ing site, has been extensively mutated and was not included in our analysis (11). Three amino acid changes were introduced into domains 2 and 4, regions that had been predicted to form
␣-helices with a coiled-coil structure (23). In each domain, three hydrophobic residues were replaced with alanine resi-dues with the aim of destabilizing coiled-coil interactions. The mutations engineered into domain 4 were identical to those made by Pelletier et al. (23). Initially, three amino acid changes were also introduced into the domain 1, including L31A. This leucine is conserved in the HCMV scaffolding protein and was shown by Wood et al. (36) to be required for intermolecular association of the HCMV scaffolding protein with itself. Sub-sequently, the corresponding single amino acid changes were made individually in domain 1. Three mutations were also made in domain 3 which, like domain 1, is conserved in beta-and gammaherpesviruses as well as alphaherpesviruses. In ad-dition to the construction of mutant genes encoding proteins with lesions in each of the four conserved domains, genes specifying proteins with mutations in more than one conserved domain were produced. A construct containing triple muta-tions in both domains 2 and 4 was made and another mutant was produced with triple mutations in domains 1, 2, and 4. Details of the mutations made in the domains, together with restriction enzyme sites used in their construction, are shown in Fig. 1.
Interaction of HSV-1 wt scaffolding protein with VZV
ho-mologue.Previous work, in which the HSV-1 and VZV
scaf-folding proteins were expressed transiently in mammalian cells
on November 8, 2019 by guest
http://jvi.asm.org/
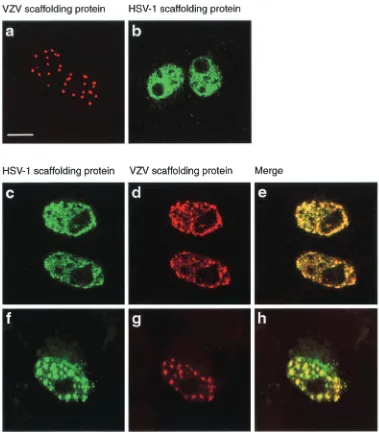
and detected using an immunofluorescence assay, showed that the VZV scaffolding protein had a different pattern of distri-bution in the nucleus from that of the HSV-1 protein (13, 26) (Fig. 2a and b). The VZV scaffolding protein was present in localized regions, probably containing large aggregates of the protein, whereas the HSV-1 counterpart was usually dispersed throughout the nucleus and had a granular appearance. This observation was exploited to develop an assay for scaffolding protein interaction. Vero cells were cotransfected with plas-mids containing the HSV-1 and VZV genes under the control of the HCMV major immediate-early promoter, and the ex-pressed proteins were detected by indirect immunofluores-cence. In cells containing both proteins, the pattern of
[image:4.587.102.481.72.504.2]distri-bution of the VZV scaffolding protein generally resembled that of the HSV-1 protein when expressed on its own (Fig. 2a and c to e). Occasionally, the VZV protein gave a spotty fluorescence pattern, and in such cases the HSV-1 scaffolding protein always colocalized with the VZV protein, suggesting that the two proteins interacted with each other (Fig. 2f to h). To confirm that the VZV scaffolding protein was associated with the HSV-1 protein, insect cells were coinfected with re-combinant baculoviruses expressing the two scaffolding pro-teins and analyzed by immunoelectron microscopy. Thin sec-tions of baculovirus-infected cells embedded in acrylic resin were incubated with mouse monoclonal antibody 5010, which is specific for the HSV-1 scaffolding protein, and a rabbit
FIG. 2. Colocalization of VZV and HSV-1 scaffolding proteins. Digital confocal images of transfected Vero cells expressing the VZV scaffolding protein (a), the HSV-1 wt scaffolding protein (b), or VZV and HSV-1 proteins (c to h). Bound rabbit polyclonal antibodies (specific for the VZV protein) were visualized with Cy5-GAR IgG (red), and mouse monoclonal antibodies (specific for the HSV-1 protein) were detected with FITC-GAM IgG (green). In both sets of three images for cells dually expressing the VZV and HSV proteins, the image in the right panel represents the merged images in the left and middle panels. Bar, 10m.
