0022-538X/82/070234-07$02.00/0
Capping of Viral RNA in Cultured Spodoptera
frugiperda
Cells Infected with
Autographa californica Nuclear
Polyhedrosis
Virus
QINJUN-CHUANtANDROBERT F. WEAVER*
Departmentof Biochemistry, University ofKansas, Lawrence, Kansas 66045
Received1
February 1982/Accepted
25March 1982Viral RNA from fall
armyworm(Spodoptera
frugiperda)
cells infected
withAutographa californica
nuclear
polyhedrosis virus contains
capstructures.
Mostof the
caplabeled in vivo with
[3H]methionine
or 32pI cochromatographed onDEAE-cellulose with the -5
charge
marker;
aminor
comonent
appeared at
-4 netcharge. The former is
probably
a cap1
structure(m
GpppXmYp),
and
the latter isprobably
acap
0(m7GpppXp).
On the basis of relativelabeling
of the two capswith
[3H]adenosine
and
[
H]guanosine,
weconcluded that each
capcon-tained
atleast
oneadenosine residue in addition to
guanosine and, therefore,
that cap0contained
m7GpppAp. Cleavage of [3H]methionine-labeled viral RNA
withtobacco acid
pyrophosphatase
released
alabeled
component that
cochromato-graphed
onpolyethyleneimine-cellulose
with
m7Gp, confirming the m7GpppX
linkage in the
cap.These
results
werealso
seenwith host
polyadenylated
RNA.
The
capsappeared
in
nearly equal
abundance in viral
polyadenylated and
non-polyadenylated
RNAs. The ratio of
32p;
incorporated into the
capto
thatincorporated into mononucleotides
suggested
averagelengths for the
polyade-nylated and
non-polyadenylated
RNAs of
1,800
and
1,200 nucleotides,
respective-ly.
Since the
cap structurewasfirst identified
by
Furuichi (7)
atthe 5'
terminus
of
mRNA from
cytoplasmic
polyhedrosis virus,
capshave been
reported in
mosteucaryotic
mRNAs.
RNAsfrom certain animal (2, 6) and
plant
(4) viruses
and lower
eucaryotes(5, 20) contain
a cap0
structure
(m7GpppXp)
(19). Other viral mRNAs
(8, 22) and mRNAs from
higher
eucaryotescontain
acap 1with
oneadditional methyl
group(m7GpppXpYp)
(1).
Mammalian mRNAs have
aminor
amountof
afurther methylated
capspe-cies,
cap 2(m7GpppXmY'Zp)
(3). These capspecies all resist hydrolysis by alkali
or RNaseT2
because of the unusual
5'-5' triphosphatelinkage
andthe2'-O-methylation
on the X and Yresidues
that prevents formation of the 2',3'cyclic
hydrolysis
intermediate. These earlystud-ies
are reviewed in reference 19.Nuclear
polyhedrosis virus
(NPV) is abacu-lovirus
whose members have been isolated fromhundreds
of insect species. Its virions arecon-tained in
polyhedral inclusion bodies that arecomposed
mainly of a viral structural proteincalled
polyhedrin.
The alfalfa looper(Autogra-pha californica)
NPV is easily grown in cultured fall armyworm (Spodoptera frugiperda) cells (9)and
is therefore
a convenient model for studyingt Permanent address: Department of Virology, Wuhan Uni-versity, Wuhan, People's Republic of China.
all of
the NPVs. From the host
cells,
weob-tained three forms of RNA
polymerase which,
after
purification, had the characteristics of the
classic
polymerases
I,II, and III (11).
Using
isolated nuclei from infected cells,
wehave
shown that
mostof the late
viral RNA synthesis
is
notinhibited by
a-amanitin and is therefore
not
effected by
host RNApolymerase
II(10).
This unique finding raised the question of
whether
the bulkof the viral RNA is capped, ascapping
occursprimarily
on mRNA andits
precursors, which are normally made by poly-merase
II. The
presentstudy shows for the firsttime that NPV
RNAis
indeed fully capped andelucidates
some characteristics of the caps.MATERIALS AND METHODS
Biochemicals. Tobacco acid pyrophosphatase (TAP)
was purchased fromBethesda Research
Laboratories,
Inc. RNase T2(Aspergillus oryzae) grade VI was from
Sigma Chemical Corp.
32p,
(25 mCi/ml, carrier free)and
L-[methyl-3H]methionine
(12 Ci/mmol) were fromNew England Nuclear Corp. [8-3H]guanosine (14
Ci/mmol) was from ICN, Chemical & Radioisotope Div.
