0022-538X/83/040125-06$02.00/0
Copyright01983,AmericanSocietyforMicrobiology
Sequence Determination of the (+) Leader RNA Regions of
the
Vesicular Stomatitis Virus Chandipura, Cocal, and Piry
Serotype Genomes
COLOMBAGIORGI,tBENJAMIN BLUMBERG, AND DANIEL KOLAKOFSKY* DepartmentofMicrobiology, University of Geneva Medical School, 1205 Geneva, Switzerland
Received 30 November 1982/Accepted 12 January 1983
Using 3'-end-labeled genome probes, cells infected with vesicular stomatitis virus Chandipura, Cocal, and Piry serotypes were shown to contain (+) leader
RNAsof approximately 50 nucleotides in length.Thenucleotidesequenceofthe
leader RNA regions ofthese genomes was determined and compared with the
previously reported sequences of both the (+) and (-) leader RNA regions of
other vesicular stomatitis virus serotypes. Regions ofstrong conservation of nucleotide sequence among the various vesicular stomatitis virus serotypes suggest those nucleotides thought to be involved in control functions during
vesicular stomatitis virus replication.
Vesicular stomatitis virus (VSV), a rhabdo-virus,
contains
allof
itsgenetic information
(11to 12
kilobases)
inasingle
RNAchain ofnega-tive
polarity
(-). The VSV (-) genome codes for five viralproteins
which areexpressed
via fivemonocistronic mRNAs. These mRNAs aretranscribed in the order 3' N, NS, M,
G,
andL5' both in vivo and in vitro, and the relative molar abundanceof each mRNA is determined by its
proximity
to the 3' end of the genome,which is thoughttobe theonly siteatwhich the viral polymerasecan enterthe
genofpe temnplate
(1, 21). The first RNA transcribed from the (-)genome
template
is not thefirst
mRNA but ashort,
47-base(+)
leaderRNA,
which iscomple-mentary tothe
precise
3' endof the (-)genome(4).This leader RNA hasrecently been shownto
contain the nucleation site for the initiation of
nucleocapsid
(NC) assembly
(la-3). Onepossi-ble function
of
the (+) leader RNA is thus todetachthis nucleation site from the
body
of the first mRNA so thatthis mRNAwould notalso beencapsidated.
A model for VSV
replication
hasrecently
been advanced in which the
binding
ofNprotein
to the nascent leader RNA chain modulates
mRNAtranscription andgenomereplication (3, 11). In this model, the termination site forthe
leaderRNAis
analogous
totheattenuatorof the tryptophanoperonof bacteriaand thebinding
ofNproteintothe nascentleaderchain would both
promote readthrough of the leader termination
signal
andsimultaneously begin
theencapsida-tion of the nascent
antigenome
strand. Thus,tOn leave fromLaboratoriodiVirologia,IstitutoSuperiore diSanita,Rome, Italy.
manyof the features which control VSV
replica-tion
arethoughttobe encoded in the relatively short leader RNAregion.
In aneffort tobetter understand how these control processes mightoperate, wehave
sequenced
the leader regions of thePiry
(Pi),Chandipura (Ch),
andCocal(Co)serotypes of VSV.
