JOURNALOFVIROLOGY, July 1981, p.87-92
0022-538X/81/070087-06$02.00/0 Vol.39,No.1
Characterization
of the
Coat Protein
mRNA
of
Southern Bean
Mosaic Virus and Its
Relationship
to
the
Genomic
RNA
AMIT GHOSH,* TINEKERUTGERS, MANG KE-QIANG, ANDPAUL KAESBERG
Biophysics Laboratory, GraduateSchool,andDepartmentof Biochemistry, College of Agricultural and Life Sciences, University ofWisconsin-Madison, Madison, Wisconsin 53706
Received20February 1981/Accepted13April1981
RNA isolated from southernbean mosaicvirions contains, insmallamount,a
subgenomic RNA (molecular weight, 0.38x106) thatservesinvitroas anmRNA
for southern bean mosaic viruscoat protein. The RNA has a5'-linked protein
indistinguishable from the protein linked to the 5' end of
full-length
genomicRNA. Its basesequence, determined to91bases from the 3' end, is identicalto
the 3'-terminalsequenceofthe genomic RNA. The resultssuggestthat thecoat
protein messenger sequence exists as a "silent" cistronnear the 3' end ofthe
genomic RNA.
The RNA contained in southern bean mosaic
virus
(SBMV) virions is heterogeneous (16, 21);
RNAs
of
manylengths
exist
inaddition
tofull-length RNA
(Mr,
1.4x106).
Four
major
proteins
could be translated from these RNAs in vitro
(17):
(i)
tworelated
proteins of molecular
weight
105,000
(P1) and
75,000(P2) induced by
full-length RNA;
(ii)
coatprotein
(P3; molecular
weight, 29,000) induced
by
a sucrosegradient
fraction
containing predominantly RNAs
inthe
molecular-weight
rangeof
0.3x106
to0.4x106;
and
(iii)
a14,000-molecular-weight
protein (P4)
induced
by RNAs of all size
classes, including
full-length
RNA and the
coatprotein
messengerfraction. Diener found that
SBMV
full-length
RNA
is
infectious
(4).
This
implies that the
information for all four
proteins
mustbe located
in the
full-length
RNA and
that the
coatprotein
gene must
be
present as a"silent" cistron. In
this
reportweshow that the
coatprotein
cistron
is
located in the
full-length
RNA
nearthe 3' end
and that it is
expressed
by
asubgenomic
mRNA
which has
thesameprotein
linked
toits 5' end
as
does
full-length
RNA.
MATERIALS
AND METHODSIsolationof SBMV and its RNA. SMBV
(bean
strain) wasisolated from Phaseolus
vulgaris
L. cv. Bountifulessentially
bythe method ofHull(7).
ExtractionofRNAwasdoneaccordingtoZimmern (22). The RNAwas purifiedfurtherbydensity gra-dient centrifugation. The region of the RNA that contained thefull-lengthRNAof molecular
weight
1.4 x 106 and the region that contained RNA able to induce coat protein synthesis in vitro werepooled
separatelyand concentratedbyethanol
precipitation
(16).
Invivo
labeling
of virus.Thepreparation
of3P-labeled virus andRNA hasbeen described before(6).
Enzymedigestion conditions. Enzyme digestion
conditionshavebeen described before (6). Proteinase K (Boehringer Mannheim Corp.) digestion was as describedbyFlanegan et al. (5).
Fingerprinting ofSBMV RNAs. Fingerprinting
wasdone by standard procedures (1). The first-dimen-sion electrophoresis was done on cellulose acetate at pH3.5. The second dimension was homochromatog-raphyon aDEAE-cellulose thin-layer plate (CEL 300 DEAE/HR 2/15). The homochromatography mixture used was a3%solution, in 7 M urea, of yeast RNA thathad beenhydrolyzed for 25 min.