[2,8-3H]adenosine
(33 Ci/mmol) was fromSchwarz/Mann. DEAE-cellulose (fine) was from
Sig-ma. Polyethyleneimine (PEI)-cellulose F (precoated
thin-layer chromatography plastic
sheets;
layerthick-ness, 0.1 mm) was from Brinkmann Instruments, Inc.
Oligodeoxythymidylate [oligo(dT)]-cellulose (type 3)
234
on November 10, 2019 by guest
http://jvi.asm.org/
wasfromCollaborative Research, Inc. m-Aminoben-zyl-oxymethyl-cellulose was from Miles-Yeda Ltd.
Completecocktail 3a70forliquidscintillation counting
wasfrom ResearchProducts International Corp. All
otherchemicalswere reagent grade.
Viruses and cells. The continuous S.frugiperdacell
lineIPLB-SF-21 wascultured and infected by
nonoc-cluded virions of A.californicaNPV on a
supplement-ed TC-100 msupplement-edium asdescribed previously(10).
Labelingof RNAinvivo. Forlabeling hostpolysomal
RNA, S.frugiperda cells were grown to near
con-fluency(ca. 2 days after cells weretransferred). The
medium was removed, and each plate of cells was
labeledfor 4 h with 100,uCiof[methyl-3H]methionine
in 5 ml ofmethionine-freemedium, 2.5 mCi of
32Pi
in 5ml of phosphate-free medium, or 100
pLCi
of[8-3H]guanosineor[2,8-3H]adenosine in 5 ml ofnormal
medium.Afterbeinglabeled,thecellswereharvested
and the RNA was isolated. ForlabelingtheviralRNA,
the cells wereinfected at amultiplicity of infection of
10to 20
50%o
tissue cultureinfective doses per cell 24to 48 h after being seeded. From 1.0 to 1.5 h was
allowed foradsorption. Infected cellswerelabeled as
described above for 4 h (16 to 20 h postinfection). Polyhedralinclusion bodies were clearly visible in the
infectedcells under a light microscope. At 24 h
postin-fection, about
90%1o
of thecells containedpolyhedralinclusion bodies.
Isolationof host
poly(A)+
RNA. Thepolysomeswereisolated from the cells by the procedure of Palmiter
(16), modifiedasfollows. Labeledcellsfrom 10 plates
were harvested by scraping and then centrifuged at
4,000rpmin aBeckmanJS13 rotorfor 5 min. Thecell
pellet
wassuspendedin 8 mlof bufferA(25mMNaCl,5mMMgCl2,25 mMTris-hydrochloride[pH 7.5],2% Triton X-100, 1 mg of heparin per ml) and homoge-nized with 10 passes of a type A rod in a Dounce
homogenizer. The extract was then centrifuged at
8,000rpm(JS13rotor)for 20 min. Thesupernatantwas
collected,mixed with anequalvolumeof bufferB(25
mMNaCl,200 mMMgCI2,25mM
Tris-hydrochloride
[pH 7.5], 2% Triton X-100, 1 mgofheparin per
ml),
andthen put on icefor1h. Thepolysome
suspension
waslayered slowlyover8 mlof buffer C(25mM
NaCl,
5mMMgCl2,25 mMTris-hydrochloride [pH
7.51,
1 Msucrose)and spun downat8,000rpm
(JS13
rotor)for50 min. All ofthe processes described above were
performed
at0to4°C.
Thesupernatantwasdecanted,
and thepelletofpolysomeswasdissolved in 4 ml of
NETS (10 mM
Tris-hydrochloride [pH 7.5],
0.1 MNaCl, 0.5% sodium
dodecyl
sulfate[SDS],
5 mMEDTA).This
polysome
preparation
wasusedimmedi-atelyorstoredat
-70°C
untilneeded.Beforeextraction,3 Msodiumacetate(pH 5.2)was
added to thepolysome solutionto afinal concentration
of 0.05M.Theextractwasmixedwith 4 ml of
phenol
saturated with NETS in0.05 M sodium acetate and
blendedwithaVortexmixer for 3 min.Next,4 ml of
Sevag solution
(chloroform-isoamyl
alcohol, 24:1)wasadded, and the mixture was blended with a Vortex
mixer for 3 min and then
centrifuged
at8,000rpminthe JS13 rotor for 10 min. The aqueous
phase
wascollectedandextractedonce more asdescribed above. Itwasthenextractedonce with
Sevag
solutiononly
and twice with ether. The residual etherwasremoved
with a stream ofnitrogen. One-tenth volume of 3 M
sodium acetate
(pH
6.0)
was added to the aqueousphase, and the RNA wasprecipitatedwith 2volumes
of95% ethyl alcohol (EtOH) at -20°Covernight.The
pellet was collected by centrifugation at 8,000 rpm
(JS13 rotor) for 20min. It waswashed with cold 95%
EtOH anddried under vacuum. Thepolysomal RNA
was dissolved in 1 ml of TE buffer (10 mM
Tris-hydrochloride[pH 7.5], 1 mM EDTA).