MATERIALS AND METHODS
Viruses and theirgrowth. VSV Indiana(In; Mudd-Summers strain)was obtained from Donald F. Surn-mers, University of Utah School ofMedicine, Salt LakeCity. VSVCh and Cowereobtained fromCraig Pringle,Institute of Virology,Glasgow,Scotland,and VSV Pi was obtainedfrom DanielleTeninges, Labora-toire Genetique des Virus, Gif-sur-Yvette, France. Each serotype was plaque purified twice in
succes-sion,and asingle plaque was thenused to generate virusstocks on five 10-cm dishes of BHK cells. Virus
wasgrown oneither BHK orLLCMK2 cells (10 PFU/
cell) and harvestedat18 hpostinfectionfor all
sero-types exceptCo (36 hpostinfection).Radioactive virus wasgrownin the presenceof 20 ,iCi of[3H]uridineper ml and 0.5
"ag
ofactinomycinD per ml. The viruswasthenpelleted through30%oglycerol, suspended in TNE
buffer,banded onaglycerol-tartrate gradient(16), and then recoveredbypelleting.Asodiumdodecyl
sulfate-polyacrylamidegelelectrophoresispatternoftheviral
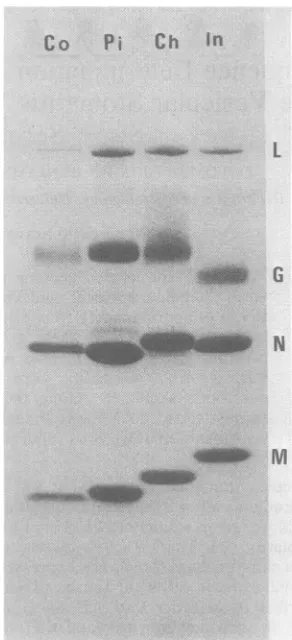
proteinsof each serotype is shown inFig. 1.
Preparation of3'-end-labeled virion RNA (genome probe).Purified virus (1 to 3mg/ml) was incubated for 20minat37°C with0.5% sodium dodecyl sulfate and 500 ,ug of proteinase K per ml and then directly
centrifuged on 5 to23% sucrosegradients (15). The 42S RNA was localized by counting samples of the gradient for13H]RNA andrecovered by two ethanol precipitations. Virion 42S RNA (5 to 10
Rg)
was then 3' endlabeled with32P-labeledpCp andRNAligase ina 20-j, reaction as previously described (15) and 125
on November 10, 2019 by guest
http://jvi.asm.org/
chromatographed on Sephadex G-50, and the excluded RNA was ethanol precipitated. The precipitate was dissolved in 25 ,ul of 10 mM Na2PO4, denatured with 40 mM CH3HgOH, and then diluted to 100 ,ul with TNEcontaining 100 mM BME and recentrifuged on a sodium dodecyl sulfate sucrose gradient. RNA which sedimented at 42S was again selected and recovered by ethanol precipitation.
Sequence determination. (i) Purine reactions. Partial digestion with ribonucleasesT1 and U2 was carried out inbuffercontaining 20 mM sodium citrate (pH 5.0 for
T1,pH 3.5for U2), 7 M urea,1mMEDTA, 0.25 mgof tRNA perml, 0.25% xylene cyanol,andbromphenol
blue (5,12). Theend-labeled RNAwasaddedtothis buffer, and thereaction mixture wasdistributed into three 20-,ul portions in siliconized Eppendorftubes. The tubes were heated at50°C for 5 min and quick
chilled in ice. A 1-,ulportionof ribonucleaseT1(10-1 U/ml)orU2(10-2U/ml)wasaddedtothefirst of the three tubes, and a single 10-fold serial dilution was
carriedoutinto the second tube. The third tubewas
incubated without enzyme. The tubeswereincubated for 15 minat50°C,chilled,and loadedonto12or20%
sequencinggels.
Fortheladder, 1.5 ,ul of end-labeled RNAplus2,ul of tRNA(2,ug/ml)wasmixed with 15 ,ul offormamide. Thereactionwasincubatedat100°Cfor 10to25 min.
(ii) Pyrimidinereactions. Forthe Ureaction,the 3' labeled RNAwaslyophilizedinthe presence of tRNA
carrier, dissolved in 50%ohydrazine-50%o water(vol/
vol), andincubated on ice for 10to15min(17). TheC> Ureaction wascarried outin 3 M NaCl-hydrazine incubatedonice for 20to30min(17).The
chemicallymodified RNAwasprecipitatedtwice with ethanol in the presenceof0.3Msodiumacetate.Itwas
thendissolved in 20 ,ulof 1.0 M aniline-acetatebuffer,
pH4.5, and incubated in the darkat60°Cfor 20 min. The reaction was terminated by freezing at -70°C.