on November 8, 2019 by guest
http://jvi.asm.org/
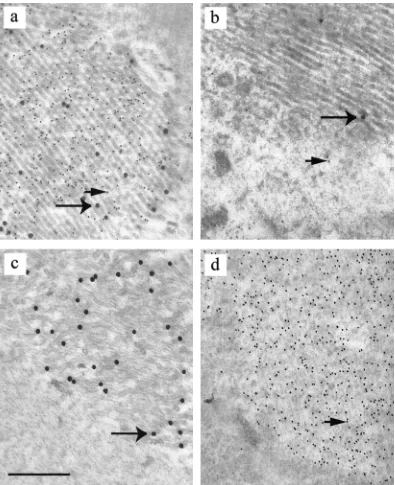
antiserum, which is specific for the VZV homologue. The sections were subsequently treated with GAM IgG coupled to 10-nm colloidal gold and GAR IgG coupled to 30-nm colloidal gold. As a control, mouse monoclonal antibodies specific for an unrelated HSV-1 protein (VP16) and preimmune rabbit serum were used in place of the primary antibodies. Both 10-and 30-nm gold particles were concentrated over aggregates of scaffolding protein in samples treated with antibodies specific
[image:5.587.95.490.71.556.2]for HSV-1 and VZV scaffolding proteins but not in prepara-tions incubated with control antibodies. This result suggested that the HSV-1 and VZV scaffolding proteins were both present in the aggregates (Fig. 3 a and b). Instead of being concentrated in regions containing scaffold-like particles, the HSV-1 antibodies were mainly found clustered over areas con-taining tubular structures, which were uncommon in insect cells expressing the HSV-1 protein on its own but frequently
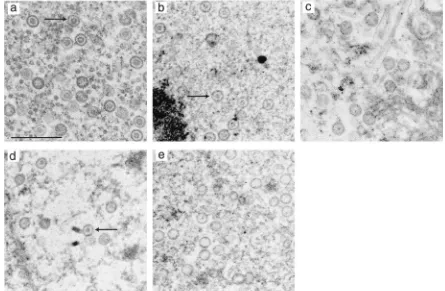
FIG. 3. Identification of aggregates of HSV-1 and VZV scaffolding proteins by immunoelectron microscopy. Thin sections of sf21 cells infected with baculoviruses expressing VZV and HSV-1 scaffolding proteins (a and b) and cells singly infected with baculovirus expressing the VZV scaffolding protein (c) or the HSV-1 scaffolding protein (d) were prepared. The samples were incubated with rabbit antiserum specific for VZV scaffolding protein and mouse monoclonal antibody specific for the HSV-1 homologue (a, c, and d) or control antibodies (b) and subsequently treated with GAM IgG conjugated to 10-nm gold particles and GAR IgG conjugated to 30-nm gold particles. Bar, 0.5m; large arrow, 30-nm gold particle; small arrow, 10-nm gold particle.
on November 8, 2019 by guest
http://jvi.asm.org/
observed in cells expressing the VZV scaffolding protein on its own. It is therefore likely that the VZV scaffolding protein can influence the structure that the HSV-1 protein adopts. To confirm that the HSV-1 and VZV specific antibodies did not cross-react with the VZV and HSV-1 scaffolding proteins, re-spectively, thin sections of insect cells containing only the VZV or HSV-1 protein were treated with both primary antibodies and subsequently both secondary antibodies. No cross-reaction was observed between the HSV-1 specific monoclonal anti-body and VZV scaffolding protein and, similarly, between the VZV rabbit antibody and HSV-1 protein in the immunoelec-tron microscopic analysis (Fig. 3c and d) and in the immuno-fluorescence assay used to detect the VZV and HSV-1 scaf-folding proteins (data not shown). In addition, no cross-reaction was observed between the GAM IgG and the rabbit polyclonal antibody and with GAR IgG and the mouse mono-clonal antibody.
Interaction of mutant HSV-1 scaffolding proteins with VZV
homologue.The ability of mutant scaffolding proteins
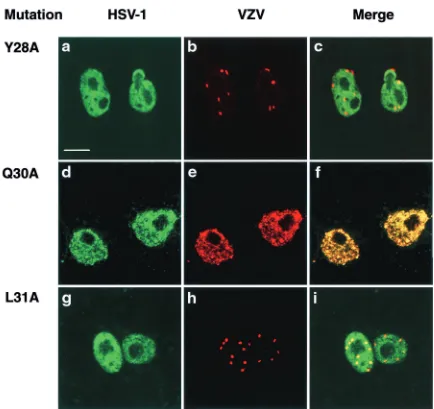
contain-ing three mutations in the conserved domains 1, 2, 3, or 4 or within both domains 2 and 4 to associate with the VZV coun-terpart was investigated using a transient-expression assay (Fig. 4). In Vero cells expressing the mutant proteins alone, the appearance of the protein was similar to that of the wt HSV-1 protein except that the domain 2 mutant had an increased tendency to form aggregates in comparison to the wt protein (data not shown). In cells containing the VZV homologue and the mutant HSV scaffolding protein with multiple mutations in domain 2 (Fig. 4d to f), 3 (Fig. 4g to i), or 4 (Fig. 4j to l), the distribution of the VZV protein was similar to the mutant HSV-1 protein, suggesting that the VZV scaffolding protein was able to associate with the mutant proteins. When the mutant scaffolding protein with the triple mutation in domain 1 was coexpressed with the VZV protein, most of the HSV-1 mutant did not colocalize with the VZV protein (Fig. 4a to c). The pattern of the two proteins resembled the distribution observed in cells singly expressing the wt proteins (Fig. 2). This finding strongly suggested that disruption of domain 1 de-creased the ability of the HSV-1 scaffolding protein to interact with its VZV counterpart. To determine which of the three amino acid changes present in the domain 1 triple mutant was responsible, mutant scaffolding proteins containing the single-amino-acid changes Y28A, Q30A, or L31A were constructed and assayed for association with the VZV scaffolding protein. In cells expressing the HSV-1 mutant Y28A or L31A proteins and the VZV homologue, the two herpesvirus proteins had different patterns of distribution from each other (Fig. 5a to c and g to i), in contrast to cells containing the VZV protein and the mutant Q30A (Fig. 5d to f). Therefore, the conserved amino acids Y28 and L31 are likely to be important in the
interaction of the VZV scaffolding protein with the HSV-1 homologue.