ThepolysomalRNA was boiled in a water bath for
60 sand thenchilled in ice water. It was thenbrought
to0.4M in NaCl by theaddition of a 5 M saltsolution
andpoured onto a 0.2-mloligo(dT)-cellulose column.
Theflow-through was collected, and the column was
then washed with 1 ml of wash buffer (100 mM
Tris-hydrochloride [pH 7.5], 0.4 M NaCl, 1 mM EDTA,
0.2%
SDS). The flow-through and wash, combined,comprised the non-polyadenylated
[poly(A)-]
RNAfraction. Thecolumn was washed three more times
with 0.5 ml of washbuffer. The polyadenylated
[po-ly(A)+] RNAwas then eluted with 0.3 ml of elution
buffer (10 mMTris-hydrochloride[pH7.51,0.1%SDS,
1mMEDTA). The column was cleaned with 1 ml of
0.1 N NaOH and washed twice with 1 ml of sterile
water and once with 1 ml of wash buffer. Thepoly(A)+
and
poly(A)-
RNAs were purified again by beingpassed through the cleanedoligo(dT)-cellulose column
and precipitated with 2 volumes of 95% EtOH as
described
above but with anEppendorf centrifuge at15,000 rpm for 10min.
IsolationofviralmRNA. Thepolysomal RNA was
isolated frominfected cells at 20 hpostinfectionand
separated into poly(A)+ and poly(A)- fractions as
described
above. Virus-specific RNA was purifiedfrom these fractions by hybridization to viral
DNA-cellulose
as follows. ViralDNA-cellulose
was pre-pared as described by Noyes and Stark (14). The A.californica
NPV DNA was isolated from inclusionbodies as described by Miller and Dawes (13). The
viral DNA-cellulose containedabout 10 ,ug of DNA
per mg. The
pellet
ofpolysomalRNA from infectedcells
was dissolved in 600p.l
ofhybridization buffer(0.1 MTris-sulfate [pH
7.4],
10mMEDTA,0.1%
SDS,0.6 MNaCl,
50%o
formamide)ataconcentrationof 0.5to 1mgof RNA per
ml.
This solution wassupplement-ed with 20
ILI
ofpolyadenylate (10p.g/p.l
ofsterilewater) and thenmixed with 12 to 24 mg of viral
DNA-cellulose and heatedat80°Cfor 1 min.
Hybridization wasperformedfor 24 hat
37°C;
thecellulose
waskeptinsuspension
byrocking.Tostophybridization, the cellulose was spun down in the
Eppendorf centrifuge for 5 min. The
unhybridized
RNAwasthenremovedbytwowashes with
hybrid-ization buffer andtwowashes with cold 2x SSC
(0.15
MNaCl,0.015 M sodium citrate
[pH 7.5]).
Thevirus-specific RNA was then eluted
by
suspending
theDNA-cellulose twice in 150
p.l
of99%o
formamide-0.1%SDS solution andheatingat600C for1 min. The
eluted viral RNAwasdiluted with sterilewaterto
give
66% formamideand
precipitated
withEtOHat-200C.
Theefficiencyofhybridization
was60to80%.Thin-layer
chromatography. To detect theblocking
terminalnucleotideof thecap
(m'Gp),
welabeledhostpoly(A)+ and
virus-specific
RNAs with[methyl-3H]methionine
invivo,
isolated them as describedabove, and then
hydrolyzed
themby
TAP in assaybuffer (50 mM sodium acetate
[pH
5.4],
10 mM2-mercaptoethanol,
1mMdisodiumEDTA).