3'- and 5'-endlabeling ofSBMV RNA. 3'-end labeling was done with cytidine 3',5'-[5'-3P]bisphos-phate ([5'-3P]pCp) (2,000 Ci/mmol; New England Nu-clearCorp.) byuseof T4 RNA ligase, as described by Dasguptaetal. (2).
5'-Endlabelingwith
[y-3P]ATP
andT4 polynucle-otide kinasewasdoneby the method ofSilberklang et al.(18).Reductivemethylation.RNAfrom thecoat pro-teinfractionwastreated with['4C]formaldehyde(45.8
mCi/mmol; New England Nuclear Corp.), using the
procedures of Rice and Means (15) and Means and
Feeney(9).
Polyacrylamide gel electrophoresis.Terminally
labeled RNAs from thecoatproteinfraction or ribo-some-selected RNAs (10 to20
gg)
orboth wereana-lyzedon8%polyacrylamidegels containing 7 M urea.
Theelectrophoresiswasfor48hat12.5V/cm,
accord-ingtoPeacock andDingman (13).
To elute the RNAs, bands were cut from the gel, homogenized withasterile siliconized glass rod, and takeninto siliconizedvialscontainingtheappropriate volume of0.25 MNaCl. Themixturewasmade 0.1% with sodium dodecyl sulfate. The vials were shaken gently at room temperature for several hours. The mixture was then filtered through membrane filters (0.45 nm, type HA; Millipore Corp.) to remove gel particles. An appropriateamountofcarrier calf liver tRNA was added to the filtrate, and the RNA was precipitatedwithethanol.
87
on November 10, 2019 by guest
http://jvi.asm.org/
Analysis ofdifferentlylabeled RNAson a5%
poly-acrylamide gel after glyoxal denaturation was done
essentially as describedby
Desselberger
andPalese(3). Acridine orangestainingofgelswasdonebythe procedure of McMaster andCarmichael(8).
Polyacrylamide-sodiumdodecylsulfategel
electro-phoresis (19) of in vitro-labeled proteins has been described before(6).
Translation. In vitrotranslation,measurementof
aminoacid incorporation, and electrophoretic analysis
of the synthesized proteins were performed as de-scribedby Salerno-Rifeetal.(17).Incaseof
transla-tionofgel-elutedRNAswhichhad beencoprecipitated
withcalf livertRNA,the reactionmixtureswere cor-rected for thatamountof tRNA.
Ribosomecaptureof mRNA. Labeled RNAwas
incubated in the rabbitreticulocyte cell-free transla-tion system understandard reaction conditions. After 10 minofincubationat30°C,the reactionmixturewas diluted withanequalvolume of ice-cold 10 mM Tris-acetate (pH 7.6)-100 mM potassium acetate-8 mM magnesium acetate. Thediluted reaction mixturewas
layeredonto 2ml ofsucrosesolution(30%sucrose,10
mMTris-acetate,[pH7.6],100 mMpotassiumacetate,
4mMmagnesiumacetate) andcentrifugedfor4hat 36,000 rpmand40C inaSpinco40 rotor. The ribo-somal pellet was dissolved in 0.2% sodium dodecyl
sulfate-0.1 M Tris. RNA was then extracted three timeswith water-saturatedphenol.The final aqueous phase was made0.2 Mwith sodiumacetateand the
RNAwasprecipitatedwith2volumes ofethanol.The
majority of the ribosomal RNAwasremovedby su-crosegradientcentrifugation (16). Fractions contain-ing labeled RNAwere pooled and concentrated by
ethanolprecipitation.
Control experiments indicated that RNAs in the coatprotein fractionareunable topenetrate the su-crosecushion in the absence ofribosomes,i.e.,unless ribosome bound.
Determinationof the 3'-end sequences. RNase
digestions and gel electrophoresis were done as de-scribed before(2).