Participation of mutant scaffolding proteins in capsid
as-sembly.To determine whether the mutant scaffolding proteins
could bring about the formation of sealed capsids, sf21 cells were infected with a baculovirus expressing VP5, VP23, and VP19C together with one expressing the mutant or wt scaf-folding protein. Numerous capsids were observed in the nuclei of cells expressing the wt protein or domain 1, 2, 3, or 4 mutant scaffolding proteins in the presence of the outer shell capsid proteins (Fig. 6a to e). Capsids containing either domain 2 and 4 mutants showed more variation in size than capsids contain-ing wt scaffolds or the other mutant scaffolds, suggestcontain-ing that there may be more disruption to the scaffold. For this reason and because a previous study using the yeast two-hybrid system had implicated domains 2 and 4 in self-interaction (23), a mutant with defects in both domains 2 and 4 (referred to as domain 2⫹4 mutant) was constructed. In cells containing the domain 2⫹4 mutant protein, incomplete capsid shells were detected, suggesting that lesions in both these domains se-verely affected the ability of the scaffolding protein to partici-pate in the assembly of intact capsids (Fig. 6f). To confirm that this mutant was unable to direct the formation of sealed cap-sids, insect cells were infected with two types of baculoviruses, one expressing the wt or mutant scaffolding protein and the other expressing the HSV-1 capsid shell proteins, and the cell extract was layered onto a sucrose gradient. After the material had been centrifuged, a band of light-scattering material cor-responding to HSV-1 capsids was observed in the wt scaffold-ing protein sample, but no obvious capsid band was observed in the sample containing the domain 2⫹4 mutant scaffolding pro-tein (data not shown). Successive fractions from each gradient were collected, the proteins present in the fractions were an-alyzed on an SDS-polyacrylamide gel, and the proteins were detected by silver staining. Examination of the pattern of pro-teins in the gradient containing the wt scaffolding protein showed that the capsid proteins were concentrated in the re-gion of the gradient containing the light-scattering material as indicated in Fig. 7a. By contrast, in the gradient containing the mutant 2⫹4 scaffolding protein, no peak of HSV-1 capsid shell proteins was observed (Fig. 7b). Western blotting was carried out to examine the distribution of the scaffolding protein in each fraction from the gradients (Fig. 7c and d). As expected, the peak fraction of wt scaffolding protein coincided with the peak fractions of HSV-1 capsid shell proteins whereas the domain 2⫹4 mutant protein was found at the top of the gra-dient. These results show that the domain 2⫹4 mutant was unable to form a stable complex with the capsid shell proteins under the conditions used to purify capsids and confirm that
FIG. 4. Interaction of HSV-1 mutant scaffolding proteins with VZV homologue. Vero cells were transfected with a plasmid expressing a mutant HSV-1 scaffolding protein together with a construct containing the homologous VZV gene, and the proteins were detected using an indirect immunofluorescence assay. Digital confocal images in groups of three represent cells containing the VZV scaffolding protein plus domain 1 triple mutant protein (a, b, c), domain 2 mutant (d, e, f), domain 3 mutant (g, h, i), domain 4 mutant (j, k, l), and domain 2⫹4 mutant (m, n, o). Bound mouse antibodies (specific for the HSV-1 protein) were visualized with FITC-GAM IgG (green), and bound rabbit antibodies (specific for the VZV protein) were detected with Cy5-GAR IgG (red). In each set, the image in the right panel represents the merged images in the left and middle panels. Bar, 10m.
on November 8, 2019 by guest
http://jvi.asm.org/
on November 8, 2019 by guest
http://jvi.asm.org/
this mutant scaffolding protein was unable to bring about the formation of sealed capsids.
Effect of proteolytic cleavage of mutant scaffolds within
cap-sids. In wt HSV-1-infected cells, cleavage of the C-terminal
end of the scaffolding protein by the UL26 product releases the scaffold from the inner capsid shell wall and leads to the gen-eration of capsids containing a small internal core provided there is no DNA packaging (25). Although the scaffolding portion of the UL26 protease (preVP21) can substitute for the UL26.5 product in capsid assembly (30), it does not form scaffold-like particles when expressed on its own or in the presence of the protease domain VP24 (24). Furthermore, capsids assembled in the presence of both the UL26 protease and the scaffolding portion of the protease in insect cells in-fected with recombinant baculoviruses expressing the HSV-1 proteins either lacked an internal core or contained
incom-plete scaffolds (data not shown). To investigate the effect of proteolytic cleavage of the mutant scaffolds within the capsid, insect cells were multiply infected with recombinant baculovi-ruses expressing wt virus or mutant scaffolding protein, the essential outer capsid shell proteins, and UL26. Small cored capsids were observed only in cells expressing the wt scaffold-ing protein or domain 1 or domain 3 mutant protein, suggest-ing that the mutations in domain 2 or 4 affected the ability of the protein to retain or form a distinct spherical structure inside the capsid after cleavage (Fig. 8).