The RNAsweredissolved in 30
P,u
of assaybuffer,
mixed with 12on November 10, 2019 by guest
http://jvi.asm.org/
236 QIN AND WEAVER
100
I -1 I~~~
4 4 ~444
-z -3-4 -5-6 a
so8-60
40
20 y
-w-40 80 120
Fraction Nl
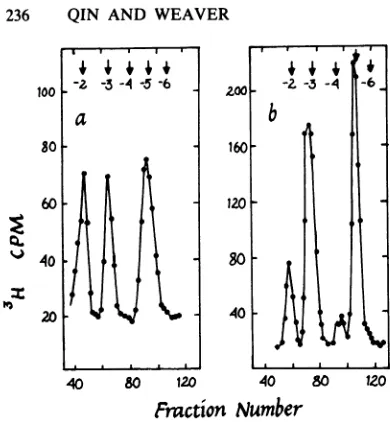
FIG. 1. Isolation of putative c
hostand viralRNAs.Hostpoly(A)
viral polysomal RNA (b) were lal
3H]methionine invivo,purified, ba
chromatographed on DEAE-cellul
thetext. Arrowsmark thepositior
ersof the indicated charges.
UofTAP,andincubated at37°Cfc
thereaction was stopped byboilin
min.Thetotaldigestswere thenal
ofPEI-cellulosethathadbeenrun(
water. GMP and m7Gp were use
parallellaneandvisualized under
ofPEI-cellulose was washedfor 1 dried, and subjected to ascendir
with1Naceticacidupto4 to5cr
MLiClupto15to16cm(18). Afi
was divided into 1-cm-wide secti cellulosefromeachsectionwasscr into2.5mlof3a70scintillationflu DEAE-cellulose chromatography
viralRNAsweredigested with0.3
20h.The digest waNs neutralizedv
acid, and after removal of the ii
perchloratebycentrifugation, the I
ed10timeswith10mMTris-hydro
M urea and mixed with theunlab
adenylate [oligo(A)]. The sample
DEAE-cellulose column (0.6 by 2
with 10 mM Tris-hydrochloride bi
taining7Mureaandthenelutedir (270 ,ul)with50-mllineargradients
inthesame buffer(21).
Absorbanceat254nm wasmoni
continuously, and the radioactivit
wascountedinacompletescintill.
RESULTS Detection ofa putative cap !
and host RNAs. Polysomal R
cells were labeledwith
[methy
and viral RNAwas purifiedby
viral
DNA-cellulose.
Host RNA
waspurified
4+
4
4
-6from
similarly
labeled,
uninfected
cellsand
sep-arated
into
poly(A)+
and
poly(A)
fractions
by
b
oligo(dT)-cellulose
chromatography.
All three
classes of RNAwere
then
hydrolyzed
with 3
MKOH,
and the
digests
werechromatographed
onDEAE-cellulose. The viral RNA
digest
showed
four
well-defined,
methylated
peaks
comigrating
with
markers with
charges
of
-2, -3,
-4,
and
-5
(Fig.
lb).
The
peak
atcharge
-5
was mostprominent
and eluted in
aposition
consistent
with its
assignment
as acap
1(m7GpppXpYp)
(19).
Identification of a
cap
1 structure on viralRNA.
The
peak
eluting
in the -5
position
wasalso
prominent in the host
poly(A)+
RNA
digest
40 80
120o
(Fig. la).
This
suggested
that
it
was a cap 1umber
structure,
because
cap1predominates
in mRNA
of higher
eucaryotes(19). This
wasconfirmed
by
ap structures from
chromatographing
L-cell
[32p,]
poly(A)+
RNA,
i+
RNA (a) andtotal
which is
known
tocontain mostly
cap 1(17);
in
beled with [methyl- our
system
it eluted
close
tothe -5
marker,
just
tse hydrolyzed, and as
the
putative
viral and host
cap structuresdid
lose
as describedin (Fig. 2). This in turn indicated that the peak atis ofoligo(A)mark- -4 was probably a cap 0. The peak at -2
probably
represented methylatedmononucleo-tides
(mXp) in which
thebase
is
methylated,
and
the
peak
at-3
probably
represented
internal
2'-gr3.5
th, atuer
which
O-methylation,
which
gives
rise
toXpYp
uponigthe mixturefor3
pplied
tothinlayers
hydrolysis.
earlier with distilled
Typical
cap structurescontain
m7G
linked
to-d as markers in a the
penultimate
nucleotide
through
atriphos-UV light. The strip
phate
group.The
presenceof such
astructurein
o0
min
inmethanol,viral RNA
wasverified
asfollows:
viraland
host
ng chromatography
[methyl-3H]methionine-labeled
RNAs weredi-m and then with 0.3 gested with TAP, and the products were subject-ter drying, the
strip
ed to thin-layer chromatography on PEI-cellu-aopnesdfarnomthesPheet
lose. Unlabeled markers (GMP andm7Gp)were-iand
frcounted.
run in aparallel
lane. Bothviral
andhost RNAs
Labeled
host
orshowed
peaks
ofradioactivity
that
coincided
M KOH at
37°C
forwith the
position
of the
m7Gp
marker
(Fig.
