CopyrightX 1993,American SocietyforMicrobiology
The Human
Immunodeficiency
Virus
Type
1
Long
Terminal
Repeat
Specifies Two Different
Transcription Complexes,
Only
One of Which Is
Regulated by Tat
XIAOBIN LU,TERRY M.WELSH,ANDB. MATIJAPETERLIN*
HowardHughes Medical InstituteandDepartments ofMedicine andofMicrobiologyandImmunology,
University of
California,
SanFrancisco,
SanFrancisco,
Califomia
94143-0724Received 18 September1992/Accepted 15 December 1992
The human immunodeficiency virus type 1 long terminal repeat sets up two different transcription complexes, which have been called processive and nonprocessive complexes. By mutating and substituting cis-actingsequences,wemappedelements of the humanimmunodeficiencyviruslongterminalrepeatthatare
responsible forcreatingeachtranscription complex.Whereasprocessivecomplexesareefficientlyassembledby upstreampromoter elements in the absence of theTATAbox, nonprocessive complexesabsolutely requirethe TATAbox.Moreover,the TATA box alonecansetupthesenonprocessivecomplexes, andnonprocessivebut notprocessivecomplexesaretransactivatedbyTat.Finally,astrongDNA-bindingsitebetweentheTATAbox andtrans-activation-responsive regioninterferes with either theassemblyormovement of thesenonprocessive complexes and diminishes the effects of Tat.Thus, Tataffectsa criticalstepinthe formation of elongation-competenttranscription complexes.
Human immunodeficiency virus type 1 (HIV-1) is the etiologic agent of AIDS. Besides structural proteins and
enzymes, its genome codes for six other proteins, two of which, Tat and Rev, are potenttrans activators that affect quantitativeandqualitative expression,respectively,ofviral proteins (8, 9, 30, 35).Botharealsorequiredforhighlevels ofproductiveviralreplicationand forcellularcytopathology (10, 13).Tat interacts withanRNAstem-loopfoundatthe5' end of all viral transcripts,which is called the trans-activa-tion-responsive region (TAR),togreatlyincreaselevels ofall viraltranscripts andproteins (7, 17, 34, 39).
By interacting with transcribed RNA, Tat could affect ratesofinitiation and/orelongationoftranscription, mRNA stability, and/or translation. In primate cells that support viralreplication, Tat primarilyincreases rates of transcrip-tion from the HIV-1longterminal repeat (LTR) (27, 32). In nuclearrun-on assays,effectsonboth initiation and
elonga-tion oftranscriptionwereobserved(12,18, 21). However,in
in vitro transcription systems, the effects of Tat on the elongation of RNA polymerase II predominate (20, 24). In
one such study, Tat-modified transcription complexes stalled artificially atposition +13 in TAR andacted
syner-gisticallywith transcription factor IIS (TFIIS) butnotwith TFIIF, which implies that TatactsanalogouslytoTFIIF late in theassembly ofelongation-competent transcription
com-plexes (20).
After the earliest nuclear run-on assays and elegant in vitro transcription studies, itwassuggested thattwo differ-ent types oftranscription complexes are assembled on the
HIV-1 LTR (18, 24, 25). One complex is processive and results infull-length transcripts thatarepolyadenylated (18).
The other complex is nonprocessive and results in
prema-turely terminated transcriptswhichconsist of the TAR RNA stem-loop (18). Whereas limited mutagenesis assays
sug-gested that cis-acting sequences that set up processive
complexesdonotinvolvethe TATA box (4),sequences3'to
*Correspondingauthor.
the TATA box andincluding TARweresufficienttogenerate short, nonpolyadenylated transcripts. Infact, the 85 nucle-otidesfrom positions -5 to +80werecalled the inducerof shorttranscripts (33). However,acorrelation betweenthese shorttranscriptsandnonprocessivecomplexesthataretrans activatedby Tat hasnotbeenmade.
Tocharacterize thesetwotranscription complexes in cells rather than in in vitro transcription systems and to define
sequencesin the HIV-1 LTR thatsetupeachtranscription complex,weperformedextensive mutageneses of theHIV-1 LTR.WhereasNF-KBandSP-1sequencesdirected
proces-sivetranscription in the absence ofaTATAbox,
nonproc-essivecomplexes required a functional TATA box. LBP-1 didnotcontribute to trans activation inthis invivoassay.
However, the TATA box was sufficient in the absence of
other transcription factors for setting up low levels of
nonprocessivecomplexes.Furthermore, only nonprocessive transcripts were trans activated by Tat. Finally, strong DNA-binding sites placed 3' to the TATA box interfered withtheassembly and/ormovementof thesenonprocessive complexes. Thus, sequences 3' tothe TATA boxmustbe free ofstrongDNA-binding proteins.
MATERLALSANDMETHODS
Cell culture. HeLa and COS cells were maintained in
Dulbecco's modified Eagle's medium supplemented with 10%fetalcalfserum, 100 U ofpenicillin, 100 ,ug of
strepto-mycin, and292 ,gofL-glutamineperml at370C.
Plasmid constructions. pHIVSCAT, pHIVSCATSV,pSV TAT, pSVTATZX, pHIVSYOCAT, and pHIVSTCCAT havebeendescribedpreviously (1, 18, 38). pHIVTCATand pHIVTCATSVwere derivedfrompHIVSCAT and pHIVS
CATSV, respectively, by deleting a fragment between the
EcoRV andXbaI restriction endonuclease sites which
con-tainsNF-KB andSP-1 sequences. Thesetwoplasmids
con-tain only the TATAbox, LBP-1, and TAR sequences. To construct pHIVSSVCAT, the following oligodeoxynucle-otides,which containthe72-bprepeatfrom the simian virus
1752
on November 9, 2019 by guest
http://jvi.asm.org/
HIV-1 LTR-SPECIFIED TRANSCRIPTION COMPLEXES 1753 TABLE 1. Synthetic oligonucleotides
Oligonucleotide Sequencea
H1HTH2H3 5'-CATGATGCTGC ATATAA GCAGCTGCTTTTTGCATG-3'
D1DTD2D3 5'-CTAGAAGACTC TATTA CACCCCACATTCTGCATG-3'
H1DTH2H3 5'-CTAGATGCTGC TATTA GCAGCTGCTTTTTGCATG-3'
D1HTD2D3 5'-CTAGAAGACTC ATATAA CACCCCACATTCTGCATG-3'
A1ATA2A3 5'-CTAGAAGGGGGG CTATAA AAGGGGTGGGGGCGCATG-3'
H1ATH2H3 5'-CTAGATGCTGC CTATAA GCAGCTGCTTTTTGCATG-3'
A1HTA2A3 5'-CTAGAAGGGGGG ATATAA AAGGGGTGGGGGCGCATG-3'
A1ATH2H3 5'-CTAGAAGGGGGG CTATAA GCAGCTGCTTTTTGCATG-3'
H1ATA2A3 5'-CTAGATGCTGC CTATAA AAGGGGTGGGGGCGCATG-3'
H1HTH2A3 5'-CTAGATGCTGC ATATAA GCAGCTGCTTTTTGCATG-3'
HlHtH2H3 5'-CTAGATGCTGC TTATAT GCAGCTGCTTTTTGCATG-3'
H1HAH2H3 5'-CTAGATGCTGC AAAAAA GCAGCTGCTTTTTGCATG-3'
aSequences are from 5' flanking, TATA, and 3' flanking regions of the HIV-1 LTR, DRA, and AdML promoters.