Formation of aggregates by mutant proteins.Although
[image:8.587.74.508.69.478.2]scaf-folding proteins with mutations in the individual conserved domains retained the ability to self-associate inside the capsid shell when the protease was absent, it was clear that in the presence of the protease, this ability was impaired for mutant domain 2 and 4 proteins. Experiments were therefore carried
FIG. 5. Interaction of domain 1 mutants containing single amino acid changes with VZV scaffolding protein. Digital confocal images in groups of three represent transfected Vero cells dually expressing the VZV scaffolding protein and domain 1 mutants Y28A (a, b, c), Q30A (d, e, f), or L31A (g, h, i). Bound mouse monoclonal antibodies (specific for the HSV-1 protein) were visualized with FITC-GAM IgG, and bound rabbit polyclonal antibodies (specific for the VZV protein) were identified using Cy5-GAR IgG. In each set, the image in the right panel represents the merged images in the left and middle panels. Bar, 10m.
on November 8, 2019 by guest
http://jvi.asm.org/
out to investigate the ability of the mutant proteins to self-associate in the absence of the capsid shell proteins. Extracts were prepared from the Sf21 cells infected with recombinant baculovirus expressing wt or mutant scaffolding proteins, and the material was layered onto a 10 to 40% sucrose gradient. After the samples had been centrifuged, fractions were col-lected from the gradient. The proteins were separated by SDS-PAGE, and the scaffolding proteins were detected by Western blotting (Fig. 9a). Wt protein and the UL26.5 domain 3 mutant proteins sedimented as a broad band of material with the highest amount of protein in fractions 6, 7, and 8. Electron microscopic analysis of fraction 7 from these samples revealed the presence of scaffold-like particles (Fig. 9b). These particles were not observed in the same fractions of gradients of the other mutant scaffolding proteins or in the peak fractions higher up in the gradients. Mutant proteins with triple muta-tions in domain 1, 2, or 4 gave similar banding patterns to each other, with the protein being concentrated in fractions 3 and 4. These results suggested that the mutations in these domains reduced the ability of these proteins to form scaffold-like par-ticles and large aggregates. This finding was supported by the finding that in thin sections of insect cells infected with recom-binant baculovirus expressing wt or mutant scaffolding protein, scaffold-like particles were observed under the electron micro-scope in cells containing the wt protein or domain 3 mutant protein only (data not shown). Sedimentation analysis of the
UL26.5 domain 2⫹4 mutant revealed that this protein was severely impaired in its ability to interact with itself, with the protein being concentrated in fractions 1 and 2.
Interaction of mutant scaffolding proteins with VP5.
[image:9.587.67.517.72.366.2]Previ-ous work by Pelletier et al. (23) suggested that self-association of the HSV-1 scaffolding protein was important for the inter-action with the major capsid shell protein, VP5, in the yeast two-hybrid assay. The domain 2⫹4 mutant protein, which was unable to bring about the formation of intact capsids, was therefore screened for the ability to interact with the VP5 by using the transient-expression assay in Vero cells. The domain 1 mutant was also included in this set of experiments since work by Wood et al. (36) indicated that the interaction of the HCMV scaffolding protein with the major capsid shell protein was also influenced by self-association of the scaffolding pro-tein. Uncleaved wt HSV-1 scaffolding protein and the wt cleaved protein lacking the carboxy-terminal 25 amino acids, which contains the VP5 binding site (11), were included as positive and negative controls, respectively. The results sug-gested that the mutations in domain 1 did not markedly inter-fere with the ability of the mutant protein to interact with VP5. In a cell expressing both VP5 and the mutant protein, VP5 was present mainly in the nucleus (Fig. 10m, n, and o), and this pattern was observed in cells containing both VP5 and wt scaffolding protein (Fig. 10g, h, and i). By contrast, the cleaved form of the wt scaffolding protein did not affect the distribution
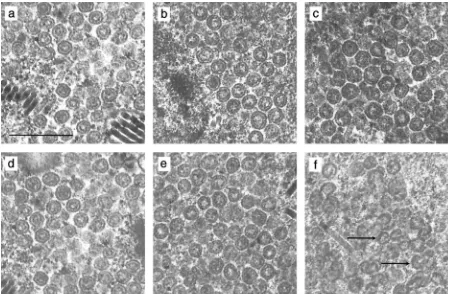
FIG. 6. Participation of mutant scaffolding proteins in capsid assembly. Insect cells were infected with a recombinant baculovirus expressing wt or mutant scaffolding protein together with a virus expressing VP5, VP19C, and VP23. Samples were harvested at 50 h postinfection, fixed, and embedded, and thin sections were examined for the presence of capsids under the electron microscope. Shown are electron micrographs of a portion of a cell expressing VP5, VP19C, VP23, and HSV-1 wt scaffolding protein (a), domain 1 mutant protein (b), domain 2 mutant protein (c), domain 3 mutant protein (d), domain 4 mutant protein (e), or domain 2⫹4 mutant protein (f). Arrows, incomplete capsids; bar, 0.5m.