The samples were lyophilized, dissolved in water, chilled at -70°C, and lyophilized again. Finally, the
samplesweredissolved in 3to5,ul of 8 M urea-20 mM
Tris-hydrochloride (pH 7.4)-i mMEDTA-0.05% xy-lene cyanol and bromphenol blue, heated 2 min at
90°C,and loadedontothesequencing gel.
Co
Pi
Ch
InL
uAIb
NFIG. 1. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the various VSV serotypes. Be-tween10to20 p.gof thevarious VSVserotypeswere
electrophoresedona 10oacrylamide-0.8% bis(acryl-amide) protein gel (13) whichwas then stained with
Coomassie brilliant blue. Theserotypesare notedat thetopof the gel. The positions of theL,G,N,and M proteinsof VSV Inareindicatedonthe right.
RESULTS
VSV Ch, Co, and Pi also contain (+) leader RNAs. To date, only the In and New Jersey serotypes of VSV have been shown toinitiate transcriptionwith a (+) leader RNA. To investi-gate whether the Ch, Co, and Pi serotypes
similarly contained leader RNAs, weprepared 42S (-)RNAfrom each of the various purified virions (Fig. 1), and these (-) genomes were
radiolabeledattheir 3'OHends with
32P-labeled
pCp andRNAligase (see above). Thepresence of(+) leader RNA in virus-infected cells was examined by annealing CsCl pellet RNA (totalcytoplasmic RNA from which viral NCs have been removedby CsCl density gradient centrifu-gation [15]) against the end-labeled genome
probes,
followed
by ribonuclease digestion todegradesingle-stranded regions ofthe RNA. As a control, a similar optical density amount of
CsCl
pellet
RNA fromuninfected
cells wasannealed in
parallel.
Thepolyacrylamide
gel of theremaining
RNase-resistant double-strandedRNAsfrom thisexperiment is shown in Fig. 2. Note that when uninfected CsCl pelletRNA is
annealedagainst the various 3' genomeprobes, only in thecaseof the Cogenome isanyof the labeledgenome RNA resistant to RNase
diges-tion. Thisshortdouble-stranded RNAprobably
represents the complementary ends of the Co
genomeitself, similarto that seenin VSVNew
Jersey (7). However, when CsCl pellet RNAs from the various virus-infected cells were an-nealedagainst their homologousgenomeprobes,
severaldouble-stranded RNAs resulted in each
case.Theresults with VSVCh andInappeared identical; three bands of decreasing intensity which migrated just in advance ofthe 75-base
4b
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.490.279.425.73.393.2]SEQUENCE
OF THE VSV (+) LEADER RNA 127A C D E F G H J K
--BPB
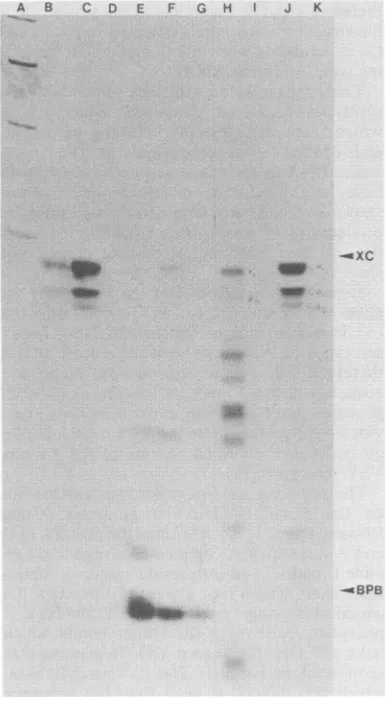
FIG. 2. Demonstrationof ()leader RNAs in the
various VSV serotype-infected cells. CsCl pellet
RNAs from the various VSV-infected BHKcells or
from uninfected cells were annealed against their
homologous 3'-end-labeled genomeprobes. The
reac-tionswerethendigestedwith 20 p.gof RNase A per ml
in 0.375 MNaCi aspreviouslydescribed(15),and the
remaining RNAwas recovered by ethanol
precipita-tion andelectrophoresed on a acrylamide-0.5%
bis(acrylamide) gel (0.4mmby20cmby40cm).Lane
A, Hinfdigest ofpBr322; lanes B to D, 0.5 and 1.5
optical density units (ODU) of VSV In-infected and
0.5 ODU of uninfected CsCl pellet RNA annealed
againstthe VSV In genomeprobe; lanes Eto G,0.5
and 1.5 ODU of VSV Co-infected and 0.5 ODU of
uninfected CsClpelletRNAannealedagainstthe VSV
Co genomeprobe;lanes HtoI, 1.25ODU of VSV
Pi-infected and 1.5 ODU of unPi-infected CsClpelletRNA
annealedagainstthe VSV Pi genomeprobe;lanesJto
K, 1.5 ODU of VSV Ch-infected and 1.5 ODU of
uninfected CsClpelletRNAannealedagainsttheVSV
Ch genome probe. XC, Xylene cyanol; BPB,
brom-phenolblue.
