JOURNALOFVIROLOGY,June1975,p.1440-1448
Copyright© 1975 AmericanSocietyforMicrobiology
Vol. 15,No. 6 Printed inU.S.A.
Interspecies Antigenic Determinants
of the
Reverse
Transcriptases and p30 Proteins
of Mammalian
Type
C
Viruses
CHARLES J. SHERR,* LOUIS A. FEDELE, RAOUL E. BENVENISTE, AND GEORGE J. TODARO
Viral Leukemia andLymphoma Branch, National Cancer Institute,Bethesda, Maryland20014
Received forpublication 7 February 1975
The major internal structural proteins (p30) of type C viruses isolated from
several mammalian species were studied by radioimmunoprecipitation and
competitive radioimmunoassays. Three antigenically distinguishable sets of interspecies determinants could be demonstrated by both methods. One setof
determinants sharedbyviruses of rodentorigin (mouseandrat) canbedetected
readily in feline leukemia viruses butnot in othertype C viral groups. The p30 proteins ol murine viruses also contain a second discrete set of antigenic
determinants related to those in infectious primate viruses and endogenous
porcine viruses, but not detected in the feline leukemia virus group. The p30 proteinsofendogenousviruses of baboons and domesticcatsshareyetathirdset
ofcross-reactive determinantsnotdetected in type C viruses isolated from other species of animals. Enzyme inhibition studies performed with antisera raised
toward thereverse transcriptases of thesesame groups oftypeC viruses showed
the same patterns of immunological cross-reactions as observed with p30
pro-teins. The antigenic cross-reactions between the homologous proteins oftype C
virus isolated from genetically distant animals mayreflect transmission oftype
Cvirusesacrossspecies barriers.
Sensitive
immunological
assaysfor the struc-tural proteins of mammalian type C viruses have becomeincreasinglyimportant inclassify-ing new viral isolates and relating them to
known groups of viruses. Agroup-specific (gs-1) antigenicdeterminantshared
by
murine typeC viruses was first describedby Gregoriades
andOld (17) and has been
assigned
to a protein ofapproximately
30,000 molecularweight (p30),
themajor internalproteinofthe virion(32, 42). Subsequent studies showed that the p30 pro-teins of murine type C viruses also share an
interspecies antigenic determinant (gs-3) with
other rodent type C viruses (rat and hamster)
and with the feline leukemia viruses (11, 13, 33,
35, 43). With the characterization of type C
virusesisolatedfromother mammalian species, it hasbecome apparent that thep30proteinsof certain groups ofisolates show only weak gs-3 reactivity, whereas other classesofinterspecies antigenic determinants can be demonstrated (14, 14a, 21, 31, 35, 36, 44, 48). The immuno-logical cross-reactions observed between dif-ferent groups of viral p30 proteins do not ap-pear to be fortuitous, since similar cross-reac-tions are observed with other viral components including reverse transcriptase (1, 26, 37) and theenvelopeglycoproteingp7O(51, 52).
Serological and amino acid sequencing
stud-ies of homologous proteins from different
spe-cies have demonstrated a correlation between
the extent of antigenic cross-reaction and the
degree of sequence homology. Such approaches canbe used to estimate theextentof
evolution-ary divergence between species and provide dataingeneral agreement with taxonomic
clas-sifications based on anatomical considerations
and the fossil record (8, 15, 19, 20, 23, 28, 411. Similar experiments have indicated that the immunological cross-reactions detected
be-tween homologous proteins of different groups
of type C viruses are due to similarities in the amino acid sequences ofthese proteins as well (12, 34). Thus, the antigenic relatedness of the
structural proteins of different type C viruses
might reflecttheir ancestral origins and evolu-tionary history in different species of animals (12, 47).
The present report describes studies of in-terspecies antigenic determinants cn the p30 proteins and reverse transcriptases of several groups ofmammalian type C viruses. The data show that at least three antigenically distin-guishable classes of interspecies determinants can be detected among the groups of viruses studied. We propose that homologies observed between type C viruses isolated from genetically
distant vertebrate species reflect trans-species infections of either somatic cells, germ cells, or
both, which have occurred in the past (7).
1440
on November 10, 2019 by guest
http://jvi.asm.org/
MATERIALS AND METHODS
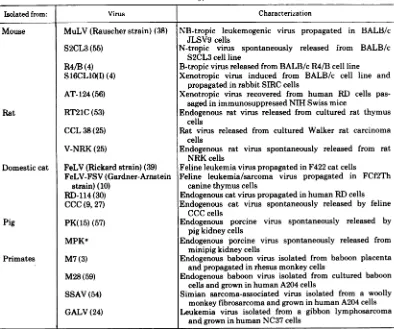
Viruses and cell lines. The mammalian type C
virusesused inthese studies are listed in Table 1 and
weregrown as described previously (49). Viruses were
concentrated from culture supernatants by
continu-ous-flowcentrifugation and were banded on sucrose at
adensity of 1.15 to 1.17g/cm3.
Purification of viral proteins. The p30 proteins of the simian sarcoma-associated virus (SSAV), the gibbon ape leukemia virus (GALV), the endogenous baboon virus M7, the endogenous domestic cat virus RD-114, the endogenous mouse virus S2CL3, feline leukemia virus (FeLV, Rickard strain), the murine leukemia virus (MuLV, Rauscher strain) were purified
by gel filtration and isoelectric focusing (49). The
p30protein of the endogenous porcine PK(15) virus
waspurifiedby column chromatographic methods as
described by Strand and August (51). All proteins had approximate molecular weights of 30,000 as
de-termined by electrophoresis in 7 and 13%
poly-acrylamide gels containing sodium dodecyl sulfate, and all were greater than 95% homogeneous as judged by multiple electrophoretic criteria (49). Viral reverse
transcriptases were partially purified as described
previously (40, 46).