on November 8, 2019 by guest
http://jvi.asm.org/
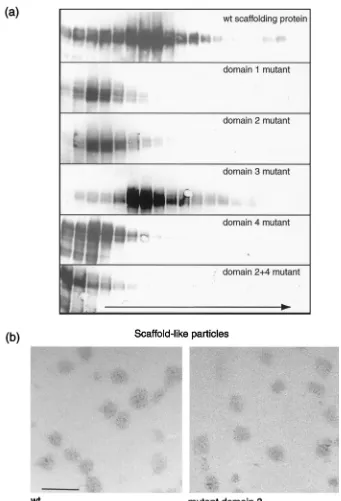
FIG. 7. Sucrose gradient sedimentation analysis of extracts from insect cells infected with recombinant baculoviruses expressing HSV-1 capsid shell proteins and either wt scaffolding protein (a and c) or mutant 2⫹4 scaffolding protein (b and d). The extracts were sedimented through 10 to 40% sucrose gradients, and successive fractions were collected. The proteins in each fractions were analyzed by SDS-PAGE and detected by silver staining (a and b), and the scaffolding protein in each fraction was detected by Western blotting using the monoclonal antibody 406 (c and d). The short arrow indicates the major baculovirus capsid protein p39 and the long arrow shows the direction of sedimentation. The positions of the HSV-1 capsid proteins are indicated on the left and in the peak fraction in the wt virus sample. On the right, the positions of theMrstandards
are given.
on November 8, 2019 by guest
http://jvi.asm.org/
of VP5, which was predominantly in the cytoplasm (Fig. 10j, k, and l). The mutant protein with lesions in both domains 2 and 4 also retained the ability to associate with VP5 and relocate it to the nucleus (Fig. 10p, q, and r). In view of the results with the domain 2⫹4 mutant and the previous results by Pelletier et al. (23), the domain 1⫹2⫹4 mutant was subsequently tested for its capacity to associate with VP5 (Fig. 10s, t, and u). When the domain 1⫹2⫹4 mutant was expressed on its own, most of the protein localized to the nucleus. In contrast to wt virus and the other mutant scaffolding proteins, however, a significant portion remained in the cytoplasm, suggesting that the muta-tions had some effect on the ability of the protein to localize to or be retained within the nucleus. Nevertheless, in cells coex-pressing both VP5 and the mutant protein, most of VP5 was present in the nuclei together with the scaffolding protein.
DISCUSSION
A major aim of the project was to examine regions of the HSV-1 scaffolding protein implicated in intermolecular self-interaction using the intact protein, and our studies have pro-vided for the first time evidence that domain 1 is likely to be involved in this process in HSV-1. This is also the first report investigating the effect of mutations in the four internal con-served regions on the assembly of scaffold-like particles and capsid formation. A summary of the results is given in Table 1. Although domain 1, 2, and 4 mutant proteins did not form detectable levels of scaffold-like particles in the absence of
other HSV-1 proteins, the lesions in domains 2 and 4 rather that those in domain 1 appeared to have the greatest effect on scaffolding protein structure. Capsids with domain 2 or 4 mu-tant proteins showed more variation in size than those con-taining wt protein or the other mutant scaffolding proteins. Furthermore, when the maturational protease was present in capsids with domain 2 or 4 mutant proteins, no distinct core was observed. These changes in the property of the scaffolding protein could be due to reduced contacts within the protein resulting in a less stable scaffold structure. Alternatively, they could be a direct consequence of disruptions to the folding of the protein, for example, if the conserved region lies in a buried hydrophobic region of the protein. Evidence from the work of Pelletier et al. (23) suggested that the domain 4 region was directly involved in self-association since a glutathionine
[image:11.587.69.514.75.366.2]S-transferase fusion protein containing a short region of UL26.5 sequences spanning domain 4 interacted with the full-length scaffolding protein in a far Western blot analysis. Fur-thermore, this association did not occur when a construct of domain 4 containing the same mutations that were used in our study was tested. Previous analysis on domain 2 was less con-clusive, and it is possible that this region is not directly involved in binding but is important for stabilizing the conformation of domain 4 (23). Interestingly, the domain 2 region although conserved in alphaherpesviruses does not have an obvious counterpart in beta- and gammaherpesviruses (data not shown). None of the mutations in individual domains affected
FIG. 8. Effect of proteolytic cleavage of mutant scaffolds within capsids. Insect cells were multiply infected with a recombinant baculovirus expressing VP5, VP19C, and VP23, a virus expressing the protease, and a virus expressing wt or mutant scaffolding protein. Shown are electron micrographs of a thin section of a cell expressing VP5, VP19C, and VP23, the UL26 gene product, and wt scaffolding protein (a) or domain 1 (b), domain 2 (c), domain 3 (d), or domain 4 (e) mutant scaffolding protein. The arrows point to capsids containing distinctive scaffolds. Bar, 0.5m.
on November 8, 2019 by guest
http://jvi.asm.org/
the efficiency of capsid assembly but the mutant containing lesions in both domains 2 and 4 failed to bring about the assembly of intact capsids. This finding suggests that limited disruption of scaffold structure can be tolerated in capsid as-sembly because there are several self-interacting domains that can function independently of one another.