pair DNA marker resulted from the annealing. These bands were in the expected position of
leader double-strandedRNA onthesegels (10). However,the presence of three double-stranded RNAbands instead ofasingleband is probably
duetothehigher resolution of the long and thin
gels used in this study. To more accurately determine the chain length of the 3' genome
RNA protected against RNase digestion by
these leader RNAs, the RNA was recovered from each of the three In and Ch bands, and a
sample of these RNAs was partially digested with RNase T1. The partial digests were then
electrophoresed along with untreated band
ma-terial andaformamide ladder of the homologous end-labeled genome probes on a7 M urea
se-quencing gel (data not shown). From the posi-tion of the guanine nucleotides in the sequence
relative to the ladder, we estimate that the lengths of the 3'-end-labeled genome RNAs were42 + 43bases for the bottom band,44+45
bases for the middleband, and 47+51bases for thetopband(bases 48to51 ineachsequence are
purines and cannot be cut by RNase A; see
below).
Theremarkably
similar pattern between VSV In and Ch shown inFig. 2parallelsaverysimilarsequence atthe 3' end of theirgenomes;
only 2outof the first 51 nucleotidesaredifferent between these two serotypes (see below). A
similar length
heterogeneity
of theIn(+)leaderRNA has recently been demonstrated directly by Schubert et al. (19)
by
directly labeling invitro transcripts with [3- 2P]ATP. Thus, VSV Ch-infected cells, like VSV In-infected cells, contain (+) leader RNAs which are 42 to 47
bases inlength.
The annealing of both Pi and Co CsCl pellet
RNA against their homologous genomeprobes also resulted in multiple double-stranded RNA bands. In the case of VSV Co, two major double-stranded RNA bandsare seeninaddition
to the putative complementary ends ofthe Co
genomeitself; thetopbandmigrates just slightly slower thanthe topbandof theInandChseries, and the faster band inaseparateexperimentwas
estimated to be
approximately
26 basepairs
long. The reason why the shortest band
in-creases in
intensity
onannealing with
infected cell RNAisnotclear,
but itmaybe dueto athirdleader RNA of this size. In the case of VSVPi, atleast nine bandscouldroutinely bedetected, of which the topband migrated similarlytothe topbandofthe In andCh series. VSVCo-and
VSV Pi-infected cells therefore also contain
leaderRNAs,but these leader RNAs are
appar-ently heterogeneous over a wider
length
than their In andChcounterparts. It should benoted, however,thatalthough theinfectingstocks usedin these experiments were prepared from plaque-purified virus, the presence of small
VOL.46,1983
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.490.47.240.94.445.2]A B C D E F G H J
... ... " ,
..
-A.
so
0 -EUXC
-
-l0.
[image:4.490.67.233.76.465.2]
--...9,N
...