Antisera. Antiseratop30proteinsand viral
polym-erases wereraised in New Zealand white rabbits (37,
46, 49). Animals received primary immunizations in
complete Freund adjuvant divided between the four footpads and two intradermal booster injections in
incomplete adjuvant. Intramuscular immunizations
were continued at 3-week intervals, and testbleeds
were obtained from an ear artery 7 days after each
booster injection.
Groups of two to three animals were immunized
with the same antigen. By the schedule described
above,maximumantibody titers top30proteins were
routinely obtained after four to five immunizations. Sera obtained at this time were titered by
immuno-precipitation withhomologous andheterologous
radio-labeledp30 proteins. No significantdifferenceswere
observed in the relativetiters of cross-reacting
anti-bodiesinindividual sera from eachgroup of animals.
Antisera to certain viral polymerases (RD-114, MuLV, FeLV) were raised in single rabbits, whereas
sera directed toward other viral enzymes [SSAV,
GALV, M7, PK(15)] were raised in groups of two to
three animals. In cases where comparisons were
possible, sera from different animals showed the same
patterns of cross-reactivity when used to inhibit
enzymes from heterologous viruses.
TABLE 1. Mammalian type C viruses studied
Isolatedfrom: Virus Characterization
MuLV(Rauscher strain) (38)
S2CL3 (55)
R4/B(4) S16CL10(I) (4)
AT-124(56)
RT21C(53)
CCL 38(25)
V-NRK (25)
FeLV(Rickardstrain)(39)
FeLV-FSV(Gardner-Arnstein
strain)(10)
RD-114(30)
CCC (9,27)
PK(15) (57)
MPKa
M7(3)
M28(59)
SSAV(54)
GALV(24)
NB-tropic leukemogenic virus propagated in BALB/c
JLSV9 cells
N-tropic virus spontaneously released from BALB/c
S2CL3cell line
B-tropic virus releasedfromBALB/cR4/Bcell line
Xenotropic virus induced from BALB/c cell line and
propagatedinrabbitSIRC cells
Xenotropic virus recovered from human RD cells
pas-sagedinimmunosuppressedNIHSwissmice
Endogenous rat virus released from cultured rat thymus
cells
Rat virus released from cultured Walker rat carcinoma
cells
Endogenous rat virus spontaneously released from rat
NRKcells
Feline leukemia viruspropagatedin F422catcells
Feline leukemia/sarcoma virus propagated in FCf2Th
caninethymuscells
Endogenouscatviruspropagatedinhuman RD cells Endogenous cat virus spontaneously released by feline
CCC cells
Endogenous porcine virus spontaneously released
by
pigkidneycells
Endogenous porcine virus spontaneously released from
minipigkidney cells
Endogenous baboon virus isolated from baboon placenta
andpropagatedinrhesusmonkeycells
Endogenous baboon virus isolated from cultured baboon
cells and growninhumanA204cells
Simian sarcoma-associated virus isolated from a
woolly
monkey fibrosarcoma andgrowninhumanA204cells
Leukemia virus isolated from a gibbon lymphosarcoma
and growninhumanNC37 cells
aM.M.Lieber,C. J.Sherr,R. E.Benveniste,andG.J.Todaro, Virology,inpress.
Mouse
Rat
Domesticcat
Pig
Primates
15, 1441
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.502.59.453.322.651.2]SHERR ET AL.
Radioiodination. Purified p30 proteins (10 gg)
were radiolabeled with 125I by the chloramine T
method to approximately equivalent specific activi-ties (5
mCi/mg)
(16).Labeledtest antigenswere stored in 0.05 M phosphate buffer (pH 7.5) containing 1%bovine serum albumin and were used within 8 days
afterlabeling. More than90% ofeach labeledantigen
could be specifically precipitated bythe homologous
antiserum.
Radioimmunoassays. Antisera titrations and
com-petitive radioimmunoassays were performed by a
"double-antibody" method (35, 46). Competition
as-says were initiated with 50% of the iodinated test antigen bound, anddetergent-disrupted viruses were used ascompetingantigens.Immunecomplexeswere precipitated with a titered excess of anti-rabbit 7S
globulin raised in sheep (Pocono Rabbit Farm and
Laboratories, Canadensis, Pa.). Supernatants were
counted by liquid scintillation to a +3% error, and
counting efficiency was monitored by external
stan-dardization.Thedataareplottedaspercent
displace-ment of the test antigen from immune complexes
(ordinate) versus micrograms of competing protein
(abscissa). Pointsonthe competitioncurvesrepresent
the average ofthree to six determinations for each
competingantigen (standarddeviation < 8%). Inthe
assays shown, no significant antigenic differences
wereobserved between the p30proteins ofthesame
typeCviruspropagatedincellsfromdifferentspecies
or between different isolates from the same type C
viral group(cf.Table 3).
Polymerase inhibition studies. All studies were
performed with immunoglobulin G (IgG) purified
from sera by salt fractionation and chromatography
onDEAE-cellulose.Reversetranscriptaseassayswere
performed,asdescribedpreviously (37), witha
polyri-boadenylate templateandanoligodeoxythymidylate
primer. Reactions were initiated with a mixture of
template, primer, and substrate.Detergent-disrupted
viruses(0.01 ml)wereusedas a sourceofenzyme,and
IgG was added prior to the addition ofenzyme. At
least 70,000 counts of [3H]thymidine
5'-monophos-phate per min wereincorporatedinto
acid-precipita-ble polydeoxythymidylate product after 60 min at
37C in theabsence ofimmune
IgG.
Incorporation
of[3H]thymidine 5'-triphosphate waslinear for 60 min
intheabsenceofspecific antibodies. Thequantitiesof
immune IgG required for 30% inhibition ofenzyme
activitywerecalculatedfrominhibitioncurves
devel-oped with serial dilutions of antibody. The actual
amounts ofIgGrequired for these levelsofinhibition
in homologous antibody-antigen systems are
pre-sented inthe legend to Table 4. The data havebeen
normalizedby assigninga value of 1.0 for the homolo-gous system.