RESULTS
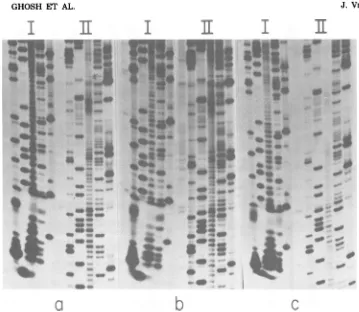
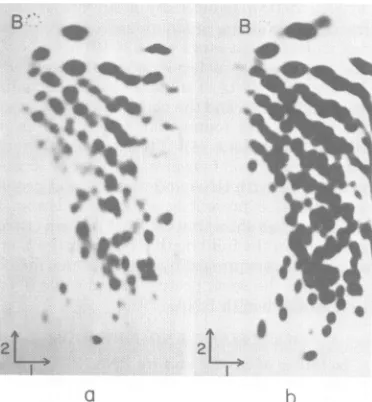
Fingerprint analysis of the
coatprotein
messenger
fraction.
The
sucrosegradient
fraction
ofSBMV RNA that
induced the
syn-thesis of both coat
protein
and P4(16)
in the in
vitro
translation
systemscontains
aconsiderable
number of different RNA
species (16).
Todeter-mine thenature and origin of these
RNAs,
uni-formly
32P-labeled
coatprotein messenger
frac-tion was
digested with RNase T1,
anditsfinger-print was
compared
with thatobtained
frompurified
1.4 x106-molecular-weight
full-length
RNA.
Theresulting autoradiograms
(Fig. la andb)
show that all of theoligonucleotides
from thedigest
of the coat proteinmessenger fraction
were present in
the
digest of
full-length
RNA.
This
indicated that
thesequences
ofthemajor
RNAspecies
inthe
coatproteinmessenger
frac-tion were subsets
of the
full-length
RNAse-quence.
Identification
ofcoatprotein mRNA
and P4mRNA.
Todetermine which of the RNAs
inthe coat
protein messenger fraction served
asthe
mRNA's
for the coatprotein
andfor
protein
P4,
the coatprotein messenger fraction
wasla-beled at the 3' end
with
[5'-32P]pCp, using
T4RNA
ligase.
TheRNAs
wereseparated
on an 8%polyacrylamide gel. Three major and
severalminor
bands
were visible. Of thethree
major
bands,
thelargest
one(designated 1)
wasmoreintense
than the others(designated
2 and3)
(Fig. 2,
lanea).
The materialfrom the
major
bands was
eluted,
andtheextracted RNAs
wereadded
separately
tothe
reticulocyte translation
system.
Only
theRNA
eluted from band 1in-duced coat
protein synthesis
(Fig. 3). The RNA
from this
band also induced the
synthesis of
asmall amount of protein P4. In contrast, RNA
eluted from bands 2 and 3 induced synthesis
only
ofprotein
P4.To
eliminate
thepossibility that translation
wasinduced by
aco-migrating nonlabeled RNA,
aribosome
captureexperiment
wasperformed
(see above)
with the3'-labeled
coatprotein
mes-senger
fraction. The
ribosome-selected RNAs
were
analyzed
again by
polyacrylamide gel
elec-trophoresis.
Bands of
labeled
RNA
appeared
in
a
b
FIG. 1.
Fingerprint
analysis of32P-labeledSBMVRNA fr-om the coatProteinfraction ofthe sucrose
densty radentrun and itscomparisonwith thatof
32P-lbeled genomic
full-length
RNA. See text forfurther explanations. (a) Coatprotein
fraction;
(b)full-length
RNA. Thefirstdimnension
waselectropho-resisoncelluloseacetate,pH 3.5; the second
dimen-sionwas
homochromatography
onaDEAE-cellulosethin-layerplate.
B,Positionofbromophenol
blue. J. VIROL.on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.493.262.448.357.558.2]VOL. 39, 1981
2-
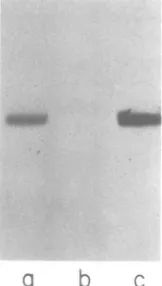
3-FIG. 2. Analysis ofSBMV coatprotein fr-action
RNA on a denaturing 8%polyacrylamide gel. (a)
Coatproteinfr-actionRNAlabeledatthe 3' ends with
32P; (b) coatproteinfr-action RNA labeled in vitro with ['4CJformaldehyde; (c) coat protein fr-action
RNA labeled in vitro with [14C]formaldehyde
fol-lowedby proteinaseKdigestion.