[image:12.587.121.462.69.570.2]Work from Wood et al. (36) showed that the amino-terminal conserved domain in the HCMV scaffolding protein, homolo-gous to domain 1 of HSV-1, was required for the association of the protein with itself. The finding that the HSV-1 domain 1 was important in a heterologous interaction between the HSV-1 and VZV proteins in an immunofluorescence assay
FIG. 9. Aggregates of wt and mutant scaffolding proteins. Insect cells were infected with a single recombinant baculovirus expressing the wt protein or a mutant scaffolding protein and harvested at 72 h postinfection. Extracts were prepared, and the clarified extracts were layered onto 10 to 40% sucrose gradients and centrifuged. Fractions were collected, starting from the top of the gradient. (a) The proteins from each gradient were separated on an SDS–10% polyacrylamide gel, and the scaffolding protein was detected by Western blotting, using monoclonal antibody to the UL26.5 protein. The arrow indicates the direction of sedimentation. (b) The material from selected fractions from each gradient was negatively stained and examined under the electron microscope. Scaffold-like particles were observed in the peak fraction (fraction 7) from gradients of wt protein and domain 3 mutant scaffolding protein only. Bar, 100 nm.
on November 8, 2019 by guest
http://jvi.asm.org/
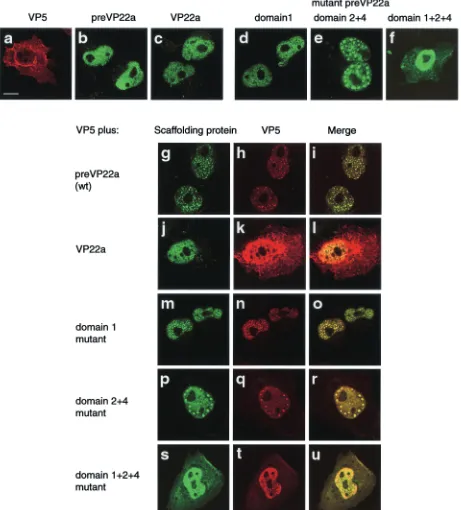
FIG. 10. Interaction of VP5 with mutant scaffolding proteins. Vero cells were cotransfected with a plasmid expressing VP5 and another expressing the wt protein or a mutant HSV-1 scaffolding protein. Transfections with individual plasmids were also carried out. Proteins were detected using an indirect immunofluorescence assay. The top six digital confocal images represent cells expressing VP5 (a), wt scaffolding protein (b), VP22a (c), domain 1 mutant containing three mutations (d), domain 2⫹4 mutant (e), and domain 1⫹2⫹4 mutant (f). The subsequent confocal images, in series of three, represent cells containing wt scaffolding protein (preVP22a) and VP5 (g, h, i), VP22a and VP5 (j, k, l), domain 1 triple mutant and VP5 (m, n, o), domain 2⫹4 and VP5 (p, q, r), and domain 1⫹2⫹4 and VP5 (s, t, u). Bound mouse monoclonal antibodies (specific for the HSV-1 protein) were visualized with FITC-GAM IgG, and bound rabbit polyclonal antibodies (specific for VP5) were identified using Cy5-GAR IgG. In each set of three images of dually expressed HSV proteins, the image in the right panel represents the merged images present in the left and middle panels. Bar, 10m.
on November 8, 2019 by guest
http://jvi.asm.org/
suggests that it is likely to be involved in intermolecular self-association of the HSV-1 protein. This idea was strengthened by the observation that the same conserved leucine residue that was important for interaction of the HCMV scaffolding protein was required for the association of the HSV-1 protein with the VZV homologue. The failure of the HSV-1 domain 1 mutant to form scaffold-like particles and large aggregates provides further evidence that domain 1 is involved in self-association of the HSV-1 scaffolding protein. However, in con-trast to the findings with the HCMV scaffolding protein (36), mutation of conserved amino acids within this domain did not prevent the intact protein from interacting with itself. More-over, because the domain 2⫹4 mutant protein was severely impaired in its ability to self-associate, it is likely that the HSV-1 domain 1 interaction is relatively weak. Although there is reasonable conservation between VZV and HSV-1 scaffold-ing proteins in all the domains analyzed, only domain 1 was shown to be important for the association of the two proteins in the immunofluorescence assay. One explanation for this observation is that the requirements for interaction within do-main 1 are less stringent, with more changes of amino acid residues tolerated, than binding within the other domains. Al-ternatively, the interaction between domain 1 regions of the HSV-1 scaffolding protein may not be as strong as the associ-ation of other domains, allowing the region to be more acces-sible for interaction with the VZV protein. It is unclear why the yeast two-hybrid study by Pelletier et al. (23) did not identify domain 1 in self-interaction of the scaffolding protein. The association may have been too weak to be detected; the bind-ing site may have been affected by deletions elsewhere in the scaffolding protein or destroyed in the formation of the UL26.5 hybrid protein.
The findings from the yeast two-hybrid study on self-inter-action of the HSV-1 scaffolding protein suggested that there were several sites involved in self-association, and our results are consistent with this idea since none of the mutations in the individual domains abolished self-interaction of the protein (23). Recently, further evidence in favor of multiple interac-tions within the scaffold itself has been obtained from a study of the structure of HSV-1 procapsids by using cryoelectron microscopy. Radial density profiles from three-dimensional density maps revealed three peaks of density in the region corresponding to the scaffolding protein, which was interpreted to represent regions of condensed scaffolding protein domains separated by flexible linkers (21).