Proteindetermination.The concentrations of pu-rified rabbit IgG were determined from absorbance
measurements at 280 nm (E"'-m = 14). All other
proteins were quantitated bythe method ofLowry et
al. (29), withbovine serumalbumin as astandard.
RESULTS
Although a spectrum of different antigenic determinants is probably present in most type
C viral proteins, three general classes of deter-minants have been described: type-specific
de-terminants, whichdistinguishdifferent isolates from the same type C viral group (e.g.,
Rauscher and Kirsten strains of MuLV);
group-specific determinants shared by a group of viruses isolated from the same species (e.g., MuLV's); andinterspecies determinants shared
by viruses isolated from different species (e.g.,
MuLV's and FeLV's) (la). In general, antisera raised to p30 proteins are directed
primarily
toward group-specific determinants,
although
manyseracontainantibodies of lower titer and
avidity which react with interspecies
determi-nants aswell.
Table 2 shows the results of a series of experiments in which antisera to p30 proteins
weretitrated against radiolabeled p30 proteins purified from the same or different type C viruses. The resultsareexpressedasthe recipro-cal oftheserumdilution requiredtobind20%of the labeled test antigen. An antiserum to the p30 protein of FeLV bound 20% of the 1251_ labeled FeLVtestantigenata serumdilutionof 1:400,000. At a serum dilution of 1:20,000, equivalent binding was obtained with the p30
proteins oftwomurinetypeC viruses(Rauscher
MuLV [Table
21
and the endogenous BALB/c N-tropic virus S2CL3 [not shown]). The cross-reactions between the p30 proteins of FeLV's and MuLV's reflect the presence of the gs-3determinant (11) and have beenobserved with
many sera prepared from several species of
animals (11, 13, 33, 35, 43, 44). By contrast, equivalentbinding of the p30 proteins ofSSAV, GALV, the endogenous porcine PK(15) virus, the endogenous feline virus RD-114, and the endogenous baboonvirus M7 wasonlyobserved atsignificantly lower dilutionsof theanti-FeLV
p30 serum. A lower-titer antiserum to the p30
protein ofR-MuLV showedareciprocalpattern
ofcross-reaction with the p30 protein of FeLV
and didnot reactstronglywiththep30 proteins
of theothertypeCvirusesstudied.These results
show that theinterspeciesdeterminants shared
by FeLV and MuLV p30 proteins are not as readily detected on the p30 proteins of several
othergroupsofmammalian typeC viruses.
At least twoother major classes of interspe-cies determinants can be identified on type C viral p30 proteins (Table 2). An antiserum to the p30 protein of SSAV reacts strongly with both the SSAV and GALV p30 proteins (cf. 14, 21, 36, 49, 60) andcross-reacts to alesserextent with the p30 proteins of the porcine PK(15)
virusandRauscher MuLV. An antiserum tothe
p30 proteinofthePK(15) virus also cross-reacts with SSAV, GALV, and MuLV p30 proteins.
1442 J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
TABLE 2. Antigenic cross-reactions betweenp30proteins of type C viruses as measured by immunoprecipitationa
I125-labeledp30 Relative antibody titer
protein Anti-FeLV Anti-R-MuLV Anti-SSAV ] Anti-PK(15) Anti-RD-114 Anti'M7
FeLV 400,000 4,000 400 100 100 400
R-MuLV 20,000 40,000 3,000 3,000 200 300
SSAV 100 200 200,000 5,000 <50 100
GALV 50 200 200,000 10,000 <50 <50
PK(15) 50 < 50 8,000 50,000 < 50 < 50
RD-114 800 <50 400 50 15,000 2,500
M7 1,600 400 400 .50 5,000 8,000
aPurified
p30
proteins were radiolabeled with 1251 and titrated with antisera raised toward homologous andheterologous p30 proteins. The results are presented as the reciprocal of the serum dilution required for 20%
binding of the labeled test antigen. Results obtained with the homologous antibody-antigen systems are
underlined andreflect the titers of the sera employed.
Neitherserum, however, reacts strongly to the p30 proteins of FeLV, RD-114, or M7. These
results suggest thatthe p30 proteins of SSAV,
GALV, PK(15), and MuLV share an interspe-cies determinant different from that shared by FeLV and MuLV. By contrast, antisera raised tothe p30proteins ofRD-114 andM7show that thep30proteins ofthesetwoviruses arehighly related (cf. 48, 49), whereas neither serum reactsstronglywith thep30proteins oftheother
isolates.
Competition radioimmunoassays were used to further study the interspecies determinants ofp30proteins in an attempt togeneralize these conclusions to other viral isolates. When such
assays are designed with an antiserum and homologous test antigen, the assays are
initi-ated withrelativelyhigh dilutionsofantisera so
that antibodiesbinding group-specific
determi-nants are preferentially selected (cf. Table 2). Cross-reactions
.with
unrelated groups of typeCviruses arethenrarely detected by competition.
In contrast, competitive radioimmunoassays employing heterologous combinations of anti-sera and labeled test antigens are initiated at lower serum dilutions, permittingthe detection
ofp30 proteinsfromdifferent but relatedgroups ofviruses.
Figure 1Ashows the results of acompetition
assay utilizing an antiserum tothe p30 protein
of FeLV and an iodinated p30 protein from Rauscher MuLV. Disrupted viruses were used
as competing antigens. Both FeLV and
Rauscher MuLV competed efficiently for the labeled test antigen; competition was also
ob-served with the
endogenous
rat viruses RT21C and CCL-38. Similar results have been seenwith endogenous murine type C isolates of various host range types [ATP-124, S2CL3,
S16CL10(I),
R4/B],
with another endogenousrat virus (VNRK), and with other strains of
FeLV. Only low levels of competition were
observed with SSAV, GALV, PK(15), RD-114, and M7 viruses or with other viruses of these groups(cf. Table 3). These results confirm that
the
p30
proteins ofFeLV and viruses ofrodent origin (rat and mouse) share interspeciesanti-genic determinants that are not readily
de-tectedby these methods inseveral othergroups of type C viruses.