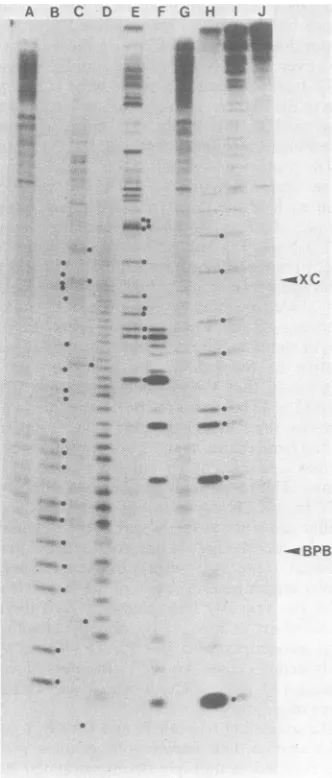
FIG. 3. Sequence determination of3'-end-labeled
VSV Pi genome.The conditions for partialdigestion at specific bases are described in the text. Lane A,
formamideladder (10 minat100°C);lane B, chemical reaction for U; lane C, chemical reaction for C> U; laneD, formamide ladder (25 min at 100°C); lane E, 10- Uof RNase U2; lane F,10-2Uof RNase U2; lane
G, formamideladder (10 minat100°C), lane H,10' U of RNase
T1;
lane I,10-2Uof RNaseTj;
lane J, no enzymeadded. BPB and XC on the right-hand margin show thepositions of the bromphenol blue and xylene cyanol dye markers, respectively. Dots on the right-hand side of given bandsindicate those bands which determine the sequence. The positions of the chemicalcleavageproducts containing 5' phosphates relativeto
theladder bands containing5'OH groups were deter-mined by sequencing VSV In on a separate gel. The dots show the sequence from the G at position 2 (lowest band, lanes H andI)tothe AAAAsequence at
positions 46 to 49, except for the U residues at
positions39, 40, 42, 43, and44whicharedetectableon
the film but not on the photograph of this
20%o
amountsof defective interferinggenomes
gener-ated inourinfections cannot be excluded. It is thereforepossiblethat some of the smaller dou-ble-stranded RNA bands detected in the Pi and Coexperimentswereduetoincomplete
anneal-ingwith (-)leader RNA.
Thus,cellsinfected with each of various VSV serotypes examined contained leader RNAs
which were complementary to the precise 3' ends oftheir(-)genometemplates. Thelargest
leader RNA in each case wasapproximately 50 baseslong.Inaddition,results identical to those shown inFig. 2 were obtained by annealing the products of in vitro virion transcription reac-tions to their homologous genome probes (not
shown).
Sequence determination of the (+) leader
re-gionsofVSVCh, Co, andPi. To date, only the
(+) leader regions
of
the In and New Jerseyserotypesof VSV have been sequenced. It was therefore of interest to determinethenucleotide sequence at the 3' ends ofthe Ch, Co, and Pi genomes,sinceannealing experiments have pre-viously shown that there is onlylimited homolo-gy between the coding regions of the various VSVserotypes (18).
The sequence determination was carried out on the 3'-end-labeled (-) genomes, using RNasesT1 andU2 to determine the position of G andArespectively, chemicalcleavageto deter-mine C andU, andformamide laddersto deter-mine chainlength (see above). Toposition the chemical cleavage products which contain a 5' phosphate relative to the ladder bands which have a 5' OH, the known VSV In genome was sequenced in parallel. The 3' terminal base, which was difficult to read from the sequence gel, was determinedby nearestneighbortransfer after alkalinehydrolysisand foundtobe U in all cases. Anexample of the sequencing gels, that for VSVPi, is shown inFig. 3. Thesequence of the 3' ends of VSVCo, Ch,and Piareshown in
Fig.4alongwithpreviously reported sequences
of VSV In and VSV NewJersey, and also the (-) leader regions of three In variants [which represent the 3' region of the (+) antigenome
strand].