_lm
-CPJo
_NQ
-P4
a
b
C dFIG. 3. Polyacrylamideslab gel electrophoresisof translationproducts, in the rabbit reticulocyte
sys-tem,oftheRNA bandseluted fromthe gel in Fig.2.
Lanes: (a-c) productsmade by RNAs1 through3of
Fig.2;(d) products obtainedwhencoatprotein frac-tionRNAwastranslatedunderidentical conditions.
CP,Coatprotein;P4, proteinP4.
positions, 1, 2, and 3, and their translation
con-firmedourprevious results. On the basis of these
results, we identified the band 1 RNA as the
mRNA forcoatprotein.
The molecular weights of the mRNA's in
bands 1, 2, and 3 were estimated from a gel
electrophoretic analysisafterglyoxal
denatura-tion of the RNAs (3, 12) in the presence of
molecular-weight
standards. The molecularweights of RNAs in bands 1, 2, and 3 were
determinedtobe
approximately
0.38x106,
0.19x
106,
and 0.17 x106,
respectively (data notshown). The 3'-labeled RNAs from bands 1, 2,
and3weresequenced.Thesequencesofthe first
91 nucleotides from the 3' ends of the RNAs
from bands1,2,and3wereidentical(Fig. 4)and
were the same as that found in the 3'-end se-quenceof thefull-lengthRNA(6).Theseresults,
togetherwith the
fingerprint
data(Fig. 1),indi-SBMV COAT PROTEIN mRNA 89
cated that the
geneticinformation
for bothcoat
protein
andprotein
P4is
located
inthe 3' part
ofthefull-length
RNA.5' terminus of
coatprotein mRNA. To
investigate
thenature of the 5' terminus of coatprotein mRNA,
the coatprotein messenger
frac-tion was labeled
either
with[5'-32P]pCp, using
T4 RNA
ligase,
or, after aphosphatase
treat-ment,
with
[y-32P]ATP,
using
polynucleotide
ki-nase.
The RNAs
werethenanalyzed
on an 8%polyacrylamide gel. It
wasfound that
only band
1RNA, which
was anefficient substrate for
T4ligase, could
notbe
labeled by
polynucleotide
kinase at all. This
indicated that
the coatprotein
mRNA
probably had
ablocked 5' terminus.
Attempts
tolabel the
coatprotein
mRNA,
using guanylyl
transferase and[a-32P]GTP
(10)
after
periodate
oxidation and
fl-elimination
toremoveany
putative 5'
pm7G,
werealso
unsuc-cessful.
A
further
attempt toidentify the 5'-blocking
group was
made
by
complete
hydrolysis of
uni-formly 3P-labeled
coatprotein
fraction RNA
with a
mixture of RNases
T2,
T1, and
Afollowed
by
analysis of the
products
by
thin-layer
chro-matography, according
tothe
method of
Nishi-mura(11). In
addition
tofour major
mononu-cleotides, only
twoother spots,
onedue
tofree
phosphate and the other
atthe
origin,
werefound. There
were noother
spotscharacteristic
of
anm7GpppX-like
structure(data
notshown).
The
translational
studies, like
thestructural
studies, also suggested that
theblocking
group atthe
5'terminus of
the coatprotein mRNA
was not acap structure.
Translation of the
coatprotein
messengerfraction in
thereticulocyte
lysates
inthe
presenceof
thecap
analog
7-meth-ylguanosine-5'-monophosphate
(m7Gp)
resulted
in
about 30%
stimulation of
the aminoacid
in-corporation
activity, whereas
thetranslation
ofbrome mosaic virus RNA
4(a
capped mRNA)
showed
a90% inhibition
(Fig.