In an assay with an antiserum to the p30 protein of the porcine PK(15) virus and a
radiolabeled SSAV p30 protein (Fig. 1B),
SSAV, GALV, and endogenous porcineviruses
all competed with identical slopes for the la-beled test antigen. All of the mouse viruses
tested cross-reacted inthis assay, as evidenced
by
the reducedslopes
of thecompetition
curves, indicating that MuLV's contain p30 proteins with interspecies determinants
par-tially
related to those found inSSAV,
GALV,
and endogenous porcine viruses. In contrast,only limited competition was observed in this
assay with FeLV or with the endogenous rat virusesRT21C, VNRK, and CCL38.
By
replac-ing the labeled SSAV test antigen with a labeled
p30
protein from the endogenous mu-rinevirus S2CL3 (Fig.1C),
it was againshown that thep30
proteinsfromSSAV,
GALV,
por-cine, and murine viruses share antigenic deter-minants not found in FeLV orendogenous
rat viruses. Thus, thep30
proteins of murine vi-ruses contain at least twodistinguishable
classes of interspecies determinants, one set
on November 10, 2019 by guest
http://jvi.asm.org/
SHERR ET AL.
80
6
uZ 60_
4 -J 40
20
0.003 0.01
z
g
n
MICROGRAMS VIRAL PROTEIN
01 10
MICROGRAMS VIRAL PROTEIN
100
p
z
Lfi J.Lf
n
Ca
80 60
20
UM
MICROGRAMS VIRAL PROTEIN
FIG. 1. Competitiveradioimmunoassays for p30proteins. Detergent-disruptedtype C viruseswereusedas
sourcesofcompeting antigens.Allassayswereinitiatedwithantiseratoonep30proteinandapurified, labeled p30proteinfromaheterologousvirus. Theamountofcompetingviralprotein requiredtoinitiatedisplacement
of the labeled test antigen reflects both the degree ofpurity of the competing virus preparation and the
sensitivityoftheassaysystem.Intheassaysshown,a20%displacement ofthelabeledtestantigenisobtained
withIto2ngofunlabeled p30proteinidenticaltothetestantigen.Thereagentsemployed in the variousassays were:(A)Anti-FeLVp30: I25I-MuLVp30; (B) Anti-PK(15)p30:I25I-SSA Vp3O; (C) Anti-PK(15)p30:125I-S2CL3
p30; (D) Anti-RD-114 p30:I25I-M7p30. Symbols: MuLV (Rauscher strain); A, S16CL10(I) (endogenous
virusfrom BALBIcmouse); A,AT-124(endogenous virus from NIH/Swissmouse);*, CCL38(endogenousrat
virus); 0,RT21C (endogenous ratvirus); 0,SSAV (infectious primate virus); 0, GALV (infectious primate
virus);V, PK(15) (endogenousporcinevirus); x,FeLV (Rickard
strain);O,
RD-114(endogenous domesticcatvirus);*,M7(endogenous baboon virus).
shared with FeLV's andendogenousrat viruses
(Fig. lA), and a second setshared with SSAV, GALV,andendogenous porcine viruses(Fig. 1B
and C).
By using an antiserum to the p30 protein of
theRD- 114 virus andalabeledp30 proteinfrom
the M7 baboon virus (Fig. 1D), it was shown
that the RD-114 and M7 viruses share
interspe-cies determinantsnotfound in thep30 proteins
of other type C viral groups (49). Identical
re-actions also were seen with other endogenous
feline and baboon type C isolates (CCC and
M28, respectively).Thisassay, then,definesyet
another set of interspecies determinants that
distinguish endogenousbaboon andendogenous
feline type C viruses from the other groups
ofviruses studied.
Table 3 summarizes the resultsobtainedwith
radioimmunoassaysforp30proteins.The slopes of the competition curves generated with
ho-mologous competing antigens are assigned
val-ues of 1.0, and the degree of antigenic
cross-reactionseenwithheterologous competing
anti-gens has been determined by a slope-ratio
method (49). The results generalize the data
showninTable2toadditional viral isolates and
show that at least three distinguishable sets of interspecies antigenic determinants can be
found on p30 proteins of mammalian type C viruses. Goldbergetal. (14a)have alsoreported
similar patterns ofcross-reactivity between the
p30 proteins of these different groups of type C viruses.
To determine whether the cross-reactions
observed between p30 antigens could also be
demonstrated with anothertypeC viralprotein, the immunological properties of the reverse
transcriptasesfromtheseviruseswerealso
stud-ied. Table 4 shows the results of studies in which antisera were used toinhibit the polym-C.
100
a
Z 60
-J 40
20
0.
I I
)- D. _4- 4
n)arrl r) I n If
).003 001
1444 J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.502.63.453.54.328.2]TYPEC VIRAL ANTIGENS
erases from various viruses. The results are
presented as the relative quantity of immune
IgG required for 30% inhibition of enzyme
activity. Although only data obtained with
respresentative viruses and sera are shown,
quantitatively similar results have been ob-tained with the other isolates from these same
virus groups (Table 1) and with other antisera
preparations.