3).
In
vith
70% perchloricfact,
this
wasthe
only significantradioactive
nsoluble potassium peak aside from that of the residual RNA at the
products
were dilut- origin. The radioactivity in these peaks relative )chloride (pH 7.5)-7 to that in the residual RNA peaks was about half)eled marker
oligo-
what would
beexpected from
the quantitative20
cm)equilibrated
recovery ofasingle
methyl
groupin
m7Gp.
uffer (pH 7.6) con- This
conclusion is
derived from
aninspection
nsix-drop fractionsof
the
relative
amountsof label
inthe
various
of 0 to 0.35 M
NaCI
peaks
shown in Fig. 1; these data are shown in Table1,
along with data on host poly(A)- RNA. itored and recorded Host cap1 contained 45% of the methyl label inty in each fraction host
poly(A)+
RNA, whereas caps1 and 0 fromation fluid
(3a70).
viral RNA contained 41 and 4%, respectively(Table
1).
Because the
m7Gp
portion of cap
1 would contain only half of the methyl label in the structure in viral cap (the other half being in the 2'-O-methyl NAs in infected group on residue X), the expected percentage of1l-3H]methionine,
methyl label inm7Gp
after TAP digestion would e hybridization to be about 20% of the total, assuming aon November 10, 2019 by guest
http://jvi.asm.org/
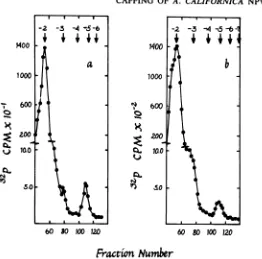
[image:3.504.53.248.56.267.2]CAPPING OF A. CALIFORNICA NPV RNA 237
4t
16
K
CA. ti
Q.
60 50100 120 60 80 100 120
[image:4.504.140.402.63.322.2]Fraction Number
FIG. 2. Host and L-cellpoly(A)+RNAs containingcapstructureswith identicalchromatographicproperties.
RNAswerelabeled with32Piin vivoand purified, basehydrolyzed, and chromatographedonDEAE-celluloseas
described inthetext.(a)Hostpoly(A)+RNA; (b) L-cellpoly(A)+RNA. Arrows show thepositions of oligo(A)
markers of the indicated charges.
tive
yield. In fact,
it constituted 10 and8%,
respectively, in host poly(A)+
and viral RNAs.These
numbers
mayreflect incomplete
hydroly-sis of RNA
by
TAPorthe trapping of
m7Gp in theresidual RNA
atthe
origin.
Detection of
adenosine and guanosine in the viral caps. Host and viral RNAs were labeledwith
[3H]adenosine
or[3H]guanosine,
hydro-lyzed, and subjected
tochromatography
on280
30 20 to
5 10 15
DEAE-cellulose
asdescribed above.
Theradio-activity incorporated by the
totalviral
RNAwas6.1
x105
and 2.2
x10'
cpmof guanosine and
adenosine, respectively. Cap
1 waslabeled with
adenosine and
guanosine (210 and 65
cpm,re-spectively), demonstrating the
presenceof both
in
the
cap.Cap 0
wasalso labeled
with
adeno-sine
(65 cpm), suggesting that
atleast
one capstructure in
viral RNA is
m7GpppAp.
Therela-b
GMP
mTGp
145
30 10 I
10 15
Oustance
ton
orngin
(Cm)
FIG. 3. Identificationof theblockingnucleotidein hostpoly(A)+and total viral polysomal RNAs.TheRNAs
werelabeled with [methyl-3H]methionine invivo, purified, hydrolyzedwith TAP, and subjectedtothin-layer
chromatographyonPEI-celluloseasdescribedin thetext. Thepositions of the markers (GMP andm7Gp)are
shown withhorizontal bars.(a)Hostpoly(A)+ RNA; (b)totalviralpolysomal RNA. 1400
I-qz~
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.504.103.417.497.635.2]TABLE 1. Distribution ofmethylatedcomponents in host and viral RNAs methyl-3Hincorporation (cpm)a into species of charge: RNA
-2 -3 -4 -5
Hostpoly(A)+ 325(27) 335(28) 545 (45)
Host
poly(A)-
59,800 (13) 384,000 (87) 1,840 (0.03)Total viral 325
(11)
1,320 (44)
120(4) 1,200 (41)aTheradioactivityin each
peak
shown inFig.