40 (SV40) transcriptional enhancer, were annealed and
li-gated(45): AvaI
5'-CCGAGGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCA-3' 3'-CCCACACCTTTCAGGGGTCCGAGGGGTCGTCCGTCTTC-S'*
*5'-GAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACC-3'
3'-ATACGTCOGTACGTAGAGTTAATCAGTCGTTGGTCCA-5'*
*5'-AGGTGTGAAAGTCCCCAGGCTCCCAGCAG(GC(AGA-3'
3'-CACCTTTCAGGGGTCCGAGGGGTCGTCCGTCTTCAT-'5*
*5'-AGTATG}CAAAGCATG}CATCTCAATTAGTCAGCAACCAC-3'
3'-ACGTTTCOGTACGTAGAGTTAATCAGTCGTTGGTGGGCC-5' SmaI The ligated fragment was then inserted into pHIVSCAT between the AvaI and SmaIrestriction endonuclease sites. All replacements of the HIV-1 TATA box and flanking sequenceswere done byreplacing the region between the XbaI(at position -40)andSphI(atposition -10) restriction
endonuclease siteswith thesyntheticoligodeoxynucleotides
listed in Table 1.
Transient transfections and CAT assays. COS and HeLa cells whichweregrownin 150-mm and 100-mmpetridishes,
respectively, were transfected with 15 or 5 ,ug of plasmid DNA,respectively, by theDEAE-dextran transfection
pro-tocol (1, 18, 38). Transfected cells were harvested, and
chloramphenicol acetyltransferase (CAT) enzymatic assays were performed as described previously (1, 18, 38). Each
experimentwasrepeatedatleast fourtimesinduplicatewith similarresults.
RNaseprotection assays. From 5 to10 ,ug ofcytoplasmic
RNAwasassayed exactlyasdescribedbySelbyetal. (38).
The probes used in theseexperiments have been described
previously (38) and are depicted in Fig. 2A. For RNase
protection assays with
pHIVTCATSV
and pHIVDRCAT SV, gels wereexposed
to an imaging screen andanalyzed
with a PhosphorImager (Molecular Dynamics, Mountain
View, Calif.).
EMSAs. For
electrophoresis mobility
shiftassays(EMSAs),
nuclear extracts from HeLa cells and purified recombinant
SP-1protein (akindgiftfrom Dr.
Tjian)
wereincubated witha-32P-labeled
double-strandedoligonucleotides
from the HIV-1 LTR and adenovirusmajorlate(AdML)
promoter, 2jig
of poly(dI-dC) per ml, and excess (100 ng) unlabeledcompetitor oligonucleotides, as indicated in the
legend
to Fig.3. TheresultingDNA-protein complexes
wereresolved onnondenaturing
4%polyacrylamide gels
andexposed
to X-ray film. These EMSA conditionswere describedprevi-ously(41).
RESULTS
Contributions
of transcriptional enhancer and promoter elementstobasalandtrans-activated levels of expression from theHIV-1 LTR. Toexamine the contributions ofcis-actingsequences in the HIV-1 LTR to the formation of different
transcription complexes, a synthetic HIV-1 LTR was cre-atedbyplacinguniquerestriction sitesbetween theNF-KB, SP-1, TATA, and LBP-1 sequences (Fig. 1). This cassette-forming approach facilitated the mutagenesis ofeach func-tional unitand the replacement of DNA-binding sites from HIV-1 withthose from other promoters (Fig. 1). For analy-ses ofexpression, this syntheticHIV-1 LTR was linked to the CAT reporter gene(pHIVSCAT). Both CATenzymatic
assays and selected RNase protection assays were
per-formed on HeLa cells and Jurkat cells transiently cotrans-fected with these synthetic HIV-1 LTR target and Tat
effector plasmids. Two different effectors were used, and
they directed the synthesis of functional and nonfunctional
Tatproteins (pSVTAT andpSVTATZ, respectively). Since thedataobtained with Jurkat and HeLa cellswereidentical,
only the results with HeLa cellsarepresented.
In these cotransfections, wild-type target and effector
plasmids yielded 600-fold-higher CAT activities than were observed with thewild-type targetplasmidalone, i.e., 600-foldtransactivation,whichwassetas100%transactivation (Fig. 1B).WhenNF-KB sitesweremutatedbytwomutations of three nucleotides, an almost identical fold increase in transactivationwasobserved(data not
presented)
(28). On the otherhand,whenthese sequenceswerereplacedby the72-bprepeatwhich is thetranscriptionalenhancer ofSV40, six-fold-higher basal levels of expression and three-fold-lower levels of trans activation were observed (Fig.
1B).
Although basal levelswere
increased,
this strongtranscrip-tional enhancer, which is active in HeLa
cells,
could not abrogate the effects of Tat. Infact,thehighestabsolute CATactivities were observed with this
target-effector
plasmid
combination.Thus,astrong
transcriptional
enhancercannot set up a transcription complex on the HIV-1 LTR that isindependent of Tat. This observation extends those for
heterologous promoters, with whichbasal levels of expres-sion andfoldtransactivationwereincreased and
decreased,
respectively, and suggests that
transcriptional
enhancersonly
partially
substitute for Tat(32, 33,
36).
To determine the effects of SP-1 and LBP-1 on trans activation by Tat,we
exchanged
SP-1 sites with those for NF-Y and theoctamerfromthe DRApromoter(Fig.
1A).