Table 4 indicates that the antisera to viral polymerases used in thesestudies react prefer-entially with group-specific determinants. Sig-nificantlygreaterquantitiesof immuneIgGare
required to inhibit the activity of heterologous viral enzymes as compared to the amounts
required for comparable levels of inhibition of the homologous polymerases. Like thep30
[image:6.502.54.451.200.391.2]anti-sera, cross-reactions withheterologous enzymes
TABLE 3. Antigenic cross-reactions betweenp30 proteins of type C viruses as measured bycompetitive
radioimmunoassaya
Cross-reaction
Virus Isolated from: Anti-FeLV: Anti-PK (15): Anti-RD-114:
MuLV SSAV M7
FeLV(Rickard) Domesticcat 1.07 0.12 <0.10
FeLV(Gardner-Arnstein) 1.05 0.11 <0.10
RT21C Rat 0.95 <0.10 0.10
CCL-38 0.95 <0.10 0.10
MuLV (Rauscher) Mouse (1.00) 0.52 0.15
S2CL3 0.97 0.51 <0.10
AT-124 0.98 0.49 0.10
SSAV Woolly monkey 0.21 (1.00) <0.10
GALV Gibbon ape 0.18 1.00 <0.10
PK(15) Pig 0.28 1.02 <0.10
MPK 0.22 1.00 <0.10
RD-114 Domesticcat 0.28 0.11 0.90
CCC 0.23 <0.10 0.98
M7 Baboon 0.27 0.10 (1.00)
M38 0.28 0.11
.99J
aAntigenic cross-reactions were quantitated by comparing the slopes of competition curves. Data were
calculated by obtaining the average slope of at least six separate competition curves for each antigen. The
averageslope of the competition curves for the homologous competing antigen in each assay was assigned a
value of 1.00. The ratio, slope of competition curve (cross-reacting antigen)/slope of competition curve (homologousantigen), isindicated in the table and is an index of the relative degree of cross-reaction between different antigens tested in the same assay (49). The homologous competing antigens are indicated by parentheses.
TAJz4. Inhibition of viralreversetranscriptaseactivity byantiseratoviralpolymerasesa
Viral Relative concentration of immuneIgG required for 30% inhibition of enzyme activity enzyme
tested Anti-FeLV Anti-MuLV Anti-SSAV Anti-GALV Anti-PK (15) Anti-RD-114 Anti-M7
FeLV |(1.0) 32 >50 >50 >15 >50 >50
MuLV l9.2 (1.0) 39 10 5.9 >50 >50
SSAV >50 >50 (1.0) 2.1 3.1 >50 >50
GALV >50 >50 1.8 (1.0) 5.4 >50 >50
PK(15) >50 >50 11 21 (1.0) >50 >50
RD-114 >50 >50 >50 >50 >15 (1.0) 2.0
M7 >50 >50 >50 >50 >15 1.6 (1.0)
aAntiseratopartially purified viral polymerasesofFeLV(Rickard strain), MuLV(Rauscherstrain), SSAV,
GALV,endogenous porcinevirus PK(15),endogenousfeline virusRD-114, and endogenous baboon virus M7
werepreparedandimmuneIgGpurified.Enzyme inhibitioncurves wereobtainedby using serial dilutions of
immune IgG and detergent-disrupted viruses as sources of enzymes. The results have been normalized by
assigningavalue of1.0(indicatedbyparentheses) forthehomologousantibody-antigen systems. The actual
amounts ofimmune IgG required for 30% inhibition of enzyme activity in the homologous systems are:
Anti-FeLV:FeLV, 0.95jg; Anti-MuLV:MuLV, 1.2pg;Anti-SSAV:SSAV, 1.0 ug; Anti-GALV:GALV, 0.81Mg;
Anti-PK(15):PK(15), 3.1 Mg;Anti-RD-114:RD-114, 0.09Mg; Anti-M7:M7, 1.0Mg.
1445
VOL.15,1975
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.502.55.453.471.586.2]SHERR ET AL.
canbedetected,and thepatternsobservedwith the reverse transcriptases are similar to those
obtained with p30 antigens (compare Tables 2 and 4).
Antiseratothereversetranscriptasesof FeLV and Rauscher MuLV showed reciprocal
cross-reactivity and did not significantly inhibit en-zymes from other type C viral groups. An
antiserum to the polymerase ofSSAV strongly
inhibited the enzyme from GALV and
cross-reacted to a lesserextent with the polymerases of the porcine PK(15) virus and MuLV. A
similar pattern wasobservedwith anantiserum
tothe reversetranscriptase ofGALV, although a greater degree ofcross-reaction was observed with the murine viral enzyme than with the
PK(15) polymerase. An antiserum to the
PK(15) enzyme strongly inhibited the polymer-ases ofSSAV andGALV and,toalesserextent,
the MuLV reverse transcriptase. Table 4 also shows that the polymerases of the endogenous felinevirus RD-114and theendogenousbaboon virus M7 are closely related antigenically
al-though neither enzyme appears immunologi-callyrelated tothe other reversetranscriptases studied. These data show that three
distin-guishablesets ofinterspecies antigenic
determi-nants can beidentifiedon thepolymerasesfrom
different groups ofmammaliantype C viruses.
DISCUSSION
Cells from a variety of mammalian species contain sequences in their DNA that can code forthe productionofendogenoustype C viruses.
These virogene sequences are thought to be present inboth thesomaticand germ cellsofall
members of the species and are inherited as
integral components ofthecellulargenome (22, 58). Endogenous virogene sequences held in common among related species should be the
direct descendants ofthe same sequence pres-ent in the most recent common ancestor. The
degree of base pair mismatching observed be-tween endogenous virogenes of tworelated spe-cies should then be a relative index ofthetime since the twospeciesdivergedfromoneanother.
For example, sequences related to endogenous
baboon type C viral RNA have been identified
inall the other Old World
monkeys,
thehigherapes, and man, and the degree ofrelatednessof such sequences to baboon type C viral RNA correlates well with the knowntaxonomic
rela-tionships ofthe primate species (2, 6).
The proteinsofendogenous virusesofrelated species, like proteins encoded byother cellular genes, areexpected to have related amino acid
sequences. Serological studies ofproteins from
different
species
have indicated thatimmuno-logical
cross-reactivity
can beeffectively
usedas ameasureof amino acid sequence
dissimilar-ity
and as an index ofthe time ofevolutionary
divergence
ofrelatedspecies (8,
12,
15, 19, 20,
24,
28, 41).
Asexpected,
theproteins
ofendoge-nous viruses of related
species
show ahigh
degree
ofimmunological
cross-reactivity.