Various programs for predicting protein secondary
struc-tures, such as Chou-Fasman (3) and Garnier-Robson (9), sug-gest that several of the conserved regions within the scaffolding protein have a high propensity for␣-helicity, notably domains 2 and 4. Pelletier et al. (23) speculated that these two regions formed short coiled coils and that the scaffolding protein as-sociated with itself via multiple weak homomeric coiled-coil interactions. In our study, the amino acid residues mutated within domain 2 or 4 were selected on that basis. However, more recent algorithms, for example, Psipred (12), do not predict any␣-helices in these regions and suggest that there is a much higher number of-sheets than previously predicted. These models have to be tested experimentally to determine the underlying structural mechanism of interaction. Whether or not domains 2 or 4 form coiled coils, the mutations intro-duced in these regions clearly had an effect on scaffold forma-tion.
Previous work with the HCMV and HSV-1 scaffolding pro-teins in the yeast two-hybrid system suggested that intermolec-ular self-interaction of these proteins was important for asso-ciation with the major capsid protein, VP5, particularly when deletion scaffolding protein mutants were assayed (23, 36). By contrast, in our study, domain 1 and 2⫹4 mutant proteins, both of which had a reduced ability to self-interact, were not signif-icantly impaired in their interaction with VP5 in a transient-expression assay. Subsequent analysis of the domain 2, 3, and 4 mutants showed that these proteins were also able to asso-ciate with VP5 and alter its intracellular distribution (data not shown). Only the domain 1⫹2⫹4 mutant had a reduced ca-pacity to localize VP5 to the nucleus, and this was probably because the mutant protein itself was transported inefficiently to the nucleus. Our results suggest that the formation of large complexes of the scaffolding protein is not required for the interaction of the scaffolding protein with VP5. These findings are consistent with previous work showing that purified VP5 preferentially formed complexes with low-molecular-weight aggregates of the purified scaffolding protein than with large oligomers (19). Our data are in keeping with a model in which the scaffolding protein interacts with VP5, forming a small complex which is transported into the nucleus where it associ-ates with the triplexes, assembling into the procapsid by the subsequent addition of further VP5-scaffolding protein com-plexes and tricom-plexes (19, 32). The ability of VP5 to interact with domain 1, 2, and 4 mutant scaffolding proteins, despite their reduced capacities to self-associate, explains why these mu-tants still functioned in capsid assembly. The scaffolding pro-tein-VP5 interaction would lead to an increased localized
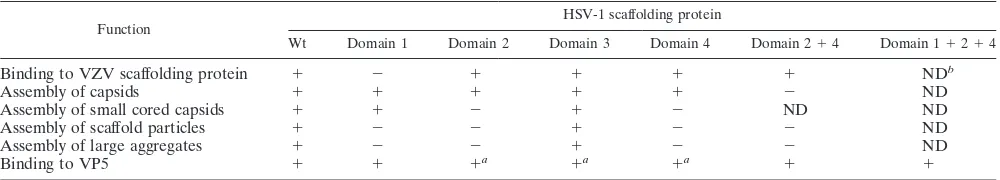
con-TABLE 1. Characterization of HSV-1 mutant scaffolding proteins
Function HSV-1 scaffolding protein
Wt Domain 1 Domain 2 Domain 3 Domain 4 Domain 2⫹4 Domain 1⫹2⫹4
Binding to VZV scaffolding protein ⫹ ⫺ ⫹ ⫹ ⫹ ⫹ NDb
Assembly of capsids ⫹ ⫹ ⫹ ⫹ ⫹ ⫺ ND
Assembly of small cored capsids ⫹ ⫹ ⫺ ⫹ ⫺ ND ND
Assembly of scaffold particles ⫹ ⫺ ⫺ ⫹ ⫺ ⫺ ND
Assembly of large aggregates ⫹ ⫺ ⫺ ⫹ ⫺ ⫺ ND
Binding to VP5 ⫹ ⫹ ⫹a ⫹a ⫹a ⫹ ⫹
aⴱ, data not shown. bND, not done.
on November 8, 2019 by guest
http://jvi.asm.org/
[image:14.587.43.543.84.174.2]centration of the scaffolding protein in the presence of the triplexes, favoring self-association of the scaffolding protein, which would promote the formation of the procapsid.
ACKNOWLEDGMENTS
We thank D. McClelland for supplying us with VP5-specific anti-body, A. C. Minson for providing us with LP1 monoclonal antianti-body, and J. D. Aitken for technical assistance. We are grateful to D. J. McGeoch and A. J. Davison for helpful comments on the manuscript.
REFERENCES
1.Addison, C., F. J. Rixon, J. W. Palfreyman, M. Ohara, and V. G. Preston.
1984. Characterization of a herpes simplex virus type-1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology138:246–259.
2.Casjens, S., and R. Hendrix.1988. Control mechanisms in dsDNA bacterio-phage assembly, p. 15–91.InR. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y.
3.Chou, P. Y., and G. D. Fasman.1978. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol.47:45–147. 4.Desai, P., and S. Person.1996. Molecular interactions between the HSV-1
capsid proteins as measured by the yeast two-hybrid system. Virology220:
516–521.
5.Desai, P., S. C. Watkins, and S. Person.1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol.68:5365–5374.