DISCUSSION
The nucleotidesequences atthe 3' end of the
(-)genome of five VSVserotypes and of the 3' end ofthe (+)
antigenome
of three variants of the Inserotypehave severalinterestingfeatures.Of theeightsequencesshown,allof whichdiffer
from eachother, there isremarkable homology
acrylamidesequencinggel. The various VSV genome sequences were alsodetermined on12% acrylamide gelstobetter resolve the bands above40nucleotides in length.
on November 10, 2019 by guest
http://jvi.asm.org/
SEQUENCE OF THE VSV (+) LEADER RNA
VSV GENOME RNA SEQUENCES
3' HO-UGCUICGUlUGUUUAAUMAAUAGUAAUUUUCCGAGUCCUCUUUGAAA
3' HO CUGUUUUGAAUGU UAUAACCGGAUCUCCCUUGAAA
3'
HO-UGCUUCUUIUGUW
UAAUUUGUC MUAUAUCAUCAUGUGC3' HO-UGiCUCWIJUUGUUUGGUUAUCGUAAGAAUAUUCUGUAUUCUUUGMAAM
3' HO-UGCUUGU1UGUUAUAUAUUUUUCCGAGUCCUCUUUGAAA
3'
HO-UlUUCUGGUGUUUlGUCUAUUUUUUAUUVY,GYGUUCUCCCAG
3' HO-U
UUCUGGUGUUUUGMCUGUUUUUUAUUUVV,V,GUUCUCCCAG
3'
HO-UfGC
UGUUGUUUUGGUCGAUUUUUUGUUUUUGGUGUCUUCCCAG...$les129.
IN MS
-NJ
-COCAL
-PIRY
-CHANDIPURA
-IN MS+
IN TSG 31 +
IN CAR4 +
FIG. 4. Nucleotide sequences at the 3' ends of various VSV genomes and antigenomes. The various serotypesarenotedatthe right-handside, where (-) and (+) denotegenomesandantigenomes, respectively.
The In(-),NJ(-), In(+), In tsG31(+),andInCAR4(+)sequences arefrom references(14,20). Theregions of
sequenceconservationatboth ends of the leader regionsareunderlined. The repetition of the hexanucleotide
UUUGGUisdottedunderneath.
within the first 18 nucleotides and then the sequencesdiverge widely. The regions of abso-lutehomology within the first 18 nucleotidesare underlined inFig. 4 and give risetothe
consen-sus sequence 3'OH-UGCUUNUNNUNN
UUUGGU,
where NrepresentseitherG, U,orC,butneverA.
Fromourpresentknowledge of VSV replica-tion,twofunctionsarethoughttobecodedfor,
atleast inpart, inthisregion. The first function is theinitiation of RNA synthesis,including both genome transcription and replication, and the second function is the nucleation site for the initiation of NC assembly, which has recently been localized within the first 14 nucleotides from the 5' end of the leader RNA(la). On the basis of lesscompleteinformation,wesuggested that the NC assembly signal was a five times repeated A residue ineverythirdpositionatthe 5' endof theleader RNA(thecomplementaryU residues in the genome are underlined above). The addition ofthe (+) leader regions of VSV Ch, Co, Pi to ourcatalog has not contradicted this hypothesis, but rather added further
sup-port. The eighth base from the 5' end of the leader strand, previously an invariant C, has now become an N inthe above consensus se-quence, since this basehas changed in both the Piand Coserotypes. In addition, A residuesare
notfound inthe first 18nucleotides from the 3'
end ofany of the genomes sequenced todate. Thus, the absence of U, the complementtothe signal base A in the NC assembly site of the leader strand, has also been confirmed by the sequencesof theCh,Co, and Piserotypes.