Y and 0 elements represent essentialcis-acting
sequences for VOL. 67,1993on November 9, 2019 by guest
http://jvi.asm.org/
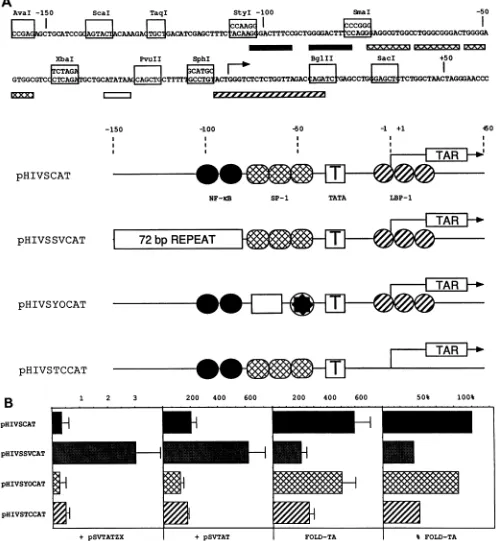
[image:2.612.61.568.92.222.2]Aval -150 ScaI TaqI StyI -100 SmaI -50
IIII
Im
I
,I
CTGATGCATCC
TAAAGAdTG GAcAA
CATTCtA
GGArCTTTC
TTCGTGGGGACT
cGsGCC
T
CGGTGGGCCGGGCGACTGGGGGA
XbaI PvuII SphI BglII SacI +50
FCTAGX
I
I
C
I
GTGGCGTCIC TGCTATAT TTTTCACTGACTGGGTCTCTCTGGTTAGAC GAGCC TCTGGCTAACTAGGGAACCC
IXx,73>s>> sZ
-150 -100 -50 -1 +1 150
I I I I
I I I I
'I
i
TARA
9~~~
pHIVSCAT
NF-KB SP-1 TATA LBP-1
pHIVSSVCAT
pHIVSYOCAT
pHIVSTCCAT
I
TAR
72bp REPEAT
I
TAR
*-I3
*-3Amh
1 2 3 200 400 600 200 400 600 50% 100%
FIG. 1. Mutations and substitutionsofsequencesinthe HIV-1 LTR.(A) Sequenceof thesyntheticHIV-1 LTRandreplacements andmutations ofcis-actingelements inthis HIV-1 LTR. At the top of thefigureis thesequenceof the HIV-1 LTR from HIV-1SF2.ChangesintheprimaryDNA
sequencethatwererequiredtointroduceconvenientrestrictionendonuclease sitesareshown in boxesabove thewild-typesequence.Onlyseven
nucleotideswerechanged, mostly with conservative substitutions. Bars underneath thesequencedenote well-characterized DNA-binding sites in
theHIV-1LTR.Solid bars, cross-hatchedbars, theopenbar, and the hatched barrepresentNF-KB,SP-1, TBP,andLBP-1DNA-bindingsites,
respectively. Below thesequenceof theHIV-1 LTRaredrawn four plasmid constructions. pHIVSCAT is the parental constructionand contains
synthetic restriction endonuclease sites. pHIVSSVCAT contains the 72-bprepeatfrom theSV40 transcriptional enhancer;pHIVSYOCATcontains
Y and 0 boxes from the human MHC class II DRApromoter; and pHIVSTRCAT contains mutations ofLBP-1 DNA-binding sites and
compensatorymutationsthat maintainbase-pairing in thestemof TAR.(B) CATenzymaticassaysfromtransient transfections of these plasmid
constructions in thepresenceoffunctionalandnonfunctional Tateffectors in HeLa cells.Target and effectorplasmidsareshowntothe right and
below the panels, respectively, and levels of expression (fold trans activation) are presented above the panels. Cotransfections with the
nonfunctional Tat(pSVTATZX)areshownfirst,followedby those with thefunctional Tat (pSVTAT). Thenextpanelrepresentsacalculation of
the increasein the level ofexpression in the presenceof the functional Tat (pSVTAT) over that in thepresence of thenonfunctional Tat (pSVTATZX) (foldtransactivation[FOLD-TA]). Finally,foldtransactivationisexpressedas apercentageof thelevel obtained with pHIVSCAT (100%). Theseexperimentswereperformedthree timesinduplicate. Standarderrorsof themean aredenotedbyerrorbars.
1754
B
pHIVSCAT
pHIVSSVCAT
pHIVSYOCAT
pHIVSTCCAT
I
I
TARJ--*-
on November 9, 2019 by guest
http://jvi.asm.org/
[image:3.612.58.557.53.594.2]HIV-1 LTR-SPECIFIED TRANSCRIPTION COMPLEXES 1755
the transcription of this human major histocompatibility (MHC) class II gene and have been studiedextensively (15,
42). As reported previously, mutations or deletions of all
SP-1 sites profoundly reduced basal levels oftranscription from the HIV-1 LTR, and little or no trans activation was
observed (data not presented) (16, 17). However, target
plasmids with Y and 0 elements instead ofSP-1 sites were
efficiently trans activated by Tat (Fig.1B). In fact, the fold
transactivation was 90% of that observed with thewild-type targetplasmid. Similar results wereobtained with mutations
of the three LBP-1 sites (Fig. 1B). Since two of these sites
are located in TAR, compensatory mutations had to be
introduced in the distal TAR to maintain base-pairingin the stem. Again, Tat functioned efficiently on this mutated HIV-1 LTR, and only twofold-lower levels of trans
activa-tion than with the wild-type targetplasmid were observed.
Thus, SP-1 sites are not absolutely requiredfor the effects of
Tat, and other transcriptional activators with disparate DNA-bindingoractivation motifs can functionallysubstitute for SP-1. Moreover, although mutations of LBP-1 sites had
deleterious effects in an in vitro transcription system (16,
17), theywerecompletely dispensable fortransactivation in
vivo. This LBP-1 phenotype has also been described
previ-ously (23). We conclude that none of the well-described and well-studied cis-acting motifs in the HIV-1 LTR, i.e., its
transcriptional enhancer (two tandemly repeated NF-KB
sites) or upstream or downstream promoter elements (SP-1 andLBP-1), areessential for trans activation byTat.
Functional TATA box is essential for trans activation by Tat. Since none of the upstream or downstream promoter elements were absolutely required for trans activation by Tat, we also replaced the HIV-1 TATA box region with
those fromthe DRA (42) and AdML (11, 43)promoters (Fig. 2A). The first TATA sequence was chosen because it does notbindTATA-binding protein (TBP) (data not presented) and because it is dispensable for DRA promoter function (2, 42a). On the other hand, the AdML promoter TATA box is
probablythebest-studied TATA box in biology and assem-bles RNA polymerase II and associated factors by first
bindingTBP(5, 26, 44). Although initially the entire cassette frompositions-40 to + 1 in the HIV-1 LTR wasreplaced by sequences from these other two promoters, other plasmid constructions with
only
parts of these other TATA box regions were also made (Fig. 2A).To our surprise, the TATA box region from neither the DRA norAdML promoter was trans activated by Tat (Fig.
2B, D1DTD2D3 andA1ATA2A3). In the case of the DRA promoter, this defect mapped to the TATA box itself. This
informationwasobtained from two additional plasmid
con-structions. First, when sequences flanking the DRA pro-moterTATAbox were placed around the HIV-1 LTR TATA
box, trans activation levels only twofold lower than wild-typelevelswereobserved(Fig. 2B,D1HTD2D3). However, when the DRA promoter TATA box replaced the HIV-1 LTR TATA box, little to no trans activation by Tat was
observed (Fig. 2B,H1DTH2H3).The trans activation levels observed with the DRA promoter TATA box weresimilar to
those observed with plasmid constructions in which the TATA sequence was replaced by the same number of A
residues(Fig.2B,H1HAH2H30)orthe TATA sequence was
inverted (Fig. 2B, HlHtH2H3). We conclude that a func-tional TATAbox is absolutelyessential for trans activation by Tat.
A more complex picture emerged with the TATA box
region from the AdML promoter. When the entire HIV-1 TATAbox cassette was replaced with that from the AdML
promoter, the trans activation effects of Tat were 10- to 20-fold lower than with the wild-type target (Fig. 2B, A1ATA2A3). However, when we placed just the TATA sequence from the AdML promoter into the HIV-1 LTR, almost wild-type levels of trans activation were observed (Fig. 2B, H1ATH2H3). Thus, the AdML promoter TATA box is functional in the HIV-1 LTR and supports the effects of Tat. However, sequences in its flanking region abrogated the effects of Tat.
To find these deleterious sequences, four additional plas-mid constructions were made (Fig. 2A, A1HA2A3, A1ATH2H3, H1ATA2A3, andH1HTH2A3). The first con-struction (A1HA2A3) tested the AdML promoter 5'- and 3'-flanking sequences in the context of the functional HIV-1 TATA box. The second construction
(A1ATH2H3)
tested the contribution of AdML promoter 5'-flanking sequences. The third construction(H1ATA2A3)
tested the contribution of AdML promoter 3'-flanking sequences. The final con-struction(H1HTH2A3) tested the contribution of the AdML promoter initiator sequence. As shown in Fig. 2B, since A1ATH2H3 but not H1ATA2A3 approached wild-typelev-els of trans activation, AdML 3'-flanking sequences were responsible for the decreased levels of trans activation by Tat. Moreover, the AdML initiator sequence functioned better than the HIV-1 initiator sequence, which mapped the
defect tosequences between the AdML TATA box and the initiator. We conclude that whereas sequences upstream and downstream from the HIV-1 LTR can be mutated or re-placed, the TATA box is absolutely essential for trans activation byTat. Furthermore, mutations in its3'-flanking sequences can be deleterious to the function of Tat (see below).