6.DiIanni, C. L., D. A. Drier, I. C. Deckman, P. J. McCann III, F. Liu, B. Roizman, R. J. Colonno, and M. G. Cordingley.1993. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J. Biol. Chem.268:2048–2051. 7.DiIanni, C. L., J. T. Stevens, M. Bolgar, D. R. O’Boyle II, S. P. Weinheimer,
and R. J. Colonno.1994. Identication of the serine residue at the active site of the herpes simplex virus type 1 protease. J. Biol. Chem.269:12672–12676. 8.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann III, I. Deckman, and R. J. Colonno.1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol.68:3703–3712.
9.Garnier, J., D. J. Ogusthorpe, and B. Robson.1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol.120:97–120.
10.Harlow, E., and D. Lane.1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
11.Hong, Z., M. Beaudet-Miller, J. Durkin, R. Zhang, and A. D. Kwong.1996. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J. Virol.70:533–540.
12.Jones, D. T.1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol.292:195–202.
13.Kennard, J., F. J. Rixon, I. M. McDougall, J. D. Tatman, and V. G. Preston.
1995. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J. Gen. Virol.76:1611–1621.
14.Liu, F., and B. Roizman.1993. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J. Virol.67:1300–1309.
15.Liu, F., and B. Roizman.1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol.65:5149–5156.
16.Liu, F., and B. Roizman.1991. The promoter, transcriptional unit, and coding sequences of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol.65:206–212. 17.Matusick-Kumar, L., W. Hurlburt, S. P. Weinheimer, W. W. Newcomb, J. C. Brown, and M. Gao.1994. Phenotype of the herpes simplex virus type1 protease substrate ICP35 mutant virus. J. Virol.68:5384–5394.
18.Newcomb, W. W., and J. C. Brown.1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol.65:613–620.
19.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A.
Steven, F. Booy, and J. C. Brown.1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol.73:4239–4250. 20.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown.1993. Structure of the herpes simplex virus capsid: molecular com-position of the pentons and the triplexes. J. Mol. Biol.232:499–511. 21.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J.
Tenney, S. K. Weller, and J. C. Brown.2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Vi-rol.74:1663–1673.
22.Nicholson, P., C. Addison, A. M. Cross, J. Kennard, V. G. Preston, and F. J. Rixon.1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol.75:1091–1099.
23.Pelletier, A., F. Doˆ, J. J. Brisebois, L. Lagace´, and M. G. Cordingley.1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil inter-actions and promotes stable interaction with the major capsid protein. J. Vi-rol.71:5197–5208.
24.Preston, V. G., M. F. Al-Kobaisi, I. M. McDougall, and F. J. Rixon.1994. The herpes simplex virus gene UL26 proteinase in the presence of the UL26.5 gene product promotes the formation of scaffold-like structures. J. Gen. Virol.75:2355–2366.
25.Preston, V. G., J. A. V. Coates, and F. J. Rixon.1983. Identification and characterization of a herpes simplex virus gene product required for encap-sidation of virus DNA. J. Virol.45:1056–1064.
26.Preston, V. G., J. Kennard, F. J. Rixon, A. J. Logan, R. W. Mansfield, and I. M. McDougall.1997. Efficient herpes simplex virus type 1 (HSV-1) capsid formation directed by the varicella-zoster virus scaffolding protein requires the carboxy-terminal sequences from the HSV-1 homologue. J. Gen. Virol.
78:1633–1646.
27.Prevelige, P. E., and J. King.1993. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog. Med. Virol.40:206–221.
28.Rixon, F. J., A. M. Cross, C. Addison, and V. G. Preston.1988. The products of herpes simplex virus type-1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J. Gen. Virol.69:2879–2891.
29.Rixon, F. J., and D. McNab.1999. Packaging competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro assembled procapsids. J. Virol.73:5714–5721.
30.Robertson, B. J., P. J. McCann III, L. Matusick-Kumar, V. G. Preston, and M. Gao.1997. Na, an autoproteolytic product of the herpes simplex virus type 1 protease, can functionally substitute for the assembly protein ICP35. J. Virol.71:1683–1687.
31.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon.1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid forma-tion suggested by structures of recombinant particles. J. Virol.73:6821–6830. 32.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown.1998. Assembly of the herpes simplex virus capsid: preformed tri-plexes bind to the nascent capsid. J. Virol.72:3944–3951.
33.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliott, and F. J. Rixon.
1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol.75:1101–1113.
34.Thomsen, D. R., W. W. Newcomb, J. C. Brown, and F. L. Homa.1995. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol.69:3690–3703.
35.Thomsen, D. R., L. L. Roof, and F. L. Homa.1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with re-combinant baculoviruses expressing HSV capsid proteins. J. Virol.68:2442– 2457.
36.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson.1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol.71:179–190.
37.Zhou, Z. H., W. Chiu, K. Haskell, H. J. Spears, J. Jakana, F. J. Rixon, and L. R. Scott.1998. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys. J.74:576–588.
38.Zhou, Z. H., S. J. Macnab, J. Jakana, L. R. Scott, W. Chiu, and F. J. Rixon.
1998. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc. Natl. Acad. Sci. USA95:2778–2783.