It is difficult to decipher the nucleotides
in-volvedin the initiation of RNAsynthesissimply
by comparing sequences. Within the first 18 bases of all thegenomesandantigenomes
stud-ied,twoblocks of nucleotidesare invariant: 3'-UGCUU(bases1to5)and 3'-UUUGGU(bases 13 to 18). Interestingly, the sequence 3'-UUUGGUisrepeatedatposition31to36of the
three In(-) leader regionsbut not inanyofthe (+) leader regions. Recently, Isaac and Keene (6) and Keeneetal.(9) have shownthat theNS
protein, which is thoughtto play a role in the
initiation of RNAsynthesis, selectively perturbs the methylation of certain nucleotides when
presentonthe NC. Thesefootprint experiments have suggested that the NS protein interacts
with bases 16 to 30 at the 3' end of the (-)
genomeand with bases 17to38atthe 3' end of
the(+) antigenome orthecopy-back defective
interfering genome. These NS interaction re-gions abut the 5' end of the first 3'-UUUGGU sequence onbothgenomesandantigenomes and completely cover the second 3'-UUUGGU on the(+) antigenome. Inaddition, all of thestrong
perturbations on the antigenome were at the ends of the interaction region (bases 17 to38), either withintherepeated hexamersequencesor directly adjacent tothem. Since previous work hassuggested that (-) leaderRNAsare
synthe-sizedmorefrequentlypermole ofgenome
tem-plate than (+) leaderRNAs(14),the hexamer
3'-UUUGGU maybe involved in the initiation of
RNAsynthesis,and itsdoublingatthe 3' end of
the (+) antigenome may explainthis enhanced
rate of (-) leader synthesis. The 3' proximal pentanucleotide UGCUU is probably involved
in boththe initiation ofRNA synthesisand the
initiation of NC assembly.
To determine those nucleotides involved in
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.490.112.394.109.239.2]terminating the (+) leader RNA chains, we
similarly examined these regions of the (-) genomes fornucleotideconservation. From the
data presented in Fig. 2, we have established that the largest(+)leaderRNAs of VSV Ch and
Pi are47base pairslong, and theCo(+)leader RNA is one or two nucleotides longer. The
nucleotidesequenceswhichappear to havebeen
conserved near the termination sites are also
underlined in Fig. 4 and leadtothe consensus sequence 3'-Y3_6NUIGAAA, where Y repre-sentspyrimidines andNis eithera U or aG. The arrowshowsthe presumed point ofchain termi-nation. Keene etal. (8) havepreviouslypointed
outthat(-) leader RNAsappear tobe terminat-edby thesequences 3'-CCCIAGAA.Thus (+)
and (-) leaderRNAtermination sequences are
related but somewhat different, and this
differ-encemayreflect the relativeeasewith which the viral polymerase reads through these
termina-tionsignalsduringgenomereplication.
In conclusion, extensive conservation of the
nucleotidesequencein theleader RNA
(noncod-ing) regions of five
different
VSV serotypes provides suggestive evidence for theinvolve-mentofthese sequences incontrolfunctions. In
addition, the lengths ofthe (+) leader RNAs appear to havebeenhighly conserved,
suggest-ing that the distances between the concerned regions may also play arole in the control of VSV transcription and replication.
ACKNOWLEDGMENTS
Wethank Rosette Bandelier andColettePasquierfor expert technicalassistance.
This work wassupportedbyagrantfrom the Swiss National ScienceFoundation.
LITERATURE CITED
1. Banerjee, A. K., G. Abraham,and R.J. Colonno. 1977. Vesicularstomatitis virus; mode oftranscription (a
re-view). J.Gen.Virol. 34:1-8.
la.Blumberg, B. M., C. Giorgi, and D. Kolakofsky. 1983. VSVNprotein selectivelyencapsidatesVSV leader RNA
in vitro.Cell 32:554-567.
2. Blumberg,B.M.,and D.Kolakofsky.1981.Intracellular vesicular stomatitis virus leader RNAs are found in nu-cleocapsidstructures.J.Virol. 40:568-576.
3. Blumberg,B.M.,M.Leppert, and D.Kolakofsky. 1981. Interaction of VSV leaderRNAandnucleocapsid protein may control VSVgenomereplication. Cell23:837-845.