1[and
inasimilarexperiment
involving
hostpoly(A)- RNA]
ispresented. Numbers inparentheses indicate thepercentages of total
radioactivity
in all of the peaks.tive
inefficiency
oflabeling
caps with[8-3H]guanosine
probably
accountsfor the lack ofdetectable
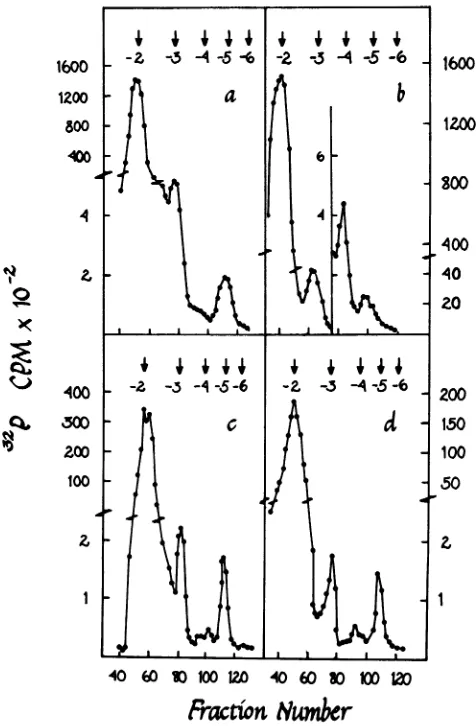
label in cap 0 with this substrate. Viralpoly(A)+
andpoly(A)-
RNAscapped.
Viral and host RNAs were labeled with 3
P,
invivo, extracted,
separated into poly(A)+ and poly(A)-fractions, hydrolyzed,
and chromato-graphed as described above. Viralpoly(A)+
andpoly(A)-
RNAs
both showed prominent cap 1peaks
andeasily detectable cap 0 peaks (Fig. 4).1600
1200 800
400
I
N1.0 -400
Ct,
300
100
1600
1200
g00
400
40
20
200
150 100
50
40 60
10 100120
40 60 301001IO
Fractio
Numbr
FIG. 4. Detection of cap structures in host and viral poly(A)+ and poly(A)- RNAs. Host polysomal RNA in
uninfectedcellswaslabeled with
32Pi,
purified, separated into poly(A)+ and poly(A)- fractions, hydrolyzed, andchromatographedonDEAE-cellulose as described in the text. Viral polysomal RNA was treated the same
way,
except that it was purified from cells at 20 h postinfection and separated into viral and nonviral species by
hybridizationtoviralDNA-cellulose after oligo(dT)-cellulose chromatography. (a) Host poly(A)+
RNA;
(b)hostpoly(A)-RNA; (c) viral poly(A)+ RNA; (d) viral poly(A)- RNA. Arrows show the positions of oligo(A) markers
of the
indicated
charges.t4
I
0
Ir-X
CZ
't I
I
1
on November 10, 2019 by guest
http://jvi.asm.org/
Host
poly(A)+
RNA also showeda cap1peak,but
cap0, if
presentatall,wasbarely detectable.The host
poly(A)-
pattern was somewhatdiffi-cult
tointerpret
because this fractioncontainedmassive quantities
of methylated rRNA andtRNA. This resulted
inalarge methylateddinu-cleotide peak appearing
atcharge -3 (Table 1)and another
major species at charge -4 (Fig.4b). The latter is
probably mostlypXpfromthe5'
terminus of
poly(A)-
RNA, because itap-peared
when
poly(A)-
RNA was labeled with"Pi
but
not[methyl-3H]methionine.
The peakatcharge
-5(Fig.
4b) wasnotaseasytoidentify.It
was methylated; it appeared in RNAlabeledwith
32p,
or methionine. It is possible that itrepresentsa cap structureinasmallproportion
of the
poly(A)-
fraction.