Forexample,
thehomologous proteins
ofendoge-nous rodent
type
C viruses areantigenically
similar
(11, 13,
14a, 34, 35,
44) as are thep30
proteins
isolated from several groups ofpri-mates
(47,
50).
However,
in certain cases, thehomologous
proteins
of viruses isolated fromgenetically
unrelatedspecies
ofanimals showanunexpect-edly
high degree
ofantigenic
relatedness. Theproteins
ofendogenous
viruses isolated fromprimates
and domesticcats exhibit anextraor-dinary degree
ofhomology (18,
48, 49, 59)
eventhough
these groups of animals have beenevolutionarily
separated
for at;least 80 million years.These resultssuggested
that theRD-114/
CCC group ofdomestic cat viruseswasderived from
endogenous
primate
viruses that infectedtheancestorofcertainFelis
species
atapoint
inrecent
evolutionary history,
a conclusionstrongly
supported
by
nucleic acidhybridiza-tion
experiments (7).
Thus,
animals of onespecies
may be infectedby
type
C virusesderived from a
second,
distantly
related group ofanimals, leading
to theincorporation
ofnewvirogenes
intotheDNA of infected animalsand,
ultimately,
totheperpetuation
ofthese genes inthe germ line.
Antigenic
cross-reactions observed betweenthe
homologous
proteins
of type C viruses iso-lated from unreiso-latedspecies
of animals may thus beexplained
by
horizontal transmission oftype
C viruses acrossspecies
barriers. We pro-pose that such events may be more common thanpreviously thought,
and thatserological
studies may offer the first clues as to the
origins
of certain type C isolates. Theinter-species
determinants sharedby SSAV,
GALV,
and
endogenous porcine
andmurineviruses sug-gest that these virusesoriginated
from acom-mon ancestor. The presence of viruses of the
SSAV-GALV group in
primates,
wherethey
are not
endogenous,
would appeartoexemplify
a
relatively
recent infection. Viruses of theSSAV-GALV groupcontain RNA genomes that are
partially homologous
to sequences in the genomes of murine type C viruses(5).
ThatSSAVand GALVmay bederived from murine viruses that infected
primates
is supportedby
theobservation that sequencesrelated toSSAV
1446 J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
and GALV RNA canbe found in murine DNA
(2) but not in the DNA of normal woolly
monkeys or gibbons (2, 45). Similarly, the
re-lationships observed betweenrodent viruses and viruses of the FeLV group, now horizontally
transmitted in domestic cats, may also reflect
infection ofcats with viruses ofrodent origin. Surveys for related groups of endogenous
virogene sequences in species of animals both
related and unrelated tothose from which type C viruses have been isolated should provide
additional insights into the validity and generality of these interpretations.
ACKNOWLEDGMENTS
We thankLinda Tolerand BarbaraDetwilerforassistance
with these experiments, and Michael M. Lieber for his
suggestions concerningthemanuscript.
Thisworkwassupported by theVirus CancerProgramof
the NationalCancerInstitute.
LITERATURE CITED
1. Aaronson,S. A., W. P. Parks,E. M. Scolnick,andG. J.
Todaro. 1971. Antibody tothe RNA-dependent DNA polymeraseof mammalian C-typeRNAtumorviruses.
Proc. Natl.Acad. Sci. U.S.A.68:920-924.
la. August, J. T., D. P. Bolognesi, E. Fleissner, R. V. Gilden, and R. C. Nowinski.1974.Aproposed nomen-clature forthe virion proteins ofoncogenic RNA
vi-ruses. Virology 60:595-601.
2. Benveniste, R. E., R. Heinemann, G. L. Wilson, R. Callahan,andG.J. Todaro. 1974. Detection of baboon
type C viral sequencesinvariousprimatetissues by molecularhybridization.J. Virol. 14:56-67.
3. Benveniste, R.E.,M. M.Lieber,D. M.Livingston,C. J.
Sherr, G.J.Todaro,and S. S. Kalter. 1974. Infectious typeCvirus isolated fromababoonplacenta. Nature (London) 248:17-20.
4. Benveniste, R.E.,M. M.Lieber,and G. J. Todaro. 1974. A distinct class of inducible murine type C viruses
whichreplicateinthe rabbit SIRC cellline. Proc.Natl. Acad. Sci. U.S.A. 71:602-606.
5. Benveniste, R. E., and G. J. Todaro. 1973. Homology betweentypeC viruses of variousspeciesasdetermined
by molecular hybridization. Proc. Natl. Acad. Sci. U.S.A.70:3316-3320.
6. Benveniste, R.E., and G.J.Todaro. 1974. Evolution of typeCviralgenes.I. Nucleicacid from baboontypeC
virus as a measure of divergence among primate species.Proc. Natl.Acad.Sci. U.S.A.71:4513-4518.
7. Benveniste, R.E., and G. J.Todaro. 1974. Evolution of typeCviralgenes.Inheritance ofexogenously acquired viralgenes.Nature(London) 252:456-459.
8. Buettner-Janusch, J.,and R. L. Hill. 1965. Molecules and
monkeys. Science 147:836-838.
9. Fischinger, P. J.,P. T. Peebles,S.Nomura, and D. K.
Haapala. 1973. Isolation of an RD-114-like oncor-navirusfromacatcell line.J. Virol. 11:978-985. 10. Gardner,M.B.,P.Arnstein, E.Johnson,R. W.Rongey,
H. P. Charman, and R. J. Huebner. 1971. Feline
sarcomavirustumorinduction incatsanddogs.J.Am. Vet.Med. Assoc. 158:1046-1053.
11. Geering, G.,T. Aoki,and L. J. Old. 1970. Shared viral antigenof mammalian leukemia viruses.Nature (Lon-don) 226:265-266.
12. Gilden,R.V.,and S. Oroszlan. 1972. Coevolutionof RNA tumor virus genomes and vertebrate host genes, p.
460-476. In Molecular studies in viralneoplasia. The
Williams & Wilkins Co., Baltimore.