4. Colonno,R. J.,and A. K. Banerjee. 1977.Mappingand initiation studiesontheleaderRNAof vesicular
stomati-tis virus.Virology 77:260-268.
5.Donis-Keller, H., A. M.Maxam, and W.Gilbert. 1977. Mapping adenines, guanines and pyrimidines in RNA. Nucleic Acids Res. 4:2527-2538.
6. Isaac, C. L., andJ.D. Keene. 1982. RNA polymerase-associated interactions neartemplate promoter sequences of defective interfering particles ofvesicular stomatitis virus. J.Virol. 43:241-249.
7. Keene, J. D., M. Schubert, and R. A. Lazzarini. 1979. Terminal sequences of vesicular stomatitis virus RNAare
both complementary and conserved. J. Virol. 32:167-174. 8. Keene, J. D., M. Schubert, and R. A. Lazzarini. 1980. Intervening sequence between the leader region and the nucleocapsid geneofvesicularstomatitisvirus RNA. J. Virol. 33:789-794.
9. Keene, J. D., B.J. Thornton, andS.U. Emerson.1981. Sequence-specificcontactsbetween the RNApolymerase ofvesicular stomatitis virus and the leader RNA gene. Proc.Natl. Acad.Sci. U.S.A. 78:6191-6195.
10. Kolakofsky, D. 1982.Isolation of vesicularstomatitisvirus defective interfering genomes with different amounts of 5'-terminalcomplementarity. J. Virol. 41:566-574. 11. Kolakofsky, D.,and B. M.Blumberg. 1982. A model for
the control of non-segmented negative strand virus genomereplication,p.203-213.InB. W. J.Mahy,A.C. Minson, and G. K.Darby (ed.),Viruspersistence, sym-posium33.Societyfor GeneralMicrobiologyLtd. Cam-bridgeUniversityPress.
12. Krupp, G.,and H.J.Gross.1979.RapidRNA sequenc-ing: nucleases fromStaphylococcusaureusand Neuros-pora crassa discriminate between uridine and cytidine. Nucleic Acids Res. 6:3481-3490.
13. Laemmli, U. K. 1979. Cleavage of structural proteins during the assembly of the head ofbacteriophage T4. Nature(London)227:680-685.
14. Leppert, M., andD.Kolakofsky.1980.Effect ofdefective interfering particles on plus- and minus-strand leader RNAs invesicular stomatitis virus-infected cells. J. Virol. 35:704-709.
15. Leppert,M.,L.Rittenhouse, J.Perrault,D. F.Summers, andD. Kolakofsky. 1979. Plus and minus strand leader RNAsinnegativestrandvirus-infectedcells.Cell 18:235-245.
16. ObUeski,J. F.,D.H. L.Bishop,F. A.Murphy,andE. L. Palmer.1976.Structural proteinsof La Crosse virus. J. Virol. 19:985-997.
17. Peattie,D. A.1979.Direct chemical methodfor sequenc-ingRNA. Proc.Natl. Acad.Sci. U.S.A. 76:1760-1764. 18. Repik, P., A. Flamand, H. F. Clark, J.F.Obijeski, P.
Roy,andD. H. L.Bishop. 1974.Detection ofhomologous RNA sequences among six rhabdovirus genomes. J. Virol. 13:250-252.
19. Schubert,M., G.G.Harmison, J.Sprague,C. S.Condra,
and R. A.Lazzarni.1982.In vitrotranscriptionof vesicu-larstomatitisvirus: initiation with GTPat aspecificsite
withintheNcistron.J. Virol.43:166-173.
20. Semler,B.L.,andJ. J.Holland. 1979. Persistent vesicular stomatitis virus infection mediates base substitutions in viral RNAtermini.J.Virol.32:420-428.
21. Wagner,R. R.1975.Reproductionofrhabdoviruses,p.
1-94.InH.Fraenkel-Conrat andR.Wagner(ed.), Compre-hensive virology, vol.4.PlenumPublishing Corp.,New York.