SP-1 binds to 3'-flanking sequences of the AdML TATA
box. To identify the nature of the 3'-flanking defect of the
AdML promoter, we performed EMSAs with nuclear
ex-tracts from HeLa cells with oligonucleotides that
corre-sponded to the HIV-1 and AdML TATA box regions. Althoughfive Tresidues are found at this site, we reasoned
that they were insufficient to cause appreciable bending of
the DNA. Rather, we expected that an essential protein
which is absent inAdML binds to these sequences from the
HIV-1 LTR and thatthis protein helps to set up a
transcrip-tion complex that is responsive to Tat. To our surprise, just
the opposite was observed. Whereas no protein bound to
this site with oligonucleotides from the HIV-1 LTR, very
strongbinding wasobserved with oligonucleotides from the
AdML promoter (Fig. 3A, lane 2). Additionally, excess unlabeledoligonucleotides from the AdML promoter but not
from HIV-1 competed for this binding (Fig. 3A, lanes 3 and
4). Furthermore, oligonucleotides which contained se-quences that supported trans activation by Tat did not compete for thisbinding (Fig. 3A, lanes 5 and 6).
Interest-ingly,unlabeledoligonucleotides which contained three SP-1 sitesbut notthree LBP-1 sites also competed for this binding
(Fig. 3A, lanes 7 and 8).
Toprove that thisprotein was indeed SP-1, two additional experimentswereperformed.First,excess unlabeled AdML oligonucleotide competed specifically for the binding of
HeLa nuclear extracts to SP-1 sequences from the HIV-1
LTR(Fig. 3B, lanes 4 and5). Second, purified SP-1 madein HeLacells with thevaccinia virus expression systembound
specifically
tothis site in theAdML promoter (Fig. 3C,lanes2 through 7). We conclude that SP-1 binds to sequences between the TATA box and the initiator in the AdML
promoter and interferes with either the assembly or move-ment of transcription complexes along the DNA template. VOL.67, 1993
on November 9, 2019 by guest
http://jvi.asm.org/
A
TCAGATGCTGC ATATAA GCAGCTGCTTTTTGCCTGTACT CIAGATGCTGC ATATAAGCAGCTGCTTTTICTGAGTACT
CATGATGCTGC ATATAAGCAGCTGCTTTTTGCATGCACT
f.TLAMGACTC TATTACACCCCACATTCTGCATGACT
CTAGATGCTGC TATTAGCAGCTGCTTTTTGCATGCACT
C.TAGAAGACTC ATATAACACCCCACATTCTGCATGCACT
,CAAGAAGGGGGGCTATAAAAGGGGTGGGGGCGCATGCGTC
C2TGATGCTGC CTATAAAAAGCTGCTTTTTGCATGCACT
IA1AGGGGGGATATAAAGGGGTGGGGGCGCATGCGTC IAG^AGGGGGGCTATAAAAAGCTGCTTTTTGCATGCACT
C.TGATGCTGC CTATAAAAGGGGTGGGGGCCATGCGCT
CTAGATGCTGCATATAAAAAGCTGCTTTTTGCATGCGCT
100 200 300
TCAGATGCTGC TTATATGCAGCTGCTTTTTGCATGCACT
TCAGATGCTGCAAAAAAGCAGCTGCTTTTTGCATGCACT
200 400 600 800 50 100%
+ pSVTATZX + pSVTAT I FOLD-TA % FOLD-TA
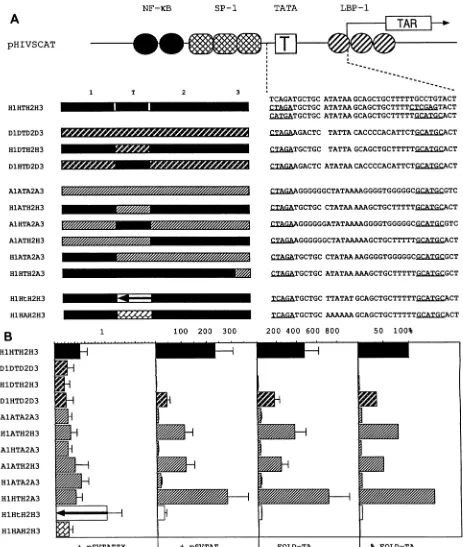
FIG. 2. Substitutions of the TATA boxregionin the HIV-1 LTR. (A) Diagrammatic representation ofsubstitutions and sequencesof substituted TATA boxregions.At the topof thisfigureisdepictedthesyntheticHIV-1 LTRwithwild-typesequencesflankedbyconvenient restriction endonuclease sites(see Fig. 1A).TheTATA boxregion has been divided into four convenientsections, labeled 1, T, 2, and 3.
Whereas Tdenotesthe TATA boxitself, regions 1, 2,and 3contain 5'- and3-flankingsequencesandthe initiatorsequence, respectively.
Thesemotifs in theHIV-1 LTRareshownassolid bars;those from the DRA and AdMLpromotersareshownascross-hatched and hatched
bars, respectively. Capitallettersdenotetheorigin of thesequence(H, HIV; A, AdML; D, DRA)andarefollowed by its location; thus, Hi
represents5'-flankingsequencefrom theHIV-1 LTR.H1HtH2H3 contains the inverted TATA box from the HIV-1 LTR, andH1HAH2H3
containsareplacementof six A residues for the TATAsequence.To theright of the bar diagramsaretheprecisesequencesof theseTATA
boxregions; restriction endonuclease sites areunderlined. Three different sequences aregiven for thewild-type plasmid construction;
however, theyallgavethesame levels ofexpression (datanotpresented). (B) CAT enzymaticassaysfromtransienttransfections ofthese plasmidconstructionsin thepresenceoffunctional andnonfunctional Tat effectorsin HeLacells.Panelsare asdescribed in the legendtoFig. 1B.Different barshadingsreflect the donorof theTATAboxregion, and onlythewild-type HIV-1LTR isdepictedas asolid bar. These
experimentswereperformeduptofive times and intriplicate. Standarderrorsof themean aredenotedbyerrorbars. 1756
pHIVSCAT
H1HTH2H3
D1DTD2D3
H1DTH2H3
D1HTD2D3
T 2 3
A1ATA2A3
H1ATH2H3
A1HTA2A3
A1ATH2H3
H1ATA2A3
H1HTH2A3
HlHtH2H3
H1HAH2H3
B
H1HTH2H3 D1DTD2D3 H1DTH2H3 D1HTD2D3 A1ATA2A3 H1ATH2H3
A1HTA2A3
A1ATH2H3 H1ATA2A3 H1HTH2A3 HlHtH2H3 H1HAH2H3
M-MENEM
moor-4;----KV4939zm
on November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.82.545.59.606.2]HIV-1 LTR-SPECIFIED TRANSCRIPTION COMPLEXES 1757 B
a".1i.a
<-.s -
r6-) ,- -Z:6-N
:4 i 6 X
6-\>
-_....~
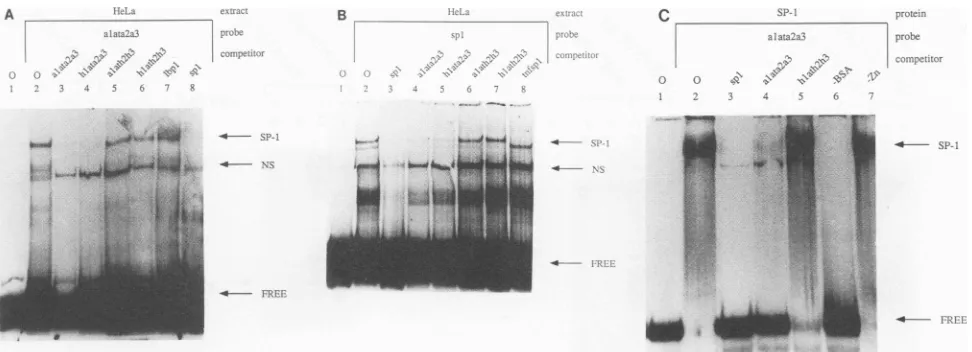
FIG. 3. EMSAs with nuclearextractsfromHeLa cells, purified SP-1, and oligonucleotides fromthe HIV-1 LTR and AdMLpromoter.(A)
EMSA with the oligonucleotide containing the TATA box region and nuclearextractsfromHeLacells. The nuclearextract, radiolabeled
oligonucleotide, andexcessunlabeledcompetitor oligonucleotides usedaregiven above the gel.Lane 1, noextract; lane 2,nocompetitor
oligonucleotide;lanes 3to9,100ngofcompetitoroligonucleotide containing only AdMLsequences(lane 3),HIV-1 5' and AdML TATA box