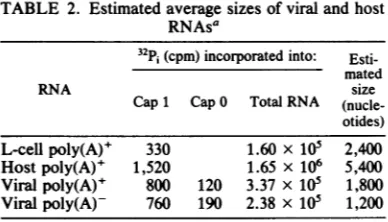
Length estimation ofhost and viral RNAs. The
amount
of
label in each
cappeak shown in Fig.2, 4c,
and 4d
wasdetermined.
The ratio of theradioactivity incorporated
into the cap to thatincorporated into mononucleotides
(Table 2)enabled
us toestimate
the maximum averagelength of the RNA species
in each classassum-ing that
all of the
RNAspecies
werecapped.
Ifsome werenot
capped, then the
average lengthwould
have been smaller. For thesecalcula-tions,
weassumed that the numbers
of32P1-labeled nuclei
incorporated into
cap 1, cap 0,and the
mononucleotide
werefive, four, and
one,respectively. Using these calculations,
wefound that the
maximum
averagelengths of viral
poly(A)+
andpoly(A)-
RNAs
were1,800 and
1,200 nucleotides,
respectively,
ascompared
with the values for
L-cell and host
poly(A)+
RNA
lengths of 2,400 and 5,400 nucleotides,
respectively (Table 2).
DISCUSSION
These
studies showed
thatbothpoly(A)+
andpoly(A)-
NPV
RNAs contain structures thathave the
properties
oftypical
caps0and1. Weleft
unanswered
thequestion
ofthe exactchemi-cal
nature of the bases in the caps, other thannoting
thatboth adenosineandguanosine
werepresent.Because of the
partial exchange
of3Hinthe 8
position
ofguanosine
uponmethylation
tom7G
(19)
and thepossible
differences in theintracellular
specific
activitiesofGTPandATP, the ratio of labeled adenosinetolabeledguano-sine
incorporation
cannot be takenas ameasureof the ratio of adenosineto
guanosine
inacap. Because our RNApreparations
were mixturesof different
species,
it did notseemproductive
to pursuetheidentification
ofspecific
bases.Iden-tification
will have to awaitpurification
ofmRNAs. However, it is worth
noting
that the cappeaks
seemedconsiderably
broader inhostand L-cell RNAs than in viral RNA. This
[image:6.504.265.459.76.187.2]sug-gests amore heterogeneous
population
ofcapsTABLE 2. Estimated average sizes of viral and host RNAsa
32Pi
(cpm)incorporated into:Esti-mated
RNA size
Cap1 Cap0 Total RNA (nucle-otides)
L-cell poly(A)+ 330 1.60 x 105 2,400
Hostpoly(A)+ 1,520 1.65 x 106 5,400
Viral poly(A)+ 800 120 3.37 x 105 1,800
Viralpoly(A)- 760 190 2.38 x 105 1,200
aThe data shown in Fig. 2 and 4 are presented here.
RNAsizes wereestimated as described in the text.
in the host poly(A)+ RNA than in the viral
mRNAs.
The
peaks in the host
poly(A)-
RNA
fraction
were
intriguing but difficult to interpret
confi-dently, because this fraction consists mostly of
rRNA and
tRNA, which are not capped but are
highly methylated.
This
means that the
32p;
peaks
at
charges -3 and -4 could
plausibly be
interpreted as
XpmYp
and
pXp,
respectively.
The
methyl-3H-labeled
peak at charge -5 most
likely
represents cap 1 of
poly(A)-
RNA, which is
well
known in
eucaryotes (12, 15). The only difficulty
with
this
interpretation
is the high
absolute
amount
of this species [ca. three times higher
than that
of host
poly(A)+
RNA].
As
suggested above, the calculated average
lengths
of
viral mRNAs could be
somewhat
inflated bacause of the
possibility that some
noncapped
species
were
present in the viral
RNA
populations.
However, these
calculations
yielded
reasonable values that were
consistent
with the
sizes
observed upon
electrophoresis
of
viral
RNAs (data not shown). These
experi-ments
thus
indicate that most of the viral
polyso-mal
poly(A)+
and
poly(A)-
RNAs
arecapped
and therefore
probably
mRNAs.
ACKNOWLEDGMENTS
We thank P.Bullerforhelpinculturing cells,R.Consigli
and D. Anderson forSpodoptera cells, and R. Wheeler for Trichoplusiaeggs.
This workwassupported by PublicHealthServicegrantno.
GM22127from theNational Institutesof Health anda Bio-medical SciencesSupport Grantfrom theUniversityof Kan-sas.