5). Such
stimula-tion in
incorporation
in the
presenceof
capanalogs
inthe
reticulocyte lysates
has been
ob-served for other viral
mRNA'swhich have
aprotein linked
totheir
5'ends
(14).
Since
full-length
SBMV
RNA has aprotein
linked
toits
5'end
(6)
and since
inall
plant
viruses examined
todate the
genomic
andsub-genomic RNAs of
aparticular
virus
always
have
the sametype ofstructure attheir termini
(20),
welooked for thepresence ofa
protein
onthe5'end of thecoat
protein
mRNA. Reductivemeth-ylation, using
["C]formaldehyde,
of the coatprotein messenger fraction and
subsequent
gelanalysis
resulted in theappearance
of"4C
labelat the coat protein mRNA
position
in the gel(Fig. 2, lane b). Treatment of the
'4C-labeled
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.493.101.181.68.190.2] [image:3.493.72.216.271.376.2]90 GHOSH ET AL.
I
:ii
I
:i
I
:ii
a
b
c
FIG. 4. Sequencing
gel
patternshowing
3'-endsequenceof (a)
RNAfrom band1, Fig. 2;(b)
RNAfrom
band2,Fig.2; (c)RNAfrom band3, Fig.2. The RNAslabeled at the3'endwerepartiallydigested with
differentenzymesandthenrunon a 12% thingel. Lanesfrom left toright, in eachpanel, are: untreated
control;RNAtreated withRNaseU2,RNase T1 formamide,RNasePhy M, pancreaticRNase. Twoseparate
loadingsofthesamplesweremade. Panel I is the shorterrunshowingbasesfromthe3'end; panelII is the
longer run.
coat
protein
messengerfraction with
proteinase
K, however, caused the
'4C
label
todisappear
from the
coatprotein mRNA
(Fig.
2,lane c).
Staining
of the
gel
with acridine
orangeshowed
that
the RNA itself
wasstill
intact. These results
suggested
the
presenceof
aprotein
linked
tothe
5' end
of the
coatprotein mRNA.
The
protein
wasanalyzed by gel
electropho-resis after
acomplete RNase digestion
of the"4C-labeled
coatprotein
mRNA fraction. Anelectrophoretic
comparisonof
the releasedpro-tein with the
'4C-labeled
genome-linked protein(6)
showed that the two proteins comigrated(Fig.
6,lanes
a and c). The two proteins werecompared further
by digesting thempartially
withprotease Staphylococcus aureus V8 and
analyzing
the productsformed
on apolyacryl-amide-sodium
dodecyl sulfate gel (19). They gaverise
to anidentical cleavage
pattern
con-sisting of
three bands, indicating that the twoproteins
areidentical
(data not shown). Thepresence ofa
protein
presumably
linked
tothe
5'
terminus of the
coatprotein
mRNA
indicated
that the
coatprotein mRNA is
asubgenomic
RNA,
synthesized
in
vivo and
encapsidated
in
minor
amountsin the virions.
DISCUSSION
Our results show that the
coatprotein
geneis
present as a
silent cistron in the
genomic
RNA
and that it is
expressed
viaasubgenomic
mes-senger
which has the
sameprotein
linked
toits
5' end
asthegenomic
RNA. Protein
P4could be
translated
from thecoatprotein
mRNA and
also
from
smaller RNAs
having
the
same3'-end
se-quence.
From
denaturing
gels,
the
molecular
weight
of thecoatprotein
mRNAwasestimated
to
be
0.38 x106.
This may beinsufficient
toaccommodate the information for both
thecoatprotein
(molecular weight, 29,000) and the
P4protein
(molecular weight, 14,000)
independ-ently.