and3-flanking sequences(lane 4), AdML 5'-flanking and TATA box and HIV-1 3'-flankingsequences(lane5), HIV-1 5'- and 3'-flanking
sequences and AdML TATAbox (lane 6), three LBP-1 DNA-binding sites (lane 7), and three SP-1 DNA-binding sites (lane 7). The
oligonucleotideswereidenticaltothecassettesused in the construction of the synthetic HIV-1 LTR (Fig. 2A)andaredenoted by lowercase
letters. Arrows pointtoSP-1 andanonspecific proteincomplex (NS),sonamed because unlabeled specificand nonspecific oligonucleotides
didnotcompetefor itsbinding. Free probe referstoradiolabeled oligonucleotides thatwerenotcomplexed withprotein. (B) EMSA with the
oligonucleotide containing the three SP-1 sites and nuclear extracts from HeLa cells. Lanes are labeled as in panel A. Radiolabeled
oligonucleotide containingthe threeSP-1 sites from theHIV-1 LTRwasincubatedwith nuclearextractsfrom HeLa cells (lane 2)and 100ng
of thesameunlabeledoligonucleotide (lane 3)andthosecontaining the AdML TATA boxsequences(lane 4), HIV-15'-flankingand AdML
TATA boxand3'-flanking sequences(lane 5),AdML5'-flanking and TATA box and HIV-1 3'-flankingsequences(lane 6), HIV-1 5'- and
3-flankingsequencesand AdML TATA box(lane 7), andSP-1 sites from thetumornecrosis factor alphapromoter(lane 8).Noextractwas
added in lane 1.(C)EMSA with theoligonucleotide containing theAdML TATAboxregion and purified SP-1 from HeLa cells infected with
avaccinia virus which directs thesynthesisof SP-1. Lanesarelabeledasinpanel A. SP-1 withan excessof bovineserumalbumin (BSA)
wasincubatedwith radiolabeled oligonucleotide containingAdML TATAboxsequencesandnospecific oligonucleotide (lane 2)or100ng
of thesameunlabeledoligonucleotide (lane 4)andthosecontaining three SP-1 sites (lane 3) and HIV-15'-flanking and AdML TATA box and 3-flankingsequences(lane 5).The radiolabeledoligonucleotidewasalsoincubatedinthe absence ofSP-1 (lane 1), in the absence of BSA (lane 6),which stabilized the dilute SP-1proteininsolution, and in theabsenceofZn2+ (lane 7), which didnotdestabilize this binding.
The TATA boxregionof HIV-1 functionsefficientlybecause
nonuclearproteinbindsstronglytoits3'-flankingsequences
or toTAR DNA.
TATA box alone can set up nonprocessive transcription complexes that are trans activated by Tat. Having demon-strated that whereas upstream and downstream promoter elements increaseexpressionfrom the HIV-1LTR butdo not affect the response to Tat and that the TATA box and its immediate 3'-flanking sequences areabsolutelyessential for
transactivationby Tat,wewondered whether theTATAbox by itself could assemble nonprocessive transcription
com-plexes. Previously,weand others demonstrated thepresence
oftwodifferent transcription complexesthatwereidentified bytheirhybridization topromoter-proximaland-distal frag-ments in nuclear run-on assays (12, 18, 21). In studies of steady-state RNA, they were reflected in full-length poly-adenylated and shortnonpolyadenylated transcripts,
respec-tively (18). Since the half-lives of these RNAspecies were
similar(31),weexaminedtransientlytransfected cells for the
presenceof thesetwodifferenttranscripts.Sincehigherlevels of RNA could be achieved inCOS cellsby using replicating vectors, these studies were carried out with targets and effectors that containedtheSV40 originofreplication.
SinceLBP-1 didnotcontribute totransactivation in either
HeLa or COS cells, we removed only NF-KB and SP-1
sequences from the HIV-1 LTR and cotransfected these
plasmids with functional and nonfunctional Tat effector
plasmids. As depicted in Fig. 4A, wild-type targets and effectorsgavehigh levels of short transcripts in the absence
of Tat and correspondingly high levels of full-length
tran-scriptsin thepresenceof Tat(Fig. 4A, lanes 3, 4, 7, and 8). These RNA levels approached those observed in CAT
assays (Fig. 4A). Likewise, with an HIV-1 LTR which
containedonly the TATA boxbut lackedNF-KB and SP-1
sequences,low levels of these shorttranscripts,whichwere
efficientlytrans activated by Tat,were observed (Fig. 4A,
lanes 3 and 4). However, since levels of expression were
low, these gels had to be exposed to a phosphorimaging screen to demonstrate the presence ofspecific bands (Fig. 4A). Weconclude that the TATA box alone is sufficient for setting upnonprocessive complexes that aretransactivated byTat.Thus,the main role of upstream promotersequences
istoincrease theloadingof RNApolymeraseII,which results
in higher levels oftranscriptionandtrans activationbyTat. Perhaps most importantly, these results suggest that any
TATA box withanopen3'-flankingsequencecanassemblea
transcription complexthat istransactivatedbyTat. What abouttranscription complexesthatareformed in the absence of a functional TATA box? To detect these
com-plexes, similar RNase protection assays were performed
with RNA from COS cells cotransfected with plasmids in
which the HIV-1LTRwassubstituted with the DRATATA
box region and plasmids carrying functional and nonfunc-tional Tateffectors. With this substituted HIV-1 LTR, only
A ...- C
Sl'-aIata2a3
4.v4
6t-0 0
2
pro:ein
Probe
co:npetitor
A5'
4 spI
4e - I-enr-F
VOL.67, 1993
"".4
#,....,
-4Irmi W-W6%li -4
L. A
-.4-- :,, .:
A.