13. Gilden, R. V., S. Oroszlan, and R. J. Huebner. 1971.
Coexistence of intraspecies and interspecies specific
antigenicdeterminantson themajorstructural
poly-peptidesof mammalianC-typeviruses.Nature
(Lon-don) NewBiol.231:107-108.
14. Gilden, R. V., R. Toni, M. Hanson, D. Bova, H. P. Charman, and S. Oroszlan. 1974. Immunochemical studies of the major internal polypeptide of woolly monkey and gibbonapetypeC viruses. J. Immunol. 112:1250-1254.
14a. Goldberg, R. J., E. M. Scolnick, W. P. Parks, L. A.
Yakovleva, and B. A. Lapin. 1974. Isolation ofa pri-mate type C virus from a lymphomatous baboon. Int. J. Cancer 14:722-730.
15. Goodman, M., G. W. Moore, and W. Farris. 1974.
Primatephylogeny from theperspective ofmolecular systematics. Transplant. Proc. 6:217-222.
16. Greenwood, F. C., W. M. Hunter, and J. S. Clover. 1963. The preparation of1'3I-labeled humangrowth hormone ofhigh specific radioactivity.Biochem. J.89:114-123. 17. Gregoriades, A., and L. J. Old. 1969. Isolation andsome
characteristics ofagroup-specific antigen of the murine leukemiaviruses.Virology 37:189-202.
18. Hellman, A., P. T. Peebles, J. E. Strickland, A. K.
Fowler, S. S. Kalter, S. Oroszlan, and R. V.Gilden.
1974.Baboon virus isolate M7 with properties similar tofeline virusRD-114. J. Virol. 14:133-138.
19. Henke, N., E. M. Prager, and A. C. Wilson. 1973.
Quantitative immunological and electrophoretic com-parison of primate lysozymes. J. Biol. Chem. 248:2824-2828.
20. Hermann,J., J. Jolles, D. H. Buss, and P. Jolles. 1973. Aminoacidsequenceoflysozyme from baboon milk.J.
Mol.Biol.79:587-595.
21. Hoekstra, J., and F. Deinhardt. 1973. Simiansarcoma andfeline leukemia virusantigens: isolationof species-and interspecies-specific proteins. Intervirology 2:222-230.
22. Huebner, R. J., andG. J. Todaro. 1969. Oncogenesof
RNAtumorviruses asdeterminants ofcancer. Proc.
Natl. Acad. Sci. U.S.A.64:1087-1094.
23. Hummel, B. C. W., G. M. Brown, B. R. Webster, andJ. C. Paice. 1974. Cross-reactivity of monkey pituitary
extracts in human radioimmunoassays. Gen. Comp. Endocrinol. 23:143-153.
24. Kawakami, T., S. D. Huff, P. M. Buckley, D. L.
Dungworth, S. P. Snyder, and R. V. Gilden. 1972.
C-type virusassociated withgibbon lymphosarcoma.
Nature(London) NewBiol.235:170-172.
25. Lieber, M.M., R. E. Benveniste,D. M.Livingston,and G. J. Todaro. 1973. Mammalian cells in culture
fre-quentlyrelease type Cviruses. Science182:56-59.
26. Livingston,D.M.,E. M.Scolnick,W. P.Parks,and G. J. Todaro. 1972. Affinity chromatography of
RNA-dependentDNApolymerasefrom RNA tumorviruses
on asolid-phase immunoadsorbent. Proc.Natl. Acad. Sci.U.S.A.69:393-397.
27. Livingston,D. M.,and G.J. Todaro. 1973.Endogenous typeC virus fromacatclonewithpropertiesdistinct frompreviously describedfeline type C viruses.
Virol-ogy53:142-151.
28. Lovejoy, C. O., A. H.Burstein,and K. G.Heiple. 1972.
Primate phylogeny and immunological distance. Science 176:803-805.
29. Lowry, 0. H., N. J. Rosebrough, A. L. Farr, and R. J.
Randall. 1951. Protein measurement with the Folin phenolreagent.J. BiolChem. 193:265-275.
30.McAllister, R. M., M.Nicolson, M. B.Gardner, R. W. Rongey, S. Rasheed,P. S.Sarma, R. J.Heubner,M. Hatanaka, S. Oroszlan, R. V. Gilden, A. Kabigting,
and L. Vernon. 1972. C-type virus released from
1447
on November 10, 2019 by guest
http://jvi.asm.org/
SHERRET AL.
cultured human rhabdomyosarcoma cells. Nature
(London) New Biol. 235:3-6.
31. Oroszlan, S., D. Bova, M. H.Martin-White,R. Toni,C.
Foreman, and R. V. Gilden. 1972. Purification and
immunologic characterization of the major internal
protein of the RD-114 virus. Proc. Natl. Acad. Sci. U.S.A. 69:1211-1215.
32. Oroszlan, S., C. L. Fisher, T. B. Stanley, and R. V.
Gilden. 1970. Proteins of the murine C-type RNA tumorviruses:isolationofagroup-specificantigenby
isoelectric focusing.J.Gen.Virol.8:1-10.
33. Oroszlan, S., R. J. Huebner, and R. V. Gilden. 1971.
Species-specific and interspecific antigenic determi-nants associated with the structural proteinoffeline
C-type virus. Proc. Natl.Acad.Sci. U.S.A.68:901-904.
34. Oroszlan, S., M. R.Summers, C. Foreman, and R. V.
Gilden. 1974. Murine type C virus group specific antigens: interstrain immunochemical, biophysical, and amino acid sequence differences. J. Virol.
14:1559-1574.