LITERATURECITED
1. Adams,J.M.,andS. Cory. 1975.Modifiednucleosides andbizarre5'-terminiinmousemyelomamRNA.Nature (London)255:28-33.
2. Colonno, R.J.,and H. 0. Stone. 1975. Methylationof messenger RNA ofNewcastle disease virus in vitrobya
virion-associatedenzyme.Proc.Natl. Acad.Sci. U.S.A. 72:2611-2615.
3. Cory,S.,andJ.M.Adams.1975. Themodified 5'-terminal sequence in messenger RNA ofmousemyelomacells. J. Mol. Biol.99:519-547.
4. Dasgupta, R.,F.Harada,andP.Kaesberg.1976.Blocked
on November 10, 2019 by guest
http://jvi.asm.org/
5' termini inbrome mosaic virus RNA. J. Virol. 18:260-267.
5. Dottln,R.P., A.M.Weiner,andH. F.Lodish. 1976. 5'-Terminalnucleotide sequencesofthe messenger RNAs of Dictyosteliumdiscoideum. Cell 8:233-244.
6. Dubin, D. T., and V. Stollar. 1975.Methylationof Sindbis virus "26S"messenger RNA. Biochem. Biophys. Res. Commun.66:1373-1379.
7. Furuichl,Y., and K.Miura.1975. A blockedstructureat the 5' terminus of mRNA fromcytoplasmic polyhedrosis virus. Nature(London) 253:374-375.
8. Furuichi, Y., M.Morpn, S. Muthukrishnan, and A.J. Shatldn.1975.Reovirusmessenger RNAcontainsa meth-ylated, blocked 5'-terminal structure:
m7G(5')ppp(5')G'Cp-.
Proc. Natl. Acad. Sci. U.S.A. 72:362-366.9. Goodwin,R. H.1975. Insect cell culture:improvedmedia and methods for initiating attached cell lines from the Lepidoptera. InVitro11:369-378.
10. Grula,M.A., P. L.Buller,andR. F. Weaver.1981. a-Amanitin-resistantviral RNAsynthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvaeandSpodopterafrugiperda cells.J. Virol. 38:916-921.
11. Grula, M. A., and R. F. Weaver. 1981. An improved method for isolation of Heliothis zea DNA-dependent RNA polymerase: separation and characterization of a form III RNA polymerase activity. Insect Biochem. 11:149-154.
12. Milcarek,C. 1979. HeLa cellcytoplasmicmRNAcontains three classes ofsequences: predominantly poly(A)-free, predominantlypoly(A)-containing and bimorphic. Eur. J.
Biochem. 102:467-476.
13. Miller, L. K., and K. P. Dawes. 1979.Physical map ofthe DNA genome of Autographacalifornica nuclear polyhe-drosisvirus. J. Virol. 29:1044-1055.
14. Noyes, B. E., and G. R. Stark. 1975. Nucleic acid hybrid-izationusingDNAcovalently coupledtocellulose. Cell 5:301-310.
15. OueUette, A.J. 1980. Purification bybenzoylated cellu-losechromoatography of translatable messenger ribonu-cleic acid lacking polyadenylate. J. Biol. Chem. 255:2740-2746.
16. Palmlter,R. D. 1974.Magnesiumprecipitation of ribonu-cleoprotein complexes: expedient techniques for the iso-lation of undegradedpolysomesand messenger ribonucle-ic acid. Biochemistry 13:3606-3615.
17. Perry, R. P., D. E.Keiley,K.Friderick, and F. Rottman. 1975.The methylated constituents of L-ceil messenger RNA:evidence for an unusual cluster at the 5'terminus. Cell 4:387-394.
18. Randerath, K., andE. Randerath. 1%7. Thin-layer sepa-ration methods for nucleic acid derivatives. Methods Enzymol. 12:323-347.
19. Shatlkn,A. J. 1976.Capping of eucaryotic mRNAs. Cell 9:645-653.
20. Sr{patl, C. E., Y. Groner, and J. R. Warner. 1976. Methylated,blocked 5'termini of yeast mRNA. J. Biol. Chem.251:2898-2904.
21. Tener, G. M.1967.Ion-exchange chromatography in the presence of urea.Methods Enzymol. 12:398-404. 22. Wei, C.-M., and B. Moss. 1975. Methylated nucleotides
block5'-terminus ofvacciniavirus messengerRNA. Proc. Natl.Acad. Sci.U.S.A. 72:318-322.