Thisimplies
that the
genesfor the
coaton November 10, 2019 by guest
http://jvi.asm.org/
[image:4.493.74.433.49.361.2]SBMV COAT PROTEIN mRNA 91
0
--0
c
0
0
01.-c
0
a
-o
o
c
0.5 1.0 1.5 2.0
mM pm7G
FIG. 5. Effect of m7Gp5'ontranslationalefficiency
of thecoatproteinmessengerfraction RNAin rabbit reticulocyte lysate. Reactions wereprimed with (0)
SBMVcoat proteinfractionRNAor(0) brome
mo-saic virus RNA 4at 50pg/ml. The potassium ion concentrationusedwas40mM.
a
b
c
FIG. 6. Comparison ofthe invitro-labeled protein from thecoatproteinfraction with that associated
with the fuU-length RNA. (a) SBMV-coat protein
RNA labeled with ['4CJformaldehyde; (b) SBMV-coatprotein RNA labeled with ["Clformaldehyde
foUowed byproteinaseKdigestion; (c) SBMV full-length RNA labeledwith[4Clformaldehyde.Inevery
case the sample was treated with RNase A before electrophoresis.
andP4proteinsoverlap,and since theseproteins
didnotsharecommontryptic peptides
(17), they
would have to be coded in different
reading
frames.
SBMV differsfrom most other plantviruses in
having
aprotein linked to the 5' end ofitsgenome.Unlike theviruses withgenome-linked
protein
(20),
however,
it does notsynthesizeallof itsproteins
through
apost-translationalcleav-age mechanism.
Rather,
itadopts
thestrategy
ofexpressing its internal cistronsthrough
themediationofsubgenomicRNAs.
Turnip
rosettevirus,
which hasagenomicRNAwithaproteinattached
toit,
alsosynthesizes its
coatprotein
from
asubgenomic
messenger
(B.
A. M.Morris-Krisinich,
70thAnn.Rept.,
John Innes
Institute,
Norfolk,
England,
1979).
Itthus appears that
there
is notnecessarily
acorrelation
between
the terminal structure of the
RNA
and thestrat-egy
adopted by
thevirus
intheexpression
of itsgenetic
information.
ACKNOWLEDGMENTS
We thankTerrySalerno-Rifeforhelpfuldiscussions. This work waasupportedbyPublic HealthServicegrants AI-15342 andAI-01466andCareer AwardAI-21942 from the National Institutes of Health andbygrant7800002 from the Science and EducationAdministration of theU.S.
Depart-mentofAgriculture.
LITERATURE CITED
1.Barrell,B.G. 1971.Fractionationand sequenceanalysis
ofradioactivenucleotides,p. 751-779. InG.L.Cantoni and D. R. Davies (ed.), Procedures in nucleic acid research,vol. 2.Harper&Row,NewYork.
2. Dasgupta, R.,P. Ahlquist,and P. Kaesberg. 1980. Sequenceofthe 3'untranslated regionofbromemosaic
coatproteinmessengerRNA.Virology104:339-346. 3. Desmlberger, U., and P. Palese. 1978. Molecular
weightsof RNAsegmentsofinfluenzaAand Bviruses. Virology88:394-399.
4. Diener,T.0. 1965. Isolation of infectious ribonucleic acid fromsouthernbean mosaicvirus.Virology 27:425-429. 5. Flnnegan,J.B., R.F.Petterson,V. Ambros, M. J. Hewlett,and D.Baltimore.1977.Covalentlinkageof
a proteinto adefined nucleotide sequence at the 5' terminusofvirion andreplicative intermediate RNAs ofpoliovirus.Proc. Natl. Acad. Sci.U.S.A. 74:961-965. 6. Ghosh,A., R.Dasgupta,T.Salerno-Rife,T.Rutgers, and P.Kaesberg. 1979.Southernbean mosaicvirus hasa5'linkedproteinbut lacksa3'terminalpoly(A). Nucleic Acids Res.7:2137-2146.
7. Hull,R. 1977. Thebandingbehaviour of the viruses of southembean mosaicvirusgroupingradients of
cae-siumsulphate. Virology79:50-57.