35. Parks,W.,and E.M.Scolnick.1972.Radioimmunoassay
of mammalian type C viral proteins: interspecies antigenicreactivities of themajorinternalpolypeptide. Proc. Natl. Acad. Sci. U.S.A. 69:1766-1770. 36. Parks,W.P.,E.M.Scolnick,M.C.Noon,C. J.Watson,
and T. G. Kawakami. 1973. Radioimmunoassay of
mammaliantypeCpolypeptides.IV.Characterization
ofwoolly monkey and gibbon viral antigens. Int. J. Cancer12:129-137.
37. Parks,W.P.,E. M.Scolnick,J.Ross,G.J.Todaro,and S. A. Aaronson. 1972. Immunologic relationships of reverse transcriptases from ribonucleic acid tumor
viruses.J. Virol. 9:110-115.
38. Rauscher, F. J. 1962. Avirus-induced disease of mice
characterized byerythrocytopoiesisandlymphoid leu-kemia. J.Natl.Cancer Inst.29:515-543.
39. Rickard,C.G.,J. E.Post,F.Noronha, and L. M. Barr. 1969. Atransmissible virus-induced lymphocytic leu-kemia ofthecat.J. Natl. Cancer Inst. 42:987-1014. 40. Ross, J., E. M. Scolnick, G. J. Todaro, and S. A.
Aaronson. 1971. Separation of murine cellular and
murine leukemia virus DNA polymerases. Nature
(London) 231:163-167.
41. Sarich,V.M., andH.C. Wilson. 1967. Rates of albumin
evolution in primates. Proc. Natl. Acad. Sci. U.S.A.
58:142-148.
42. Schafer, W.,F. A.Anderer,H.Bauer,and L. Pister.1969. Studies on mouse leukemia viruses. I. Isolation and
characterization ofagroup-specific antigen. Virology
38:387-394.
43. Schafer, W.,J.Lange,D.Bolognesi,F. DeNoronha,J. E.
Post,and C.Rickard.1971.Isolation and characteriza-tion oftwogroup-specific antigensfromfelineleukemia
virus.Virology44:73-82.
44. Schafer, W.,L.Pister,G.Hunsmann, and V.Moennig.
1973.Comparative serological studiesontypeCviruses of various mammals. Nature (London) New Biol. 245:75-76.
45. Scolnick,E.M., W.Parks, T. Kawakami, D.Kohne,H.
Okabe, R. Gilden, and M. Hatanaka. 1974. Primate and murine type C viral nucleic acid association
kinetics: analysis of modelsystemsandnatural tissues. J.Virol. 13:363-369.
46. Scolnick,E.M., W. P. Parks, and D. M. Livingston. 1972.
Radioimmunoassay of mammalian typeC viral
pro-teins.I.Species-specificreactions of murineand feline
viruses.J.Immunol. 109:570-577.
47. Sherr, C.J., R. E.Benveniste, and G. J. Todaro. 1974.
TypeCviralexpression in primate tissues. Proc.Natl. Acad. Sci. U.S.A.71:3721-3725.
48. Sherr, C. J., M.M.Lieber, R.E. Benveniste, and G.J.
Todaro. 1974. Endogenous baboon virus (M7): bio-chemical and immunologic characterization. Virology 58:492-503.
49. Sherr,C.J.,and G. J. Todaro. 1974.Radioimmunoassay
of the major group specific protein of endogenous
baboon type C viruses: relation to the RD-114/CCC
group and detection of antigen in normal baboon
tissues.Virology 61:168-181.
50. Sherr, C. J., and G. J. Todaro. 1974. Type C viral antigensin man.Antigensrelatedtoendogenous pri-matevirus in human tumors. Proc. Natl. Acad. Sci. U.S.A.71:4703-4707.
51. Strand, M., andJ. T. August. 1973.Structuralproteins of
oncogenicribonucleic acidviruses.InterspecII,anew
interspeciesantigen.J. Biol. Chem.248:5627-5633. 52. Strand, M., and J. T. August.1974.Structural proteinsof
mammalian oncogenic RNA viruses: multiple
anti-genic determinants of themajorinternal protein and envelopeglycoprotein. J.Virol. 13:171-180.
53. Teitz, Y. W., E. H. Lennette, L. S. Oshiro, andN. E.
Cremer. 1971.ReleaseofC-typeparticles from normal ratthymuscultures and those infected with Moloney leukemiavirus.J.Natl. CancerInst. 46:11-23. 54. Theilen, G.H., D. Gould,M. Fowler, and D. L.
Dung-worth. 1971.C-typevirus in tumortissueofawoolly
monkey (Lagothrix spp.) with fibrosarcoma. J. Natl. Cancer Inst.47:881-889.
55. Todaro, G. J. 1972. "Spontaneous" release of type C
viruses from clonal lines of "spontaneously" trans-formed BALB/3T3 cells. Nature (London) NewBiol.
240:157-160.
56. Todaro,G.J.,P. Arnstein, W.P.Parks, E. H. Lennette,
and R. J. Huebner. 1973. A typeC virus in human rhabdomyosarcoma cells after inoculation into
anti-thymocyteserum-treated NIH Swissmice.Proc.Natl. Acad.Sci. U.S.A. 70:859-862.
57. Todaro, G. J., R. E. Benveniste,M.M.Lieber, and C.J. Sherr. 1974.CharacterizationofatypeCvirusreleased
from theporcine cellline, PK(15). Virology58:65-74. 58. Todaro, G. J., and R. J. Huebner. 1972. The viral
oncogenehypothesis. New evidence.Proc.Natl. Acad. Sci.U.S.A.69:1009-1015.
59. Todaro, G. J., C. J. Sherr, R. E. Benveniste, M. M.
Lieber, and J. L. Melnick. 1974. Type C viruses of
baboons: isolation from normal cell cultures. Cell
2:55-61.
60. Tronick, S. R., J. R. Stephenson, and S. A. Aaronson.
1974. Comparative immunological studiesof RNA
C-type viruses: radioimmunoassay for a low molecular
weight polypeptide ofwoolly monkey leukemia virus.
Virology 57:347-356.
1448 J. VIROL.