.*,. .400 .4 "il
W-4 S-i
. .1 k. -lA-l -4 N' s
9 V44
on November 9, 2019 by guest
http://jvi.asm.org/
[image:6.612.69.555.78.254.2]N
-,.4
1234f
/-.
pSVTAT - + - +
5 6 7 X
5678
LT
ELI
;% -W
ST
I.L
I _ IN.I
\\
~~~~~~~~~~~~~~~~~~\~
F--- ir -i-n]* - pSVI|AT _
3
: 4 5 6 /7 8
*Av
.;=
"'I.E
*-O_I
loti Probe
-Sh
Er-=* ST L w
Slioi-1 Prombe
60 9. I transactivation
foldsby CATassays
CAT
*
22-0bpprobeprotected tlrainscripts (LTr)ong 78-80bp
RNAs sho)rt
traiiscriptts
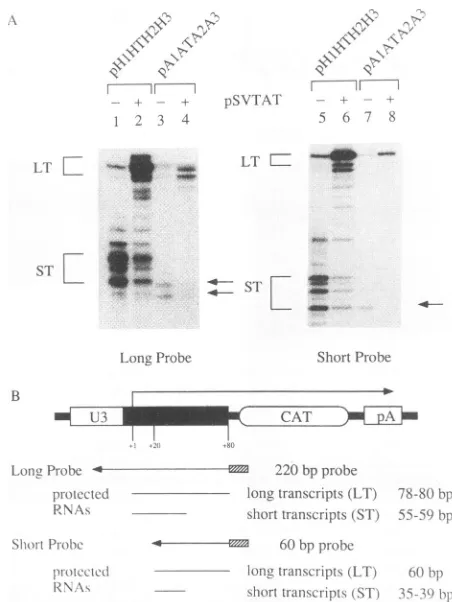
(ST) 55-59p)pFIG. 4. RNaseprotectionofRNA from COScellscotransfected withplasmids carryingtheTATA boxregionalone andplasmidsin
whichthe HIV-1 LTRwasreplaced bytheTATA box from the DRA promoter in the presence of plasmids carrying nonfunctional or
functional Tat effectors.(A) Phosphorimagesof RNaseprotectionof cotransfected COS cells. In both panels, thetargets are givenon
top, and thepresenceof nonfunctional (-)and functional(+) Tat effectors is depicted below the brackets. Foldtrans activation in CATenzymaticassaysisgivenbelow thephosphorimages.Lanes1, 2, 5,and 6 represent thewild-typesituation,inwhich theHIV-1 LTR targetswerecotransfected with both forms of Tat. In lanes 3 and 4are
similarexperimentswithplasmidsin whichsequences5'totheTATA box in the HIV-1 LTRwereremoved. In lanes 7and 8arepresented
cotransfections withplasmidsinwhich the HIV-1 LTRwasreplaced
bythe TATA box from the DRA promoter.Long(LT)and short(ST) transcripts correspond respectively to full-length polyadenylated transcripts and short nonpolyadenylatedtranscripts, which contain
onlyTARsequences and are55, 57, and 59nucleotides in length.
HIV-1 transcription initiates atallthree G residues 23to 26
nucle-otides 3'tothe TATAbox(18). (B)Diagrammatic representationof RNA probes. Below the picture of the HIV-1 LTR are given the
transcribed antisense RNAprobeand hybridizing speciesfrom the HIV-1 LTR. The probe is 220 nucleotides long and hybridizes to
full-length, polyadenylatedtranscriptsof 78to80bporthree short,
nonpolyadenylated transcripts of 55to59 bp. pA, poly(A) site.
full-length transcripts were observed, and these were not
affectedbyTat (Fig. 4A, lanes 7 and8). We conclude that whereas nonprocessive complexes require a functional
TATA box, processive complexes, which are formed by
upstream promoter sequences, do not require a functional
TATAbox and do notrespondto Tat.
SP-1placed betweenthe TATA box and TARinterfereswith assembly and/or migration of nonprocessive transcription
Probe - -- - 220bpprobe
lrtc'O t--I loneg transcripts(1LT) 78-80 hpii)
RN.As Short transcripts(Si)55}t- ( hin ShortP'robc 4*--- --- 60bpporhe
)tCLttl tranIscripts I )1|)
R \2 shorttralnl c; iti s SPi 3-'i)
FIG. 5. RNaseprotectionof the HIV-1 LTR substituted with the
TATA boxregionfrom the AdML promoter.(A) Autoradiogramsof
RNase protection of RNA from COS cells cotransfected with the
plasmidsin which HIV-1 LTRwasreplaced bytheAdMLTATA
box region and plasmids with nonfunctional or functional Tat effectors. Lanes are labeled as in Fig. 4. Lanes 1, 2, 5, and 6 represent the wild-type situation, in which the HIV-1 LTR was
cotransfected with both forms of Tat. In lanes 3, 4, 7, and 8 are
RNAs from similar cotransfections with plasmids in which the HIV-1 LTRwasreplaced bythe AdML TATA boxregion.Whereas RNAprobes in the left-hand panel were full-length, those in the
right-hand panelwere shorter by 20 nucleotides at their 3' ends because ofdigestion oftemplate DNAwithBglII. Note that two smaller bands in lanes 3 and 4arereducedtoasingleband in lanes 6and 7. Thus,the shorttranscriptsfrom theplasmidin which the HIV-1 LTRwasreplaced bythe AdML TATA boxregioninitiated
correctly and were shorter at their 3' ends. (B) Diagrammatic representation of RNA probes. Both full-length and short,
60-nucleotide-long probes are depicted, with the corresponding
ex-pected hybridizingbands.pA, poly(A)site.
complexes. Finally, we wanted to know how strong DNA-binding proteins placed between the TATA box and TAR affect nonprocessive complexes that aretrans activatedby Tat. To this end, we performed RNase protection
experi-ments with RNA from COS cells cotransfected with plas-midsin whichthe HIV-1 LTRwasreplaced with the AdML
TATA box region and plasmids carrying functional and nonfunctional Tat effectors.AsshowninFig. 5A, lanes 1, 2, 5, and6, thesamepatternsasinFig. 4A, lanes1, 2, 5, and
6, were observed with the wild-type target and effector plasmids. However, with the plasmid which contained the TATA boxregionfromthe AdMLpromoter, lower steady-state levels of short and full-length transcripts were ob-served. In addition, whereas the sizes of full-length tran-scriptswerethesame aswith thewild-type HIV-1 LTR, the
.A
60 2.5 T.-, 0
CA-FD:LpA-I -1
_ T 2
MM9 -.j
2
on November 9, 2019 by guest
http://jvi.asm.org/
[image:7.612.64.296.66.403.2] [image:7.612.319.545.79.380.2]HIV-1 LTR-SPECIFIED TRANSCRIPTION COMPLEXES 1759
short transcripts were smaller (Fig. 5A, lanes 3, 4, 7, and 8). Since the full-length transcripts were of comparable sizes, we reasoned that they initiated from the same site. By making the RNA probe shorter at the 5' end by 20
nucle-otides,we alsoobservedthat these short transcripts initiated
correctly (Fig. 5A, lanes 7 and 8). However, they were
shorter at their 3' ends. Moreover, this shorter probe re-duced two short transcripts obtained with the longer probe to one transcript (Fig. 5A, lanes 3, 4, 7, and 8). From
inspection of the stem-loop structure, which has a bulge at the bottom, we conclude that these differences between
wild-type and substituted targets can be explained if
tran-scription complexes do not migrate as far with constructs which contained the AdML TATA box region than with
thosecarrying thewild-type HIV-1 LTR. Thus, SP-1 inter-fereswith themigrationofnonprocessive transcription com-plexes; only a few reach the 3' border of TAR, and none goes beyond TAR.