8.MMaster,G.K.,and G. G.CarmichaeL1977.Analysis ofsingle anddouble stranded nucleic acidson polyacryl-amide andagarosegels by using glyozal and acridine
orange.Proc.Natl. Acad. Sci. U.S.A.74:4835-4838. 9. Means,G.E.,andR. E.Feeney. 1968. Reductive
alkyl-ation of aminogroupsinproteins. Biochemistry 7:2192-2201.
10.Moss, B. 1977. Utilization of guanylyltransferase and methyltransferasesof vacciniavirustomodify and iden-tifythe5'-terminalsofheterologousRNA species. Bio-chem.Biophys.Res.Commun. 74:374-383.
11.Nishimuras,S.1975. Minorcomponentsin transferRNA: theircharacteristiclocation andfunction. Prog. Nucleic AcidRes. Mol. Biol.12:49-85.
12. Palese, P.,and J. L Schulman. 1976. Differences in RNA patterns in influenza A viruses. J.Virol. 17:876-884.
13. Peacock,A.C.,andC. N.Dingman. 1968.Molecular weightestimationandseparation of ribonucleic acidby electrophoresis in agarose-acrylamide composite gels. Biochemistry7:668-674.
14. Pelham,H.R. B. 1979.Translation of tobacco rattle virus RNAs in vitro: four proteins from threeRNAs.Virology 97:256-265.
15. Rice,R.H., and G. E. Means. 1971. Radioactivelabeling ofproteinsinvitro. J. Biol.Chem.246:831-832. 16.Rutgers, T., T. Salerno-Rife, and P.Kaesberg.1980.
MessengerRNA for the coatprotein ofsouthern bean VOL. 39, 1981
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.493.73.221.57.210.2] [image:5.493.106.187.277.420.2]mosaic virus.Virology104:506-509.
17. Salerno-Rife, T.,T.Rutgers,and P.Kaesberg.1980.
Translation ofsouthern bean mosaic virus RNA in
wheat embryo and rabbitreticulocyteextracts.J.Virol. 34:51-58.
18.Silberklang, M.,A.Porchiantz,A.L.Haenni,andU.
L.Rajbhandary.1977. Studiesonthesequenceofthe 3'terminalregionofturnip-yellow-mosaic-virusRNA.
Eur. J. Biochem.72:465-478.
19.Swank, R. T., and K. D. Munkres. 1971. Molecular
weight analysis of oligopeptides by electrophoresisin
polyacrylamide gelwith sodiumdodecylsulfate.Anal.
Biochem. 39:462-477.
20.VanVloten-Doting,L.,andL. Neeleman.1980.
Trans-lation of plant virus RNAs,p.511-527.In C. J. Leaver
(ed.), Genome organization andexpression in plants.
PlenumPublishing Corp., New York.
21. Veerisetty, V., S. A. El-Hassan, and0.P. Sehgal.
1981. Temperature-inducedstructural stabilization of theencapsidated RNA ofsouthernbeanmosaic virus
and itsbiologic significance.Virology 108:286-296.
22. Zimmern,D. 1975.The 5' endgroupof tobacco mosaic
virus RNA is m7G5'ppp5'Gp. NucleicAcids Res. 2:1189-1201.
J. VIROL.
![FIG.2.RNARNAlowedwithCoat32P; Analysis of SBMV coat protein fr-actionona denaturing 8% polyacrylamide gel.(a) protein fr-action RNA labeled at the 3' ends with(b)coat protein fr-action RNAlabeled invitro['4CJformaldehyde;(c)coat proteinfr-actionlabeledinvitrowith [14C]formaldehyde fol- byproteinase K digestion.](https://thumb-us.123doks.com/thumbv2/123dok_us/1477665.100399/3.493.72.216.271.376/rnarnalowedwithcoat-denaturing-polyacrylamide-rnalabeled-cjformaldehyde-actionlabeledinvitrowith-formaldehyde-byproteinase.webp)