These data stronglysuggestthat nonprocessive complexes
assembled at theTATA box are sluggish and susceptible to
prematureterminationand that Tat completes the process of making these complexes processive at TAR. Furthermore, they also argue against the notion that Tat reaches back to the TATA box toincrease initiationof HIV-1 transcription,
sincelevels oftrans-activated full-lengthtranscripts with the
AdMLTATA boxregion should have more closely resem-bledthoseobservedwith the wild-type target if that were so.
DISCUSSION
Thedatafrom these studies confirm the existence of two different transcription complexes which are set up on the HIV-1 LTR. Whereas processive complexes are formed
independentlyof theTATAsequence and do not respond to Tat, nonprocessive complexes require a functional TATA sequence and relatively free 3'-flanking sequences and are transactivatedbyTat. It is veryattractivetospeculate that
the relativeamounts ofthesecomplexes determinethe level
oftransactivationby Tatand thattranscriptional enhancers
and other acidicactivators such as ElA affecttheseratios (22, 32, 33, 36).
CAT enzymatic studies were confirmed by analyses of
transcripts from the HIV-1 LTR, and short but not long
transcripts correlated with trans activation by Tat. More
importantly, when no short transcriptswere observed, Tat
hadnoeffect. Onthe otherhand,even with a minimal TATA sequence, short transcripts predominated and were trans activated by Tat. Thus, Tat affects a generic RNA poly-merase II complex that is able to move, albeit sluggishly, alongthe DNAtemplate. Thebrittlenessof thistranscription complex was confirmed by studies with the AdML pro-moter,whoseTATA sequence functionedin thecontext of the HIV-1 LTR but whose 3'-flanking sequences inhibited
trans activationby Tat. This was due to the binding of SP-1 between the TATA box and TAR. The fewer and shorter
transcripts
observed with thisstrongDNA-binding sitemost likely reflect a barrier to movement of RNApolymerase II along thetemplate. However,its effects on the assemblyoftranscription complexes cannot be ruled out. It should be noted that Tat functioned less well on targets which con-tained SP-1binding sites 3' tothe TATA box on
nonrepli-catingvectors in HeLa cells than onreplicating vectors in
COS cells (compare Fig. 2A and 5), which could be due either to amounts of SP-1 insufficient to saturate all cognate
bindingsites or toits removal from DNAduringreplication.
Furthermore, since Tat did not increase overall levels of
expressionbut only effected a qualitative change from short to long transcripts in this hybrid promoter, it is unlikely that Tatcan reach back and increase the loading of RNA
poly-merase II. Finally, since DRA promoter 3'-flanking
se-quences could functionally replace those from HIV-1, the only requirement for these sequences is their inability to bind proteins tightly.
Ourdata extend and support those of previous studies on transcriptional regulation of HIV-1 gene expression (3, 18, 20, 24, 25, 29, 33). For example, initial studies with trans-fected or intrans-fected cells and in vitro transcription systems
revealedastrongpolarityof HIV-1 transcription, which was
modifiedby Tat (12, 18, 19, 21, 24). Whether called
antiter-mination, increased elongation, attenuation, or processivity, all refer to the simple notion that although many transcrip-tioncomplexesfrom HIV-1 LTR cannot copy the entire viral
genome, this situation is reversed by the addition of Tat. What this study adds is the precise mapping of sequences in the HIV-1 LTR that specify different transcription com-plexes and the notion that any TATA sequence that can bind
TBPcan set up transcription complexes that are intermedi-atebetweenabortive initiation and competent elongation.
Recent studies on cis-acting elements in the HIV-1 LTR and the role of the TATA box in trans activation by Tat
support these conclusions (3, 29). Furthermore, our study clearlysuggests that the same transcription complexes that are sensitive to 5,6-dichloro-1-3-D-ribofuranosylbenzimida-zole(DRB)and that are trans activated by Tat in vitroresult
in theformationoftheseshort transcripts in vivo (25). Also, the inducer of short transcripts consists primarily of the TATA box and its3'-flankingsequencesthat are free or only
looselybound by DNA-binding proteins (33). We conclude
that short transcripts that are generated by the inducer of
short transcripts are also the substrate for Tat.
The situationwith the HIV-1 LTR is similar to that found in theheat shockpromoterofDrosophila melanogaster (37) and several mammalian proto-oncogenes, such as c-myc,
c-myb, and c-fos (40).In thefirstexample, RNApolymerase
II isarrested just 3' of the site of initiation of transcription and is released following heat shock. In the second case,
variousactivationandproliferation signals alter the polarity oftranscription so that sequences distal to these genes are
copied. From these and other examples, it appears that Tat
mimics a cellular protein(s) that, by associating with the
transcription complex, increasesitsabilityto elongate. Cer-tainly Tat can function after several phosphodiester bonds have beensynthesizedandaftersignificanttranscription has
occurred. Whether these effects of Tat are assimple asthe
phosphorylationofC-terminal domainsofRNApolymerase
II,which is required for thetransitionof RNA polymerase II frominitiation to elongation (6), or more complex, such as
interactionswithbasal transcription factors such as TFIIE,
TFIIF, TFIIH, and TFIIJ (14, 44), or both remains to be
established.
ACKNOWLEDGMENTS
We aregrateful to MichaelArmaninifor expertsecretarial assis-tance; Shaw-Yi Kao, Mark Selby, and Sandra Tong for helpful suggestions and criticisms; and other members of the Peterlin laboratoryforcarefulcommentsonthemanuscript.
Thiswork wassupportedbythe HowardHughes Medical Insti-tute.
REFERENCES
1. Alonso,A., D.Derse, and B. M. Peterlin. 1992. Human chromo-some 12 is required foroptimal interactions between Tat and VOL.67, 1993
on November 9, 2019 by guest
http://jvi.asm.org/
TARofhumanimmunodeficiencyvirustype1 in rodentcells.J. Virol. 66:4617-4621.
2. Benoist, C.,and D.Mathis. 1990.Regulationofmajor histocom-patibility complexclass-IIgenes:X, Y,andother letters of the alphabet. Annu. Rev.Immunol. 8:681-716.
3. Berkhout,B., and K.-T.Jeang. 1992. Functional roles for the TATA promoter andenhancers in basal and Tat-induced
ex-pression of the human immunodeficiency virus type 1 long terminalrepeat.J.Virol. 66:139-149.
4. Bielinska, A., S. Krasnow, and G. J. Nabel. 1989. NF-KB-mediated activation of the human immunodeficiencyvirus
en-hancer: site of transcriptional initiation is independentof the TATAbox.J.Virol. 63:4097-4100.
5. Buratowski, S., S. Hahn,L.Guarente, and P. A. Sharp. 1989. FiveintermediatecomplexesintranscriptioninitiationbyRNA polymeraseII. Cell56:549-561.
6. Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387.
7. Cullen,B.R. 1990.TheHIV-1Tatprotein:anRNA sequence-specific processivityfactor?Cell 63:655-657.
8. Cullen, B. R. 1991. Regulation of HIV-1 gene expression. FASEB J.5:2361-2368.
9. Cullen, B.R., and W. C. Greene. 1989. Regulatory pathways governingHIV-1replication. Cell 58:423-426.
10. Dayton, A. I., J. G. Sodroski, C. A. Rosen,W. C. Goh, and W. A. Haseltine.1986. The trans-activatorgeneofthe humanT celllymphotropicvirustypeIIIisrequiredforreplication.Cell 44:941-947.
11. Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a
solubleextractfrom isolatedmammalian nuclei.Nucleic Acids Res. 11:1475-1489.
12. Feinberg,M.B.,D.Baltimore,andA. D. Frankel.1991.Therole ofTatin the humanimmunodeficiencyvirus lifecycleindicates
aprimaryeffectontranscriptionalelongation.Proc.Natl.Acad. Sci. USA 88:4045-4049.
13. Fisher, A. G., M. B. Feinberg, S. F. Josephs, M. E. Harper, L. M.Marselle, G. Reyes,M. A.Gonda,A.Aldovini,C.Debouk, R.C.Gallo,and F.Wong-Staal. 1986.Thetrans-activatorgene ofHTLV-IIIis essential for virusreplication.Nature(London) 320:367-371.
14. Flores, O.,H.Lu,and D. Reinberg. 1992. Factorsinvolved in specific transcription bymammalianRNApolymeraseII. Iden-tification and characterization of factor IIH. J. Biol. Chem. 267:2786-2793.
15. Glimcher,L.H.,andC.J.Kara.1992.Sequencesandfactors:
a guide toMHC class-II transcription. Annu. Rev. Immunol. 10:13-50.
16. Jones,K.A., J.T.Kadonaga,P. A.Luciw,and R.Tjian. 1986. Activation of the AIDS retrovirus promoter by the cellular transcriptionfactorSpl. Science 232:755-759.
17. Jones, K.A., P. A.Luciw, and N.Duchange. 1988. Structural arrangements of transcriptioncontrol domains within the 5'-untranslated leaderregionsoftheHIV-1andHIV-2 promoters. GenesDev.2:1101-1114.
18. Kao,S.Y.,A. F.Calman,P. A.Luciw,and B.M.Peterlin. 1987. Antitermination oftranscription within the long terminalrepeat ofHIV-1bytatgeneproduct.Nature(London)330:489-493. 19. Kato,H., M. Horikoshi,andR.G. Roeder. 1991.Repression of
HIV-1 transcription by a cellular protein. Science 251:1476-1479.
20. Kato, H., H. Sumimoto, P.Pognonec, C. Chen, C. Rosen, and B.G. Roeder. 1992. HIV-1 Tat acts as a processivity factor in vitro inconjunctionwithcellularelongation factors. Genes Dev. 6:655-666.
21. Laspia,M.F.,A. P. Rice, and M. B. Mathews. 1989. HIV-1 Tat proteinincreases transcriptionalinitiation and stabilizes elonga-tion. Cell 59:283-292.
22. Laspia, M.F.,A. P.Rice, and M. B. Mathews. 1990. Synergy between HIV-1 Tat and adenovirusElA is principally due to stabilizationoftranscriptional elongation. Genes Dev. 4:2397-2408.
23. Malim, M. H., R. Fenrick, D. W. Ballard, J. Hauber, E.
Bohnlein, and B. R. Cullen. 1989.Functional characterization of a complex protein-DNA-binding domain located within the human immunodeficiency virus type 1 long terminal repeat leaderregion.J.Virol. 63:3213-3219.
24. Marciniak, R.A.,B.J.Calnan,A. D.Frankel,and P.A. Sharp. 1990. HIV-1 Tatprotein trans-activates transcription in vitro. Cell 63:791-802.
25. Marciniak, R. A.,and P. A. Sharp. 1991. HIV-1 Tat protein promotesformation ofmore-processive elongation complexes. EMBOJ. 10:4189-4196.
26. Mitchell, P. J., and R.Tjian. 1989.Transcriptionalregulationin mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378.
27. Muesing, M.A.,D. H.Smith,and D.J.Capon. 1987. Regulation ofmRNA accumulation by a human immunodeficiency virus trans-activatorprotein. Cell 48:691-701.
28. Nabel, G., and D. Baltimore. 1987. An inducibletranscription factoractivatesexpression of humanimmunodeficiencyvirus in Tcells.Nature(London) 326:711-713.
29. Olsen, H.S.,andC. A. Rosen. 1992.Contributionof the TATA motiftoTat-mediated transcriptional activation ofhuman im-munodeficiencyvirus geneexpression.J.Virol. 66:5594-5597. 30. Pavlakis, G. N.,and B. K. Felber. 1990. Regulationof
expres-sion of humanimmunodeficiency virus.NewBiol. 2:20-31. 31. Peterlin, B. M., A. F. Calman, S. Y. Kao,M. J. Selby, S.E.
Tong-Starksen, and P. A. Luciw. 1988. Activation and
trans-activation ofHIV-1, p. 45-57.InB. R. Cullen, and F. Wong-Staal (ed.), Thecontrol of human retrovirus gene expression. Cold SpringHarborLaboratory, ColdSpring Harbor, N.Y. 32. Peterlin, B. M., P. A. Luciw, P. J. Barr, and M. D. Walker.
1986. Elevatedlevels ofmRNAcan accountfor the transacti-vation of human immunodeficiency virus (HIV). Proc. Natl. Acad. Sci. USA 183:9734-9738.
33. Ratnasabapathy, R.,M. Sheldon,L. Johal,and N. Hernandez. 1990. The HIV-1 long terminal repeat contains an unusual element thatinduces thesynthesis of shortRNAs fromvarious mRNAand snRNA promoters.GenesDev.4:2061-2074. 34. Rosen, C. A. 1991. Regulation of HIV gene expression by
RNA-protein interactions. Trends Genet. 7:9-14.
35. Rosen, C.A.,andG. N. Pavlakis. 1990. Tat and Rev:positive regulatorsof HIV geneexpression. AIDS4:499-509.
36. Rosen,C.A., J. G. Sodroski, and W. A. Haseltine.1985. The locationofcis-actingregulatorysequencesinthehuman Tcell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat.Cell41:813-823.
37. Rougvie,A. E., and J. T. Lis. 1988. The RNApolymeraseII molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795-804. 38. Selby,M.J.,E.S.Bain,P.A.Luciw,and B. M. Peterlin.1989.
Structure, sequence, and position of the stem-loop in TAR determine transcriptional elongation byTatthrough theHIV-1 long terminalrepeat.GenesDev.3:547-558.
39. Sharp,P.A.,and R. A.Marciniak. 1989. HIV TAR: anRNA enhancer? Cell 59:229-230.
40. Spencer,C.A.,and M.Groudine. 1990. Transcription elonga-tionandeukaryoticgeneregulation.Oncogene 5:777-785. 41. Tong-Starksen, S.E.,T. M. Welsh, and B. M. Peterlin. 1990.
Differencesin transcriptional enhancersof HIV-1 and HIV-2. Response to Tcell activation signals. J. Immunol. 145:4348-4354.
42. Tsang, S. Y., M. Nakanishi, and B. M. Peterlin. 1990. Mutational analysisoftheDRA promoter:cis-actingsequences and trans-acting factors. Mol. Cell. Biol. 10:711-719.
42a.Voliva,C.F., and B. M. Peterlin. Unpublished data.
43. Weil, P. A., D. S. Luse, J. Segall, and R. G. Roeder. 1979. Selective and accurate initiation of transcription at the Ad2 major latepromoterin a soluble system dependent on purified RNApolymeraseIIand DNA.Cell18:469-484.
44. Weinmann, R. 1992. The basic RNApolymerase II transcrip-tionalmachinery. GeneExpr. 2:81-91.
45. Wildeman,A.G. 1988. Regulationof SV40 early gene
expres-sion. Biochem.Cell Biol.66:567-577.